Abstract
Cell-type diversity is governed in part by differential gene expression programs mediated by transcription factor (TF) binding. However, there are few systematic studies of the genomic binding of different types of TFs across a wide range of human cell types, especially in relation to gene expression. In the ENCODE Project, we have identified the genomic binding locations across 11 different human cell types of CTCF, RNA Pol II (RNAPII), and MYC, three TFs with diverse roles. Our data and analysis revealed how these factors bind in relation to genomic features and shape gene expression and cell-type specificity. CTCF bound predominantly in intergenic regions while RNAPII and MYC preferentially bound to core promoter regions. CTCF sites were relatively invariant across diverse cell types, while MYC showed the greatest cell-type specificity. MYC and RNAPII co-localized at many of their binding sites and putative target genes. Cell-type specific binding sites, in particular for MYC and RNAPII, were associated with cell-type specific functions. Patterns of binding in relation to gene features were generally conserved across different cell types. RNAPII occupancy was higher over exons than adjacent introns, likely reflecting a link between transcriptional elongation and splicing. TF binding was positively correlated with the expression levels of their putative target genes, but combinatorial binding, in particular of MYC and RNAPII, was even more strongly associated with higher gene expression. These data illuminate how combinatorial binding of transcription factors in diverse cell types is associated with gene expression and cell-type specific biology.
Cellular diversity in multicellular organisms is achieved in part by distinct transcriptional programs mediated by transcription factors (TFs). The human genome is believed to encode ∼1400 sequence-specific TFs (Vaquerizas et al. 2009). Identifying the genomic binding locations of TFs provides insights into how their activities shape gene expression. Recent studies combining chromatin immunoprecipitation of human TFs with deep sequencing (ChIP-seq) have identified tens of thousands of TF binding sites which function as promoters, enhancers, insulators, and silencers (Barski et al. 2007; Johnson et al. 2007; Ku et al. 2008; Valouev et al. 2008; Cuddapah et al. 2009; Moqtaderi et al. 2010; Raha et al. 2010; Euskirchen et al. 2011; Rada-Iglesias et al. 2011). However, such location information for any given factor is currently available for only a limited number of human cell types. There are few systematic studies identifying the genomic locations of multiple TFs across a diverse set of cell types, carried out in conjunction with gene expression studies. For most cell types in human, it is unclear how many sites on the genome are occupied by different kinds of TF, how these binding sites are distributed relative to genomic features, and how TF binding might be involved in regulation of cell-type specific gene expression.
The Encyclopedia of DNA Elements (ENCODE) Project has the goal of characterizing all functional elements in the human genome. Binding sites for many TFs were identified in the pilot phase of ENCODE, which investigated 1% (30 Mb) of the human genome (ENCODE Project Consortium 2007). The ENCODE Consortium has now scaled up to identify cis-regulatory elements genome-wide in a wide variety of cell types (ENCODE Project Consortium 2011). As part of the ENCODE project, we investigated the genome-wide binding sites of three different kinds of transcription factor, namely MYC (formerly c-Myc), CTCF, and RNA polymerase II (RNAPII) in the human genome in multiple cell types.
MYC is a sequence-specific TF that integrates diverse internal and external stimuli (Wierstra and Alves 2008). MYC has been proposed to be a “global” regulator of transcription, potentially regulating ∼15% of human genes (Dang et al. 2006; Meyer and Penn 2008) implicated in cell cycle progression, differentiation, apoptosis, DNA repair, angiogenesis, chromosomal instability, and ribosome biogenesis (Dang 1999; Adhikary and Eilers 2005; Knoepfler et al. 2006; Dai and Lu 2008). MYC is also important for lineage-specific cell growth and metabolism and thus a key contributor to cell fate decisions (Grandori et al. 2000). However, it is not clear how many lineage-specific genes are under regulation by MYC in the human genome.
The CCCTC binding factor (CTCF) is evolutionarily highly conserved and ubiquitously expressed. CTCF contains 11 zinc-finger DNA-binding domains, and is involved in gene activation as well as repression, hormone-responsive gene silencing, imprinting of genetic information, enhancer blocking, and chromatin insulation (Filippova et al. 1996; Burcin et al. 1997; Vostrov and Quitschke 1997, 2002; Hark et al. 2000). Aberrant expression of either CTCF or MYC can cause detrimental consequences such as developmental disorders, disease, and a wide range of cancers (Ohlsson et al. 2001; Ladomery and Dellaire 2002; van Riggelen et al. 2010).
