Abstract
Nucleosomes play an important role in gene regulation. Molecular studies observed that nucleosome binding in promoters tends to be repressive. In contrast, genomic studies have delivered conflicting results: An analysis of yeast grown on diverse carbon sources reported that nucleosome occupancies remain largely unchanged between conditions, whereas a study of the heat-shock response suggested that nucleosomes get evicted at promoters of genes with increased expression. Consequently, there are few general principles that capture the relationship between chromatin organization and transcriptional regulation. Here, we present a qualitative model for nucleosome positioning in Saccharomyces cerevisiae that helps explain important properties of gene expression. By integrating publicly available data sets, we observe that promoter-bound nucleosomes assume one of four discrete configurations that determine the active and silent transcriptional states of a gene, but not its expression level. In TATA-box-containing promoters, nucleosome architecture indicates the amount of transcriptional noise. We show that >20% of genes switch promoter states upon changes in cellular conditions. The data suggest that DNA-binding transcription factors together with chromatin-remodeling enzymes are primarily responsible for the nucleosome architecture. Our model for promoter nucleosome architecture reconciles genome-scale findings with molecular studies; in doing so, we establish principles for nucleosome positioning and gene expression that apply not only to individual genes, but across the entire genome. The study provides a stepping stone for future models of transcriptional regulation that encompass the intricate interplay between cis- and trans-acting factors, chromatin, and the core transcriptional machinery.
Nucleosomes are the basic unit of chromatin. Originally described for their structural role in compacting genomic DNA, they have become increasingly associated with playing a central role in regulating transcription. The yeast genome contains about 50,000 to 60,000 nucleosomes, each consisting of a complex of positively charged histone proteins around which a 147-base pair (bp) section of DNA is wrapped.
Recent advances in high-throughput technologies have dramatically expanded our understanding of genome-wide nucleosome organization (Bernstein et al. 2004; Yuan et al. 2005; Albert et al. 2007; Lee et al. 2007; Field et al. 2008; Mavrich et al. 2008; Shivaswamy et al. 2008; Kaplan et al. 2009; Zawadzki et al. 2009). Currently, the standard procedure for nucleosome profiling involves digesting genomic DNA with Micrococcal Nuclease, extracting nucleosome-bound DNA, and sequencing the resulting DNA fragments (MNase-seq). By mapping these sequence reads to a reference genome, it is possible to identify the location of nucleosomes in a particular cellular condition.
In coding regions of the yeast genome, nucleosomes are typically positioned with a spacing of ∼18 bp (Lee et al. 2007; Mavrich et al. 2008; Shivaswamy et al. 2008). In promoters, however, the so-called +1 and −1 nucleosomes flank a considerably larger opening termed the nucleosome-free region (NFR). These two nucleosomes occupy well-defined locations relative to the transcription start site (TSS), and their precise positioning is considered to be essential for successful transcriptional initiation (Lee et al. 2004; Yuan et al. 2005; Lee et al. 2007; Mavrich et al. 2008; Shivaswamy et al. 2008).
Detailed molecular studies have shown that nucleosomes play an important role in gene regulation: For genes such as PHO5 or ADH2, nucleosomes have a repressive effect, and their removal from the promoter region is sufficient to activate gene expression even in the absence of additional transcription factors (TFs) (Han and Grunstein 1988; Durrin et al. 1992; Adkins and Tyler 2006). Further, nucleosomes have been observed to relocate within these promoters upon changes in growth conditions, so providing a means for controlling expression in response to environmental stimuli.
At a genomic level, several studies have attempted to classify promoters according to nucleosome organization. Tirosh grouped promoters based on average nucleosome occupancies upstream of the TSS; however, their subsequent analysis compared the properties of only the most extreme promoters (i.e., the most occupied and most depleted promoters), and so excluded most yeast promoters from the study (Tirosh and Barkai 2008). Field used a similar classification based on nucleosome densities around TF-binding sites (Field et al. 2008). Both studies reported correlations between occupancies and additional features; for instance, housekeeping and essential genes tend to have depleted promoters, whereas those of stress-related genes are more densely occupied. The latter were also linked with properties such as high expression plasticity and noisy expression (Albert et al. 2007; Field et al. 2008; Tirosh and Barkai 2008). However, the studies considered only the promoter region typically occupied by the −1 nucleosome, thus excluding the contribution of the +1 nucleosome to promoter function. Further, the promoter classifications applied arbitrary thresholds for nucleosome occupancies and required additional information such as the presence of TF-binding sites.
Genomic studies have also reported conflicting observations about the relationship between nucleosome positioning and gene expression. On the one hand, an early study by Wyrik showed that global depletion of nucleosomes alters the expression of a quarter of yeast genes; but contrary to expectations nearly half of these genes were down-regulated instead of being up-regulated (Wyrick et al. 1999). In support, Shiwasvamy described that nucleosomes tend to be evicted from the promoters of genes whose expression levels increased during heatshock, though this was also observed among genes that remained unchanged (Shivaswamy et al. 2008). Using their promoter classification, Tirosh also stated that nucleosome occupancies are lower among highly expressed genes, although here the observed correlations were very weak (Tirosh and Barkai 2008). On the other hand, studies by Kaplan and Zawadzki reported that nucleosome occupancies remain largely unchanged in yeasts grown on different carbon sources (Kaplan et al. 2009; Zawadzki et al. 2009). A consequence of these conflicting observations is that there is no agreement about whether nucleosome positioning is a stable and inherent property of promoters (Field et al. 2008; Tirosh and Barkai 2008; Kaplan et al. 2009; Zawadzki et al. 2009), or whether nucleosomes reposition significantly according to cellular conditions through the action of trans-acting factors like remodeling enzymes (Lam et al. 2008; Shivaswamy et al. 2008). In summary, although nucleosomes are considered to be important for transcriptional control, there are still few general and consistent principles that can be applied on a global scale.
Here we utilize publicly available data to examine nucleosome-positioning behavior in the yeast Saccharomyces cerevisiae under different growth conditions. We introduce a four-state model for nucleosome architecture in promoter regions, which helps to explain several important properties of gene expression. In particular, we demonstrate that nucleosome architecture determines the switch between on and off transcriptional states, but not the actual expression level of a gene. In TATA-box-containing promoters, the architecture is also indicative of the amount of transcriptional noise. Finally, by comparing data across three growth conditions, we report that these nucleosomes display significant movement relative to the TSS, with >20% of genes altering their promoter state, and so their expression output. We show that a combination of DNA-binding TFs, chromatin-remodeling enzymes and the underlying genomic sequence help establish nucleosome architecture.