Although RNAPII is not a sequence-specific transcriptional regulator, here we consider it to be a transcription factor in the sense of being a protein complex essential for the process of transcription. It is responsible for synthesizing the precursors of mRNA, miRNA, and most snoRNAs (Sims et al. 2004). RNAPII interacts with CTCF as well as MYC, and significant co-localization of RNAPII and CTCF is observed in the nucleus (Barski et al. 2007; Chernukhin et al. 2007; Rahl et al. 2010). CTCF–RNAPII protein complexes are found in distal regions, 1.5–15 kb away from the nearest transcription start site (TSS), and remain intact until RNAPII release (Chernukhin et al. 2007). Approximately one-third of ∼20,000 CTCF binding sites are located in protein-coding regions (Barski et al. 2007). However, it is unclear how many RNAPII binding sites are co-localized with CTCF genome-wide, whether there is lineage-specific co-localization in various tissue types, and how the interaction between RNAPII and CTCF affects expression of their target genes. MYC can promote phosphorylation of the C-terminal domain of the large subunit of RNAPII as well as mRNA cap methylation (Cowling and Cole 2007). A recent study showed that a major function of MYC is release of paused RNAPII (Rahl et al. 2010).
To identify the genome-wide binding sites of CTCF, MYC, and RNAPII in diverse cell types and elucidate combinatorial TF binding effects, we performed chromatin immunoprecipitation (ChIP) followed by high-throughput sequencing (ChIP-seq). Across 11 cell types we found an average of 45,000, 30,000, and 8000 binding sites for CTCF, RNAPII, and MYC, respectively. Analysis of these binding sites in relation to genomic annotations, gene expression levels, and cell type diversity sheds considerable light on how these transcription factors of different types function individually and in combination with one another to occupy target genomic loci and shape gene expression programs in a cell-type specific manner.
Results
ChIP-seq identifies tens of thousands of binding sites for CTCF, MYC, and RNAPII
We performed ChIP-seq for CTCF, MYC, and RNAPII in 11 different human cell types including primary, disease, and cancer cells (Table 1). For RNAPII, we carried out ChIP for its large subunit (POLR2A); in the text and figures it is shown as RNAPII, referring to the holoenzyme complex. In parallel with sequencing chromatin immunoprecipitated DNA, we also sequenced an input DNA control. We generated at least two biological replicates of ChIP-seq data for each factor in all cell types except H1 ES and NHEK cells (Supplemental Table S1). ChIP-seq samples were assayed by quantitative real-time PCR as a quality control before sequencing. We used a custom algorithm to identify peaks indicating binding sites from each replicate data set of aligned sequences (Methods). In most cases, CTCF and RNAPII showed >80% overlap between replicates, indicating high consistency, and MYC exhibited moderate reproducibility ranging from 50% to 80% (Supplemental Fig. S1). Therefore for each factor–cell line combination, we combined reads from all replicates, and then generated an initial set of candidate binding locations (Fig. 1). To minimize false positives, we normalized TF binding scores with corresponding input scores and for sequencing depth. We calculated P-values, normalized scores, and appropriate thresholds for targets (Methods; Supplemental Table S2).