Results
Overview of major data sets
The study was performed using publicly available genome-scale data sets. Nucleosome-positioning data were obtained from Kaplan and Field, which presented short sequence reads from MNase-digested DNA (MNase-seq) for yeast grown in rich (YPD), ethanol- (EtOH), and galactose-containing media (Gal) (Field et al. 2008; Kaplan et al. 2009). To estimate the genomic positions of individual nucleosomes, we applied the GeneTrack software to the mapped sequence reads (Supplemental Fig. S1; Albert et al. 2008). As shown in Supplemental Table S1, the software predicted ∼53,000 nucleosomes for each condition.
Of particular interest in this study are the nucleosomes located in gene promoters. Following convention, we defined the nucleosome overlapping with the TSS of a protein-coding gene as the +1 nucleosome, and the one immediately upstream as the −1 nucleosome. The space between them is called the nucleosome free region (NFR); 1,320 divergent genes with shared promoters were excluded from the data set of −1 nucleosome properties, since both promoter nucleosomes overlap with a 5′-end of genes.
The MNase-seq data also allowed us to estimate the occupancy and focus of each nucleosome (Albert et al. 2008). The first measures the number of mapped reads at a given nucleosome position, and larger values signify higher occupancy. The second was calculated as the inverse of the spread of reads about a nucleosome position. Also called “phasing”, this measures the distribution of nucleosomes around a particular genomic coordinate in a population of cells: Larger values correspond to greater focus, meaning that nucleosomes are precisely positioned across all cells.
Transcriptional activities in the same three conditions were calculated using tiling-array data from Xu et al. (2009). Instead of examining expression levels (e.g., high, medium, and low expression), we examined the expression state of genes (i.e., expressed and unexpressed). For each condition, we fitted a two-component, normal-mixture model to the distribution of expression values: The first component encapsulated the distribution of expressed genes and the second captured the unexpressed ones (Supplemental Fig. S2). We defined a conservative threshold between expressed (ON) and unexpressed genes (OFF) as the first percentile of the first component. Table 1 summarizes the numbers of ON and OFF genes in each condition. To ensure the robustness of our analysis we have repeated all analyses with different thresholds of ON and OFF (0.05 quantile) as well as with expression data from a different data set (Nagalakshmi et al. 2008). The results did not change significantly and are shown in the Supplemental Figure S2.
Table 1.
Numbers of ON and OFF genes and the ratio between Closed and Open for each growth condition
NFR size distributions and nucleosome-binding properties indicate distinct classes of promoters
Previous genome-scale studies reported that nucleosome positions vary substantially between different genes. In particular, nucleosome occupancies were reported to range from very high to very low levels in a continuous manner; it is worth noting that these observations were generally made using average promoter-wide occupancies, rather than by examining individual nucleosome positions (Field et al. 2008; Tirosh and Barkai 2008; Kaplan et al. 2009; Zawadzki et al. 2009).
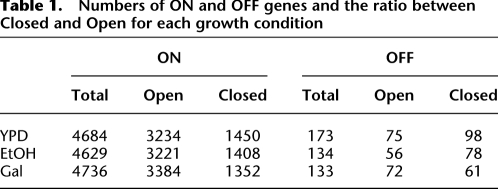
Here we demonstrate that promoters can be classified into discrete groups based on the +1 and −1 nucleosome positions. First, we examined the distribution of NFR sizes across all yeast promoters in YPD. We observe a previously unreported bimodal distribution with maxima at 30 bp and 130 bp, and a minimum at ∼80 bp (Fig. 1A). The size distribution is remarkably consistent across all three growth conditions within the Kaplan data set, and the observations are reproduced using data generated by other laboratories (Supplemental Fig. S4).
Figure 1.
Classification of promoters based on distance between +1 and −1 nucleosome position. (A) Comparison of NFR sizes for ON and OFF genes in three different conditions. Distributions of NFR sizes for ON (blue lines) and OFF genes (gray lines) in YPD1, EtOH, and Gal (from left to right). Expressed genes tend to have larger NFRs than unexpressed ones; however, for both expression states, NFR sizes fall into bimodal distributions with minima ∼80 bp (red dotted line). NFRs were classified into Open and Closed states based on this threshold. (B) Position of +1 and −1 nucleosomes relative to the TSS in Open and Closed promoters. Distribution of distances (in bp) from the TSS to the midpoints of the −1 and +1 nucleosomes in promoters of ON (blue) and OFF genes (gray) in YPD. Promoters were separated into those with Open (left) or Closed NFRs (right). The +1 nucleosome is well-positioned in ON genes compared with OFF. In contrast, the −1 nucleosome is well-defined for genes with Open/ON NFRs, but relatively poorly for genes with Closed NFRs. (C) Properties of promoter nucleosomes in Open and Closed promoters. Distributions of occupancy (top) and focus (bottom) for −1 and +1 nucleosomes in promoters with Open and Closed NFR configurations. Genes were separated into ON (blue) OFF (gray). In all cases, the +1 nucleosome is more highly occupied and better focused in the ON state. The −1 nucleosomes in Closed promoters, however, shows the opposite pattern with less occupancy and focus in the ON than in the OFF state. Wilcoxon rank sum test, P-value <0.05 (*), <0.01 (**), and <0.001 (***).
In contrast to previous studies, the bimodal distributions suggest there are two discrete populations of promoters with different nucleosome-binding conformations: those that are “Open” with large NFRs and those that are “Closed”. A natural threshold of ∼80 bp separates the two classes (ranging from ∼60 to 90 bp). As might be expected given the differences in DNA accessibility, Open promoters tend to be enriched among expressed ON genes and vice versa (Table 1; P < 1.3 × 10−9, Fisher's exact test). Surprisingly, however, both ON and OFF genes retain bimodal distributions indicating that these distinct promoter configurations are found throughout the yeast genome irrespective of the expression state (Fig. 1A).
Next we investigated further properties of nucleosome binding (Fig. 1B,C): position relative to the TSS, occupancy, and focus. In Open promoters, both +1 and −1 nucleosomes tend to occupy well-defined positions in ON genes (Fig. 1B, blue distributions) compared with OFF (gray distributions). Moreover, these nucleosomes display much stronger and focused binding in ON genes (Fig. 1C). In Closed promoters, the +1 nucleosome displays similar properties. The −1 nucleosome is a major exception as it displays less defined, weaker, and less fuzzy binding in ON than in OFF genes.
To test the robustness of the observations, we checked for sample size effects (Supplemental Fig. S5), and we examined additional MNase-seq data sets generated by different laboratories (Supplemental Fig. S4). The nucleosome positions display good correlation for data produced under equivalent cellular conditions despite differences in the specifics of the MNase-seq protocols. Moreover the bimodal distributions of NFR sizes and changes in nucleosome-binding properties are maintained throughout, indicating that the observations are very unlikely to be artifacts of the Kaplan and Fields data sets.
Four-state model for promoter nucleosome architecture
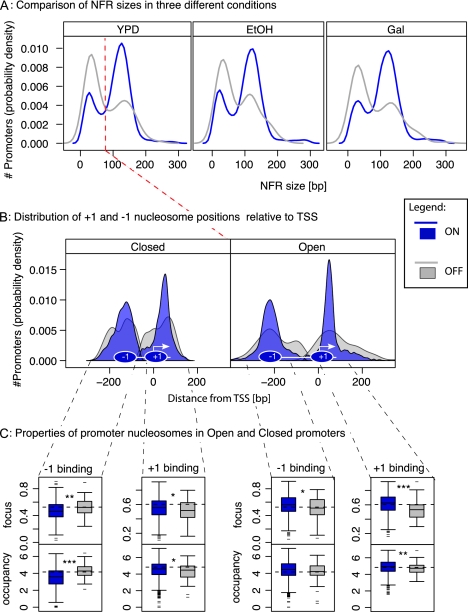
These observations allow us to propose a qualitative model for nucleosome architecture in yeast promoters (Fig. 2). The model consists of two NFR configurations, Open and Closed, and two expression states ON and OFF. Open-ON promoters have highly occupied, highly focused +1 and −1 nucleosomes flanking a large NFR; but in the Open-OFF state, both nucleosomes become weakly, and fuzzily bound. Closed promoters display similar binding for the +1 nucleosome: highly occupied and focused in the ON state, but weak and fuzzy in OFF. A major difference is the behavior of the −1 nucleosome, which is already weak and fuzzy when OFF, and becomes even weaker in the ON state.
Figure 2.
Model of promoter nucleosome architecture in yeast. Schematic of the four-state model. It summarizes the observations of nucleosome positioning, gene expression behavior, and regulatory influences.
The model encompasses the major observations made so far from the data. It also offers an explanation for the seemingly counterintuitive observation that genes lacking a proper NFR can still be expressed: The poorly positioned and weakly bound −1 nucleosome in the Closed state might allow intermittent access to the promoter by the transcriptional machinery (see below).
The classification can be applied to every single promoter in the yeast genome (Table 1). Moreover, unlike many previous studies of nucleosome positioning in promoters, the classification is not dependent on the presence of previously identified sequence motifs (e.g., TATA-box motifs), nor arbitrary thresholds of nucleosome occupancies.
Promoter states are discrete, not continuous
The bimodal distributions for NFR sizes and expression values strongly suggest that the promoter states described in the model are discrete. However, a potential concern is that promoters actually occupy a continuum and that the model depends on arbitrary thresholds.
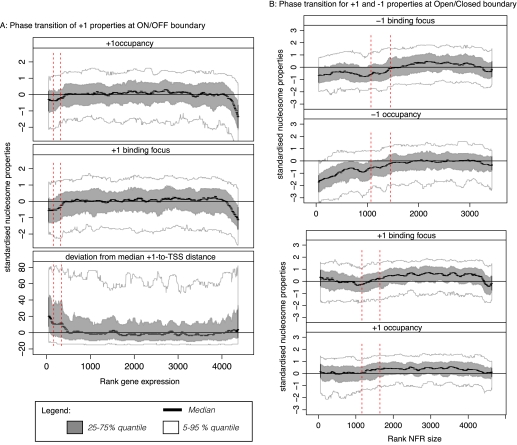
To test whether a continuous model would fit the data better, we examined how each nucleosome-binding property varies among promoters. Between ON and OFF genes, the main difference is in the behavior of the +1 nucleosome. We ranked genes according to their expression levels, and plotted them against normalized values for the relative binding position, occupancy, and focus (Fig. 3A). For all three properties, there is a clear transition in the region of the previously defined threshold between ON and OFF genes. There is a steep fall in occupancy and focus—though no corresponding change in position—among the most highly expressed genes which largely comprise ribosomal functions. This probably occurs because the +1 nucleosome is evicted at extremely high transcription rates. We suggest that these genes still belong to the Open-ON class, however, as the most important property—nucleosome position—remains unchanged, and all the downstream observations apply equally well to them.
Figure 3.
Phase transitions between promoter states. (A) Phase transition of +1 properties at ON/OFF boundary. The occupancy, binding focus, and the deviation from the median distance to the TSS (from top to bottom) for the +1 nucleosome are plotted against the rank of gene expression. The values were standardized and smoothed by a sliding window (window 200, step size 20). The median (black), 25%–75% quantiles (gray), and 5%–95% quantiles (white) are shown. A phase transition is observed at the boundary between ON an OFF genes. The red lines indicate the boundary for ON/OFF genes obtained from expression data with 1% and 5% FDR. The sudden fall in occupancy and focus toward the very highly expressed genes might suggest that nucleosomes get evicted upon very high expression. (B) Phase transition of +1 and −1 properties at Open/Closed boundary. The occupancy and focus for the +1 and −1 nucleosome are plotted against the rank of the NFR size. The values were standardized and smoothed by a sliding window, and the median (black dashed lines), 25%–75% quantiles (gray), and 5%–95% quantiles (white) are shown. A phase transition was observed for the −1 nucleosomes but not for the +1 nucleosomes. The red lines indicate the 60–90-bp window found in Figure 1A to mark the boundary between Open and Closed promoters.
The main distinction between Open and Closed promoters among ON genes is the behavior of the −1 nucleosome. Genes were ranked according to NFR sizes and plotted against the same three properties of the −1 nucleosome (Fig. 3B). Again, there is a clear phase transition around the previously defined threshold of ∼80 bp, ranging between 60 and 90 bp during which the nucleosome-binding properties undergo transitions. There is no equivalent transition for the +1 nucleosome, which is displayed as a control.
In summary, these observations support a discrete, rather than continuous, model for promoter nucleosome states.
Promoter state determines expression state and noise
Given the potential for diverse physical interactions between nucleosomes and the transcriptional machinery, it is reasonable to expect that the promoter state might influence gene expression. Previous studies reported negative correlations between promoter nucleosome occupancies and expression levels. However, these correlations were either very small (R2 < 0.04) or observed only for the highly expressed ribosomal genes (Shivaswamy et al. 2008; Tirosh and Barkai 2008).