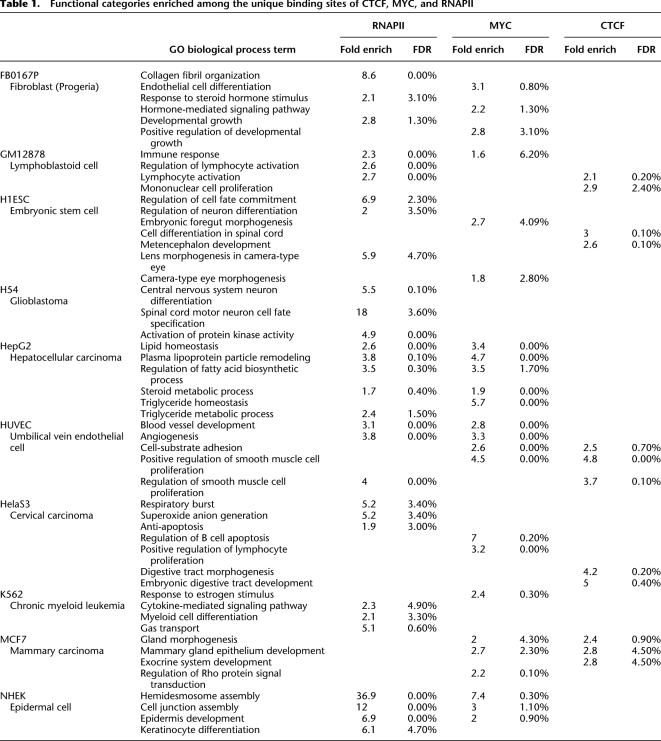
Table 1.
Functional categories enriched among the unique binding sites of CTCF, MYC, and RNAPII
Figure 1.
Overview of ChIP-seq data over a sample genomic region. Chromosome coordinates are shown on top. Data for each cell line and factor are shown as a “wiggle” track of extended reads. Gene annotations derived from the UCSC Genome Browser database are shown at bottom, with the direction of transcription indicated by arrows.
While the number of binding sites varied in different cell types, CTCF, MYC, and RNAPII had ∼45,000, 8000, and 30,000 sites in each cell type on average, respectively. These numbers include partly overlapping sites and therefore do not reflect the number of discrete binding regions across the genome or genes that are potentially targeted, particularly for RNAPII, which tended to show clusters of sites around promoters. Each additional cell line continued to show additional binding sites rather than reaching saturation (Supplemental Fig. S2), suggesting many cell-type specific binding sites exist in diverse cell types.
CTCF binding sites are prevalent in intergenic regions, while MYC and RNAPII binding sites are associated with promoters
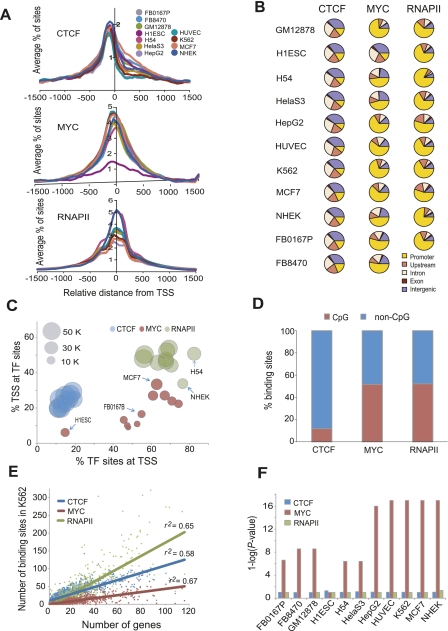
We determined the average profile of CTCF, MYC, and RNAPII occupancy around the TSS. As in previous studies (Ren et al. 2002; Tabach et al. 2007), a peak of high occupancy was seen around the TSS (Fig. 2A). The three factors often occupied TSS in combination, as illustrated in the example track image in Figure 1. None of the factors showed any bias in occupancy around the transcription termination sites (TTS) of genes (Supplemental Fig. S3). Clustering of the gene-wise occupancy signals of these three factors revealed distinct patterns. For example, CTCF binding was observed in distal regions as well as in the gene bodies of many genes, while strong RNAPII occupancy was also observed across the gene bodies of many genes. Strikingly, these occupancy patterns were maintained across all cell types for all three factors (Supplemental Fig. S4).
Figure 2.