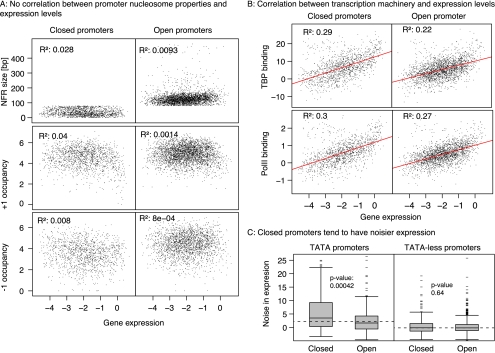
We already demonstrated that promoter nucleosomes behave differently between ON and OFF states: Here, we tested whether nucleosome-binding properties vary with the expression level of a gene once it is activated. In contrast to earlier studies, we are unable to detect significant relationships between the amount of gene expression and any features of the four-state model including NFR size, occupancy, focus, and occupancy by chromatin remodeling factors (Fig. 4A; Spearman's R2 < 0.05). Further, there are no associations between expression levels and nucleosome occupancies in the gene body. Plots for Pol II and TBP (also known as SPT15) binding are shown for comparison: As expected for components of the transcriptional machinery, they display good correlation (Fig. 4B). Therefore, the data clearly show that the pattern of nucleosome positioning indicates the transcriptional state, but not the expression level of a gene.
Figure 4.
Effects on the promoter nucleosomes on expression levels and expression noise. (A) No correlation between promoter nucleosome properties and expression levels. NFR size and occupancy of +1 and −1 nucleosomes are plotted against gene expression values (top to bottom) for Open and Closed promoters separately. No correlation is observed with R2 < 0.04. (B) Correlations between transcription machinery binding and expression levels. In contrast to nucleosome properties, the transcription machinery binding correlates nicely with expression levels with R2 between 0.2 and 0.3. (C) Closed promoters tend to have noisier expression. Distributions of gene-expression noise for TATA-containing and TATA-less promoters in Open and Closed configurations. TATA-less promoters (top panel) show significantly lower noise than TATA-containing ones (bottom panel; compare box-plots joined by red lines). Among the latter, Closed promoters show even more noise than Open promoters. There is no difference in noise levels between the two architectures among TATA-less promoters. P-values were calculated using the Wilcoxon rank sum test.
Another property of transcription is noise; that is, the amount of cell-to-cell variability in the expression level of a gene. In a single-gene study, Lam showed that noise at the Pho5 promoter rises with increased nucleosome occupancy (Lam et al. 2008). On a genomic scale, Tirosh reported that promoters with high nucleosome occupancies are enriched for the TATA-box motif and noisily expressed genes (Tirosh and Barkai 2008); similarly Field showed that gene promoters containing a combination of TATA boxes and nucleosome-occupied TF-binding sites tend to have the noisiest expression (Field et al. 2008). However, though promoter features clearly affect expression noise, it is not obvious whether the TATA box or nucleosome occupancy is responsible for this effect.
Using a genome-wide data set measuring single-cell protein expression in YPD (Newman et al. 2006), we assessed the impact of the four-state model on transcriptional noise. Figure 4C shows that, as expected, TATA-containing promoters are much noisier than non-TATA ones. In addition to the TATA effect, it is apparent that Closed promoter configurations result in greater noise than Open configurations. There is no difference among TATA-less promoters, however, indicating that the TATA box is a prerequisite for noisy expression.
In summary, these observations demonstrate that the promoter nucleosome state influences expression state and noise, but not expression levels.
Cis- and trans-determinants of promoter state
There is currently intense debate about whether nucleosome positioning is determined by the underlying DNA sequence or by regulatory proteins such as remodeling enzymes (Segal and Widom 2009; Kaplan et al. 2010). Below we examine the contributions of cis- and trans-acting factors in defining promoter nucleosome architecture.
Two sequence motifs that have previously been associated with promoter nucleosome occupancies are the TATA box and poly(dA:dT) elements. Using the genome-wide classification by Basehoar et al. (2004), we distinguished between functional TATA-box elements (which are conserved in >2 species, and/or the promoter is sensitive to mutations in the TATA-box-binding protein, TBP) and the simple presence of a TATA-sequence motif in the promoter region (which may or may not recruit TBP). There is an increased occurrence of functional TATA-box elements in Closed promoters (1.9-fold enrichment; P < 2.2 × 10−16, Fisher's exact test). Surprisingly, however, the motif sequence alone does not display a significant enrichment (1.2-fold; P < 0.24, Fisher's exact test). Among Open promoters, there is only a slight increase in the occurrence of the nucleosome-excluding 5–10-bp-long poly(dA:dT) tracts, as reported by Segal and Widom (2009) (Supplemental Table S2). In addition, we compared the occupancy of our different promoter classes with the scores of predicted nucleosomes based on DNA-nucleosome interactions (Segal et al. 2006; Kaplan et al. 2009) or DNA sequences (Xi et al. 2010) and found there was no difference between Open and Closed promoter nucleosomes in terms of predicted binding strength (Supplemental Fig. S5). These observations suggest that, although certain nucleotide compositions probably contribute to nucleosome positioning in the promoter, they are not sufficient to explain the occurrence of distinct promoter states.
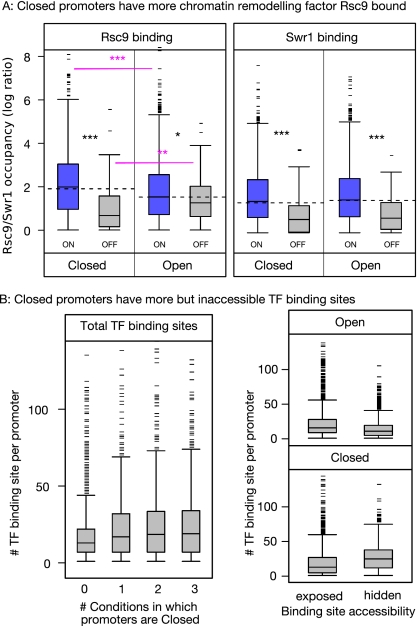
Therefore, we examined the binding pattern of two remodeling enzymes that are known to act specifically on promoter nucleosomes. Rsc9 is a member of the RSC complex involved in establishing the NFR by moving the −1 nucleosome. Swr1 is a component of the SWR complex that interacts with both the +1 and −1 nucleosomes; it is responsible for replacing the histone H2 subunit with the less stable H2A.Z variant (Mizuguchi et al. 2004; Zhang et al. 2005). Figure 5A shows the average Rsc9 and Swr1 occupancies in each promoter type. In general, both factors display greater binding among ON than OFF genes. However, there are important differences in the behaviors of the two enzymes. Swr1 binds equally to both Open and Closed promoters, indicating that it is required for switching genes from the OFF to ON states by remodeling the +1 nucleosome. Rsc9, on the other hand, displays greater occupancy at Closed promoters compared with Open; we suggest that, by associating with Closed promoters, Rsc9 helps to destabilize the −1 nucleosome and enable binding by the transcription initiation complex. Additional support for this comes from the observation that Closed promoters are more sensitive to RSC complex mutants, whereas both Open and Closed promoters are equally responsive to SWR complex mutants (P < 0.008 and 0.22, respectively, Wilcoxon rank sum test; data taken from Steinfeld et al. 2007).
Figure 5.
Different regulation of Open and Closed promoters by remodeling factors and TFs. (A) Closed promoters have more remodeling factor Rsc9 bound. Distributions of Rsc9 (left) and Swr1 (right) occupancy in Open and Closed promoters for ON (blue) and OFF genes (gray). Swr1 is present only in ON genes regardless of promoter state. In contrast, Rsc9 shows more binding to Closed promoters among ON genes and more binding to Open NFRs among OFF genes (compare box-plots joined by red lines). Wilcoxon rank sum test, P-value <0.05 (*), <0.01 (**), and <0.001 (***). (B) Closed NFRs have more but inaccessible TF-binding sites. Distributions of numbers of TF-binding sites for promoters that have a Closed NFR state in 0, 1, 2, or 3 growth conditions (left). Promoters that are never Closed have fewer binding sites than others. Distributions of numbers of binding sites that are exposed or hidden by nucleosomes in Open and Closed promoters (right). Open promoters tend to have fewer binding sites in total compared with Closed promoters; but these sites tend to be exposed.
Finally, we examined the binding pattern of sequence-specific TFs. Promoters were divided according to the number of conditions in which they assume the Closed conformation (i.e., closed in 0, 1, 2 or all of YPD, EtOH, and Gal), and then we counted the number of TF-binding sites that are present. As shown in Figure 5B, promoters that maintain an Open configuration in all three conditions have far fewer binding sites than those with a Closed conformation in at least one condition (P < 2 × 10−16, Wilcoxon rank sum test). This suggests that promoters with the potential to form a Closed NFR undergo much more complex TF-mediated regulation. In Closed promoters, most binding sites overlap with the −1 nucleosome position, indicating possible competition for DNA binding between histones and TFs; this may contribute to the weak and fuzzy binding by the −1 nucleosome. In contrast among permanently Open promoters, TF-binding sites tend to be highly exposed within the NFR; although we cannot assign causality using the current data, we suggest that TF binding may help stabilize the Open conformation.
In summary, these observations show that trans-acting factors rather that cis-elements are the most distinguishing features between the four promoter states.
Change in expression state across growth conditions is associated with nucleosome movement
Finally, we examined whether promoters switch nucleosome states across different growth conditions. Previous studies have reported apparently contradicting observations regarding the movement of promoter nucleosomes: Zawadzki stated that only 5% of genes displayed altered nucleosome binding—despite 50% of genes changing expression (Zawadzki et al. 2009); Shivaswamy reported nucleosome remodeling events also in genes that did not change expression (Shivaswamy et al. 2008). Further studies conducted in single growth conditions proposed that nucleosome binding is an inherent property of promoters (Field et al. 2008; Tirosh and Barkai 2008; Kaplan et al. 2009).
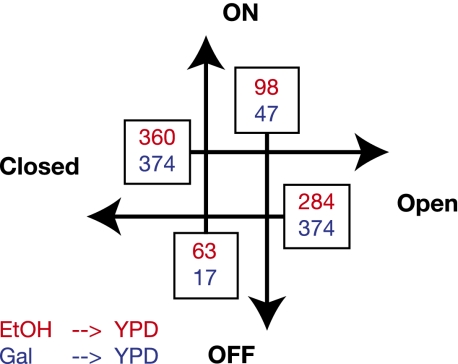
Using the four-state model, we examined the transitions of genes between the ON and OFF, and the Open and Closed states. To avoid potential artifacts in the latter, we considered only promoters in which the NFR size changed >30 bp; this threshold was based on the transition window for the Open/Closed boundary in Figure 1A as well as in the phase diagrams in Figure 3B. Figure 6 displays the number of genes that switch promoter states between the three growth conditions; a total ∼22% of yeast genes are involved, suggesting that the nucleosome state is a dynamic, condition-specific property of promoters.
Figure 6.
Promoter state transitions. The numbers of promoters that switch between the four promoter states are shown for the transitions EtOH to YPD (blue) and Gal to YPD (red).
We focused the rest of the analysis for the transitions observed between the YPD and EtOH growth conditions. Approximately one-hundred genes switch between ON and OFF expression states. As expected from the model, the +1 nucleosome is more dynamic for the switching genes: ∼50% of those in switching promoters were displaced by >10 bp (corresponding to one helical turn of the DNA) as compared to ∼15% in non-switching ones. In agreement with the statistical positioning model, the +1 nucleosome's movement propagates from the 5′-end to the 3′-end of genes (Supplemental Fig. S8). Four-hundred-and-sixty genes switch between the Open and Closed states while remaining expressed; here the main difference is in the behavior of the −1 nucleosome.
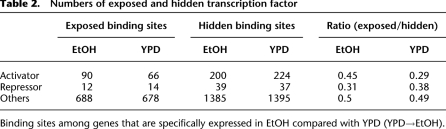
An interesting consequence of promoter state switching is the change in accessibility of TF-binding sites (Table 2). For promoters that become transcriptionally active in EtOH (i.e., OFF to ON), there is an increase in the numbers of exposed sites for ethanol-specific transcriptional activators (CAT8, HAP2/3/4/5, RDS2, SIP4, ADR1, CRZ1), and a corresponding decrease in accessibility for ethanol-specific repressors (MIG1/2, NRG1/2). In contrast, there is no change for non-condition-specific TFs.
Table 2.