Occupancy patterns of CTCF, MYC, and RNAPII relative to gene annotations. (A) Average profiles of CTCF, MYC, and RNAPII binding sites within ±1.5 kb from all annotated TSS (Transcription start site, indicated by zero on the x-axis) in 10–11 different cell types. The average percentage of binding sites within ±1.5 kb of the TSS is shown on the y-axis. (B) Pie charts show the distribution of CTCF, MYC, or RNAPII binding sites in five different genomic regions. A promoter is defined as a region within ±2 kb from the TSS of a gene, upstream is between 2 and 20 kb upstream of the TSS, and intergenic is a region excluding a promoter, upstream, intron and exon. (C) The percentage of binding sites of each TF within ±2 kb from a TSS (x-axis) is plotted against the percentage of TSS within ±2 kb from a TF binding site (y-axis). The area of the circles is proportional to the number of binding sites. Some cell types are indicated by arrows. (D) CTCF prefers to bind in non-CpG islands. The y-axis shows the percentage of binding sites in CpG and non-CpG loci for the factors indicated on the x-axis. (E) TF binding sites are positively correlated with gene density. The scatterplot shows the number of genes in each 2 Mb bin across the genome (x-axis) and the number of TF binding sites in each bin (y-axis). The three factors are shown in different colors as indicated. The lines show the linear regression fit for each factor. (F) MYC enrichment in bidirectional promoters. The y-axis shows the enrichment of binding sites in bidirectional promoters calculated as 1-log (P-value) using the hypergeometric distribution, for each of the cell types indicated on the x-axis. A value of 1 on the y-axis represents no enrichment, seen for RNAPII and CTCF.
Next, we investigated CTCF, MYC, and RNAPII binding relative to a combined gene annotation set including RefSeq, UCSC, Ensembl, and Vega annotated genes from the UCSC Genome Browser (http://genome.ucsc.edu/; Methods). In addition to annotated exons and introns, we defined a region within ±2 kb from a TSS as a promoter, between 2 and 20 kb upstream of a TSS as an upstream region, and >20 kb away from a gene as intergenic. Each factor showed a distinctive binding distribution (Fig. 2B). MYC in particular showed considerable variation in the proportion of its binding sites at promoters, ranging from 45% to 75%. The H1ES cells, however, were an outlier in this regard, with only 15% of its binding sites at promoters. While this could be partly a reflection of higher background in this data set, many characteristic MYC sites were clearly detected in H1ES cells as well (Supplemental Fig. S5). To evaluate promoter selectivity, we compared the proportion of TF binding sites that fell within promoters to the proportion of all promoters bound by a particular TF. This analysis showed that ∼70% of RNAPII binding sites were in promoters, and these may regulate up to half of all genes. About 15% of CTCF sites were in promoters, potentially contributing to the regulation of as many as a quarter of all genes (Fig. 2C). In contrast, MYC exhibited substantial variation, occupying 6%–33% of promoters, suggesting that MYC may modulate varying subsets of genes in different cell types. More than 60% of CTCF binding sites were in distal upstream and intergenic regions, consistent with previous studies of CTCF binding in individual cell types (Barski et al. 2007; Schmidt et al. 2010).
MYC and RNAPII showed significantly higher occupancy scores at promoters than in other genomic regions, but CTCF showed significantly higher occupancy signals in upstream regions rather than promoters (Supplemental Fig. S6), consistent with its role as an insulator binding protein, functioning between a promoter and an enhancer (Valenzuela and Kamakaka 2006). About half of all MYC and RNAPII sites were located in CpG islands, which are known to be associated with promoters, whereas only 12% of CTCF binding was in CpG islands (Fig. 2D). However, even CTCF was significantly enriched in CpG islands relative to background since only 0.75% of the genome is contained in CpG islands. Conversely, the binding sites of these three factors in CpG islands were enriched for promoters (Supplemental Fig. S7). Moreover, the binding sites of MYC and RNAPII in CpG islands had significantly higher occupancy scores than their sites in non-CpG-containing sites (Supplemental Fig. S8). Taken together, these results indicate that MYC and RNAPII regulate genes primarily by binding to proximal promoters, with MYC exhibiting greater diversity of gene targets across cell types, whereas CTCF modulates expression of a more conserved set of targets by associating with distal cis-regulatory elements.
Transcription factor binding sites are positively correlated with gene density across the genome
Gene density varies considerably across the human genome (Lander et al. 2001). Since all three TFs occupied distal binding sites in addition to promoters, we examined the relationship between TF binding and gene density. As shown in Figure 2E, TF binding sites were positively correlated with gene density. Even though CTCF showed a clear preference for intergenic regions over promoters (Fig. 2B), its binding was nonetheless positively correlated with gene density, consistent with previous observations (Kim et al. 2007). Interestingly, excluding CTCF binding sites within genes as well as up to 20 kb upstream of TSS did not completely abrogate the correlation between binding sites and gene density (Supplemental Fig. S9), suggesting that even the many distal CTCF binding sites may regulate gene expression at long range.