Numbers of exposed and hidden transcription factor
Interestingly, genes switching from Closed to Open in EtOH are enriched for Gene Ontology (GO) functions related to peroxisome functions that are needed for growth in ethanol (Supplemental Table S3). Genes changing in the opposite direction from Open to Closed are enriched for respiration and oxidative phosphorylation, which are necessary functions for growth on glucose. These genes do not change average expression levels between the two conditions, confirming that there is no significant relationship between promoter nucleosome positioning and the amount of transcription. However, an interesting consequence of the switch in promoter architecture is the expected amount of noise in their expression. By assuming an Open state, genes needed in a particular condition (e.g., oxidative phosphorylation during YPD) will be expressed uniformly at optimal levels across an entire population of cells. In contrast the nonuniform or noisy expression afforded by the Closed promoter state might be advantageous for genes that are not necessary for the condition in question (e.g., peroxisome function in YPD), but would allow for any sudden changes in environmental conditions among a subset of cells expressing them (Supplemental Table S3; Arkin et al. 1998; Raser and O'Shea 2005; Blake et al. 2006; Kotte et al. 2010).
Discussion
In this study, we propose a qualitative model for nucleosome positioning in yeast promoters that helps explain several important properties of gene expression. The data indicate that the +1 and −1 nucleosomes are arranged in one of four discrete configurations, and that they are repositioned upon changes in cellular conditions.
Many of the presented findings are either new, or resolve previously inconclusive observations. Of these, the most important are that (1) every single promoter in the yeast genome can be classified into one of the four discrete—not continuous—nucleosome states; (2) nucleosome positioning indicates the expression state—not the level—of a gene; (3) promoters switch states between cellular conditions and the accompanying nucleosome movement propagates down the length of the gene; and (4) state-switching is largely determined by trans-acting factors. It is known that MNase digestion can introduce a bias in the fragmentation based on the underlying DNA sequence (Travers et al. 2009). To avoid drawing conclusions from such biases we made sure either to always compare nucleosome positions across different conditions or to rely on differences that are large enough to not be affected by the bias.
A few aspects of the study will appear familiar to readers, since some of the reported observations partially overlap with previous findings (e.g., TATA-box motifs are enriched in high-occupancy or Closed promoters). These results were nonetheless included here to place them within the context of the model.
Four-state model for promoter nucleosome architecture
The four-state model we propose here provides a useful framework for understanding the role of promoter nucleosomes in the yeast genome. The +1 nucleosome distinguishes between the ON and OFF expression states, whereas the −1 nucleosome defines the Open and Closed configurations.
The +1 nucleosome is highly occupied and well positioned in ON, but weak and fuzzy in OFF genes. The −1 nucleosome is also well positioned and highly occupied in Open-ON promoters, and weak and fuzzy in the OFF promoters. However, its binding is weakest and most fuzzy in the Closed-ON states. Assuming that the OFF state reflects the “nonregulated” configuration, the model suggests that the −1 nucleosome is actively destabilized in Closed promoters.
Previous studies grouped promoters based on nucleosome occupancies and DNA sequence motifs; however, these classifications applied only to the most extreme promoters, and their relationship with transcriptional regulation remained unclear. Importantly, our classification is solely based on gene expression and nucleosome-binding data and the natural thresholds presented by them; therefore, the observations are more complete (as they apply to most or all genes) and free of bias (as they do not depend on arbitrary cut-offs). Moreover the results are robust since they can be replicated using data generated by other laboratories (Supplemental Fig. S4).
Impact of nucleosome architecture on gene expression
The data indicate that the nucleosome position explains only the transcriptional state of a gene, but not its expression level. This appears to contradict earlier studies that identified relationships between nucleosome occupancies and expression levels; however, the reported correlations were either very small (equivalent to R2 < 0.04) or applied only to ribosomal genes (Shivaswamy et al. 2008; Tirosh and Barkai 2008). As shown in Figure 2B, we do observe a small negative relationship between nucleosome occupancy and expression levels for the highly expressed genes (mainly metabolic enzymes and ribosomal genes); however, these are exceptions involving a small proportion of the yeast genome and very likely results from the eviction of the +1 nucleosome. Our genome-scale observations are consistent with an earlier molecular study demonstrating that the nucleosome occupancy determines the threshold for transcriptional activation from the Pho5 promoter, whereas the number of exposed TF-binding sites enables the scaling of expression levels (Lam et al. 2008).
Another important consequence of nucleosome positioning is the variation in transcriptional noise among TATA-containing promoters. Molecular and single-cell studies have suggested that noise arises because transcription occurs in bursts rather than as a steady stream: Increased mRNA production in each burst and longer intervals between them contribute to noisier expression (Yean and Gralla 1997, 1999; Stewart and Stargell 2001; Basehoar et al. 2004). The TATA box alters these dynamics by enabling the core transcriptional machinery to remain promoter-bound longer, so allowing multiple rounds of re-initiation instead of just one; this increases the number of transcripts produced in a single burst, and therefore noise (Zenklusen et al. 2008). Our observations suggest that a Closed promoter state adds even more noise, perhaps by introducing competition for DNA binding between histones (Miller and Widom 2003), TFs, and the initiation complex, so increasing the uncertainty in the length and interval of bursts.
Dynamic behavior of promoter nucleosomes provides additional layer of regulation
Many previous studies suggested that nucleosome-binding positions are invariant features of promoters (Field et al. 2008; Tirosh and Barkai 2008; Kaplan et al. 2009; Zawadzki et al. 2009). Instead, by comparing nucleosome positions across YPD, EtOH, and Gal growth conditions, we identified >450 promoters that switch between Open and Closed states. In agreement with our earlier observations, there was no change in the average expression level of the genes involved. Instead, we suggest that changes to the promoter state might alter the amount of noise in gene expression, though unfortunately there are currently no data available to confirm this prediction. Transcriptional noise is generally viewed as an invariable feature of particular promoters: with the amount of noise encoded in the underlying DNA sequence, such as the presence of a TATA-box motif. Promoter-state switching might allow genes to alter noise levels in a condition-dependent manner.
The potential benefits of such a mechanism become apparent upon inspection of the gene functions involved in promoter-state switching. The GO annotations indicate that Open promoters tend to occur for genes that are essential for cell survival in a given condition, whereas the Closed conformation is mostly found among genes that are less necessary. Previous studies have shown that noisy expression of nonessential genes can provide phenotypic advantage to a population of cells in case of sudden changes in environmental conditions (Arkin et al. 1998; Raser and O'Shea 2005; Blake et al. 2006; Kotte et al. 2010). Thus by altering the promoter nucleosome architecture, yeast may have evolved a mechanism to tune expression noise.