MYC is overrepresented in bidirectionally transcribed promoters
DNA binding motifs for several sequence-specific TFs have been reported to be overrepresented in bidirectional promoters (Lin et al. 2007), and we have previously reported that E2F4 binding sites are overrepresented in bidirectional promoters (Lee et al. 2011). We therefore examined whether any of the TFs showed a bias in binding to bidirectional promoters. Based on annotations for 22,279 genes in RefSeq, there are 1233 promoters corresponding to 2466 bidirectionally transcribed genes in the human genome. In the majority of cell types, bidirectionally transcribed genes were significantly overrepresented among the target genes of MYC where it bound within 2 kb of the TSS (Fig. 2F). This overrepresentation of bidirectional promoters was specific to MYC binding sites as it was not observed for CTCF or RNAPII binding sites. In most cases, the binding of CTCF, MYC, or RNAPII at bidirectional promoters activated both genes equally, regardless of the distance of its binding site from a TSS (Supplemental Fig. S10).
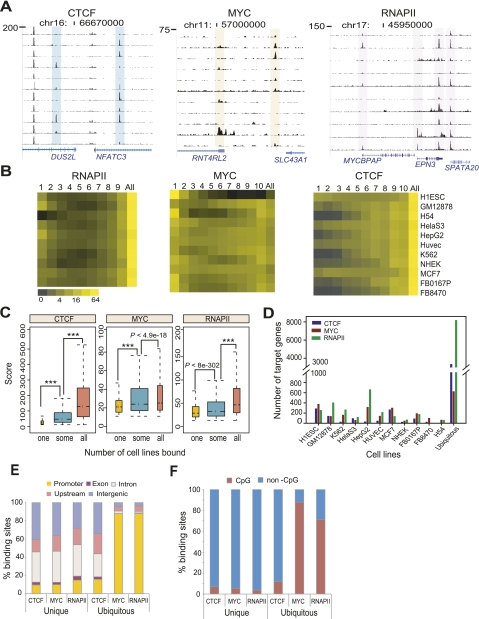
CTCF and RNAPII sites are ubiquitous, whereas MYC sites are cell-type specific
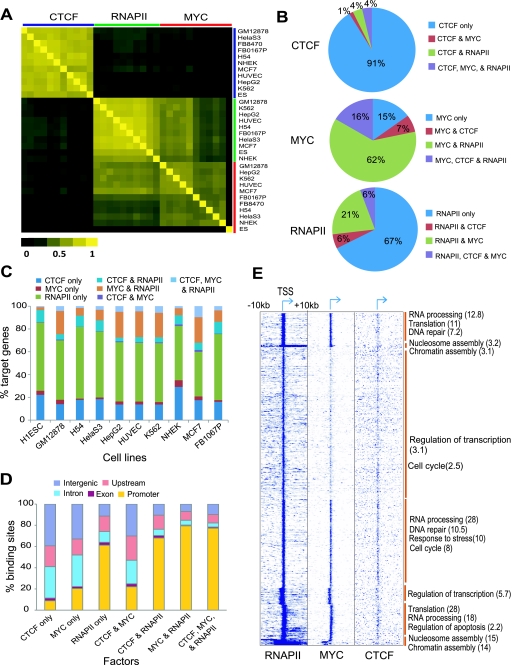
All three factors showed some cell-type specificity (Fig. 3A). To quantify the extent of cell-type specific binding of CTCF, MYC, and RNAPII, we first analyzed the overlap of their binding sites across all cell types (Methods). If a binding site overlapped in all cell types, we defined it as ubiquitous; otherwise sites were considered as cell-type specific. We further categorized cell-type specific binding sites found in only one cell type as unique binding sites. Less than 13% of MYC sites were ubiquitous, suggesting that a large proportion of MYC binding sites show some degree of cell-type specificity (Fig. 3B; Supplemental Fig. S11).
Figure 3.