Cis- and trans-determinants of promoter state
A major current debate in chromatin regulation surrounds the question of whether nucleosome positioning is determined in cis by preferential binding of histones to certain DNA sequences, or in trans by external effectors such as TFs (Zhang et al. 2009; Kaplan et al. 2010). Our observations indicate that, at least in promoter regions, trans-effects are the dominant determinants of nucleosome architecture. The main differences between promoter states are in the binding pattern of the chromatin-modifying enzymes Rsc9 and Swr1, the core transcription factor TBP, and condition-specific transcriptional regulators such as HAP2-5 for ethanol-dependent growth. In contrast, we detected only weak association between the occurrence of poly(dA:dT) sequence stretches—which are reported to exclude nucleosomes—and particular promoter states. Most significantly, though functional TATA-box elements are enriched among Closed promoters, the motif sequence alone occurs at similar frequencies among all promoter types. Finally, since many promoters switch states between growth conditions, external factors must be involved in establishing the correct nucleosome architecture in a condition-specific manner.
Methods
Nucleosome-binding data
Data were obtained from the studies by Kaplan and Field (Field et al. 2008; Kaplan et al. 2009), which used MNase extraction and Illumina sequencing to measure the amount of nucleosome binding at each base position in the S. cerevisiae genome. The data consist of mapped reads that were extended at the 3′-end by the average length of a nucleosome (147 bp). We processed these reads to obtain different types of information as described below.
MNase-seq reads are available for three cellular conditions: exponential growth in rich (YPD), ethanol-containing (EtOH), and galactose-containing media (Gal). The data set comprises six biological replicates for YPD, four for EtOH, and three for Gal. In order to maintain equivalent information content, we randomly selected three replicates for each condition for further analysis. The YPD data with six replicates allowed us to generate two independent data sets for use in control comparisons. Analyses performed using alternative combinations of replicates did not alter the results.
Nucleosome positioning
Nucleosome positioning provides the genomic coordinates delimiting the likely location of individual nucleosomes. Coordinates were calculated using the GeneTrack software (Albert et al. 2008). Briefly, the method applies Gaussian smoothing to each genomic position, i.e., each base position is represented by a normal distribution with peak height equal to the number of reads and standard deviation equal to the fitting tolerance supplied by the user. Gaussian distribution values are summed at each base position and joined together to produce a continuous line. Peaks are then identified in the continuous line using a peak-finding algorithm, which searches for non-overlapping peaks according to a user-supplied exclusion zone. These peaks correspond to the most likely positions of nucleosomes and peak heights correspond to the estimated number of MNase-seq reads present at the particular position.
For each condition, we pooled reads from the three replicates, and then applied the GeneTrack software with the following parameters: fitting window of 75 bp, standard deviation of 15 bp (i.e., the standard deviation of the normal distribution around each nucleosome read), and peak-exclusion zone of 146 bp (i.e., the minimum distance allowed between two peaks). Supplemental Table S1 summarizes the number of predicted nucleosome positions for each condition. Supplemental Figure S1 displays 10-kbp regions of the yeast genome annotated with mapped reads and corresponding nucleosomes as predicted by GeneTrack.
Nucleosome occupancy
The occupancy of a nucleosome was calculated as the logarithm of the estimated number of MNase-seq reads at the peak of a predicted nucleosome position (i.e., the peak height calculated by GeneTrack).
Nucleosome-binding focus
The binding focus—or the inverse of fuzziness—is a measure of how precisely a nucleosome binds to particular genomic coordinates in a population of cells. This was calculated as the ratio between the estimated number of reads for a given nucleosome (i.e., peak height) and the number of reads within a 73-bp region on either side of the peak. If a nucleosome binds with high focus, most reads should center at the peak, resulting in a ratio close to 1. If a nucleosome is fuzzy, only a fraction of reads should center on the peak, giving a ratio of <1.
Nucleosome classification
Nucleosomes were classified according to their binding location relative to genomic annotations. The +1 nucleosome was defined as the first nucleosome that overlaps with the 5′-end of a transcript by >10 bp. The 10-bp overlap ensures that the classification is robust. The −1 nucleosome was defined as the first nucleosome upstream of the +1, unless it is a +1 nucleosome itself (i.e., by overlapping with a divergent gene in a bidirectional promoter).
Nucleosome free region (NFR)
The nucleosome free region (NFR) of a gene was defined as the distance between the 5′-coordinate of the +1 and the 3′-coordinate of the −1 nucleosome (or +1 in bidirectional promoters). We excluded NFRs that were >500 bp since manual inspection of such regions suggested missing data.
Gene expression
Processed gene expression data were obtained from Xu et al. (2009). The data contain expression values for each gene from tiling-array hybridizations normalized to genomic DNA. The experiment was performed in six replicates for YPD and in triplicates for yeast grown in EtOH and Gal.
Classification of ON and OFF genes
We classified genes into those that were transcriptionally active (ON) and inactive (OFF) according to the expression data above. We used the YPD replicates to determine the threshold of ON and OFF by fitting a normal-mixture model composed of two normal distributions to the expression values. This was done for each replicate individually. The model fits the data very well in four out of the six replicates (see Supplemental Fig. S2). Using the replicates that fit well, we determined the OFF threshold conservatively as the value at the 0.01 quantile of the distribution describing the expressed genes in YPD (FDR of 1%, additionally analyses were repeated with an FDR of 5%; see Supplemental Fig. S2). This threshold was then applied to all conditions. Numbers of ON/OFF genes are displayed in Table 1.
Sliding window approach for testing continuous vs. discrete model
Promoters were ranked according to their gene expression levels to testing the ON/OFF threshold and their NFR sizes to test the Open/Closed threshold. Nucleosome properties (occupancy, focus, and deviation from the median distance to the TSS) were standardized to the population of promoter nucleosomes and smoothened using sliding window averaging (window size = 200 ranks, step size = 20 ranks). Figure 3A,B displays the median and interquartile ranges against the ranked gene expression or NFR size.
Judging from the transition phase in Figure 3B, we determined the error margin between Open and Closed promoters to lie between rank 1100 and 1600 (as indicated with the red dashed lines). This corresponds to NFR sizes of 54 and 88 respectively, thus resulting in an error margin of 34 bp.
Switching promoter states
Switch of ON and OFF states
We identified genes that switch expression states between ON and OFF across cellular conditions using two criteria: (1) They had to change labels between ON and OFF in the above classification, and (2) they had to change expression levels more than twofold between the two conditions.
Switch of Open and Closed states
We considered nucleosomes to switch between Open and Closed state between two conditions if they met the following criteria: (1) changed label from Open to Closed or vice versa; (2) displayed a difference in NFR size larger than the error margin identified from the Open-Close phase transition (34 bp as described above); and (3) were higher occupied than the weakest 2.5% of genome-wide nucleosomes. The additional filters (2 and 3) were used in order to avoid artifacts of marginal changes in NFR size and −1 occupancy.
Sequence motifs and binding sites in promoters
We defined promoters as the region between 50 bp downstream from and 356 bp upstream of the TSS. The upstream boundary was chosen such that it includes 95% of the 5′-coordinates of all −1 nucleosomes. We excluded divergent promoters with a shared NFR from calculations below, in order to avoid biases introduced by the missing −1 nucleosome.
TATA box
The list of functional TATA-box regions was obtained from Basehoar et al. (2004). The list of TATA sequence motifs was obtained by scanning the promoters for the TATA motif [TATA(A/T)A(A/T)(A/G)] within −200 and +50 bp of the TSS.
Poly(dA:dT) tracks
We scanned promoters for continuous Poly(dA:dT) tracks of lengths 4–11 bp (Supplemental Table S2). We tested for associations with promoter nucleosome states for TATA and non-TATA promoters separately using Fisher's exact test.
Remodeling and transcription machinery factors Swr1, Rsc9, TBP, PolII
ChIP-chip data for binding by Rsc9, Swr1, TBP, and PolII in YPD growth were obtained from Venters and Pugh (2009). For each factor, we calculated the mean occupancy at each promoter by averaging the tiling array values in the above defined promoter region. Differences across promoter types were tested using the Wilcoxon rank sum test.
Transcription factor binding sites
Transcription factor binding data were obtained from Harbison et al. (2004), MacIsaac et al. (2006), and Reimand et al. (2010).
Remodeling enzyme mutants
Data for mutants of remodeling enzymes were taken from the collection of Steinfeld et al. (2007). Data for the effects of SWR and RSC complexes were chosen based on the complex descriptions on YGD. Differences between promoter types were tested using the Wilcoxon-rank sum test.
Statistical tests
Statistical tests were performed as indicated in the main text.
Acknowledgments
We thank Kathi Zarnack, Wolfgang Huber, Maria Hondele, Aswin Seshasayee, and Lars Steinmetz for fruitful discussions and their critical reading of the manuscript. The work was funded by EMBL and the EpiGeneSys FP7 Network of Excellence.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.124099.111.
References
- Adkins MW, Tyler JK 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell 21: 405–416 [DOI] [PubMed] [Google Scholar]
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Albert I, Wachi S, Jiang C, Pugh BF 2008. GeneTrack—a genomic data processing and visualization framework. Bioinformatics 24: 1305–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin A, Ross J, McAdams HH 1998. Stochastic kinetic analysis of developmental pathway bifurcation in phage λ-infected Escherichia coli cells. Genetics 149: 1633–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL 2004. Global nucleosome occupancy in yeast. Genome Biol 5: R62 doi: 10.1186/gb-2004-5-9-r62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake WJ, Balázsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865 [DOI] [PubMed] [Google Scholar]
- Durrin LK, Mann RK, Grunstein M 1992. Nucleosome loss activates CUP1 and HIS3 promoters to fully induced levels in the yeast Saccharomyces cerevisiae. Mol Cell Biol 12: 1621–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E 2008. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216 doi: 10.1371/journal.pcbi.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Grunstein M 1988. Nucleosome loss activates yeast downstream promoters in vivo. Cell 55: 1137–1145 [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, MacIsaac KD, Danford TW, Hannett NM, Tagne J-B, Reynolds DB, Yoo J, et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Hughes TR, Lieb JD, Widom J, Segal E 2010. Nucleosome sequence preferences influence in vivo nucleosome organization. Nat Struct Mol Biol 17: 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotte O, Zaugg JB, Heinemann M 2010. Bacterial adaptation through distributed sensing of metabolic fluxes. Mol Syst Biol 6: 355 doi: 10.1038/msb.2010.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam FH, Steger DJ, O'Shea EK 2008. Chromatin decouples promoter threshold from dynamic range. Nature 453: 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-K, Shibata Y, Rao B, Strahl BD, Lieb JD 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E 2006. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113 doi: 10.1186/1471-2105-7-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Widom J 2003. Collaborative competition mechanism for gene activation in vivo. Mol Cell Biol 23: 1623–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu W-H, Sen S, Wu C 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846 [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK 2005. Noise in gene expression: Origins, consequences, and control. Science 309: 2010–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Vaquerizas JM, Todd AE, Vilo J, Luscombe NM 2010. Comprehensive reanalysis of transcription factor knockout expression data in Saccharomyces cerevisiae reveals many new targets. Nucleic Acids Res 38: 4768–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J 2009. Poly(dA:dT) tracts: Major determinants of nucleosome organization. Curr Opin Struct Biol 19: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang J-PZ, Widom J 2006. A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR 2008. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol 6: e65 doi: 10.1371/journal.pbio.0060065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfeld I, Shamir R, Kupiec M 2007. A genome-wide analysis in Saccharomyces cerevisiae demonstrates the influence of chromatin modifiers on transcription. Nat Genet 39: 303–309 [DOI] [PubMed] [Google Scholar]
- Stewart JJ, Stargell LA 2001. The stability of the TFIIA-TBP-DNA complex is dependent on the sequence of the TATAAA element. J Biol Chem 276: 30078–30084 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N 2008. Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A, Caserta M, Churcher M, Hiriart E, Di Mauro E 2009. Nucleosome positioning—what do we really know? Mol Biosyst 5: 1582–1592 [DOI] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF 2009. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res 19: 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421 [DOI] [PubMed] [Google Scholar]
- Xi L, Fondufe-Mittendorf Y, Xia L, Flatow J, Widom J, Wang J-P 2010. Predicting nucleosome positioning using a duration Hidden Markov Model. BMC Bioinformatics 11: 346 doi: 10.1186/1471-2105-11-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean D, Gralla J 1997. Transcription reinitiation rate: A special role for the TATA box. Mol Cell Biol 17: 3809–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean D, Gralla JD 1999. Transcription reinitiation rate: A potential role for TATA box stabilization of the TFIID:TFIIA:DNA complex. Nucleic Acids Res 27: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G-C, Liu Y-J, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zawadzki KA, Morozov AV, Broach JR 2009. Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae. Mol Biol Cell 20: 3503–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123: 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K 2009. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]