CTCF, MYC, and RNAPII have many cell-type specific regulatory elements. (A) Track images show examples of cell-type specific sites of each factor in different cell types. (B) Heat maps show the relative distribution of cell-type specific and ubiquitous binding sites of each factor in 10–11 different cell types. The horizontal axis represents the number of cell types sharing a binding site; thus, “1” represents “unique” sites found in only one cell type, “All” indicates “ubiquitous” sites found in all cell types, and other numbers show intermediate representation. Here we denote all sites with the exception of ubiquitous sites as “cell-type specific” sites. The color in the heatmap indicates the proportion of sites in each category, in each cell type. (C) Boxplots show the ChIP-seq score distribution of unique (“one”) and ubiquitous (“some” and “all”) sites across all cell types. Unique sites have significantly lower ChIP-seq scores than the other sites. P-values were calculated by Wilcoxon rank sum test. Asterisks indicate a calculated P-value of zero. (D) The number of unique and ubiquitous target genes of CTCF, MYC, and RNAPII in diverse cells. The downstream genes of TF-bound promoters (within ±2 kb from TSS) were considered as target genes. (E) The distribution of unique and ubiquitous binding sites of CTCF, MYC, and RNAPII across all cell types, in five different genomic regions. The x-axis represents each factor in either unique or ubiquitous sites. The y-axis shows % binding sites in the genomic regions. (F) Percent CpG and non-CpG sites in unique and ubiquitous binding sites across all cell types. The x-axis represents each factor in either unique or ubiquitous sites. The y-axis indicates percent binding sites of these three factors in CpG or non-CpG sites.
In contrast to MYC, more than half of all CTCF binding sites were ubiquitous across the 11 cell types. More than 75% of CTCF binding sites occurred in at least seven cell types, with <3% of CTCF sites unique in any of the cell types we analyzed, except ES (6.4%) and MCF7 (3.4%) (Fig. 3B). Similarly, RNAPII also exhibited a strong preference for ubiquitous binding; however, unlike CTCF, a significant proportion of RNAPII binding sites were unique to a single cell type (an average of 7.7% across cell types). Repeating this analysis using only the subset of binding sites that occurred at promoters (which had higher occupancy) and showed >80% replicate reproducibility, gave essentially the same results. These results suggest that MYC binding predominantly regulates unique cell type functions whereas CTCF's regulatory role is largely consistent across diverse cell types. Interestingly, the unique binding sites of CTCF, MYC, and RNAPII had lower occupancy scores compared with their ubiquitous binding sites (Fig. 3C).
By assigning the gene downstream from a TF-bound promoter as its target gene, we found an average of 91, 169, and 233 unique targets and 3321, 621, and 8167 ubiquitous target genes, respectively, for CTCF, MYC, and RNAPII (Fig. 3D). Most ubiquitous sites of MYC and RNAPII occurred in promoters, while their unique sites were in distal regions and introns (Fig. 3E), suggesting that the unique sites of MYC and RNAPII may function as distal regulatory elements like enhancers. Cell-type specific sites of CTCF, MYC, and RNAPII lacked CpG islands compared to their ubiquitous sites (Fig. 3F).
We used the Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al. 2010) to examine biological functions targeted by the unique and ubiquitous binding sites of the three TFs. Unique sites frequently targeted genes in functional categories relevant to the biological characteristics or tissue of origin of a cell type (Table 1). Interestingly, even though the unique sites of MYC tended to have the lowest occupancy scores in our overall analysis, this class of site targeted genes in meaningful functional categories more frequently than CTCF unique sites, which had higher scores. Moreover, the unique sites of RNAPII and MYC frequently targeted overlapping functional categories in several cell types including GM12878, HepG2, HUVEC, and NHEK, suggestive of combinatorial usage of these two factors in specifying cell function (Table 1). Ubiquitous MYC binding site target genes showed moderate enrichment in translational elongation (Supplemental Table S3), consistent with previously reported functions for MYC in regulating translation and cell growth (Boon et al. 2001; van Riggelen et al. 2010).
MYC and RNAPII co-localize in many promoters
To examine relationships between genes potentially targeted by TFs, we calculated correlations between target gene sets and clustered them. CTCF targets generally correlated well with each other across cell types, with RNAPII targets showing lower correlations, followed by MYC (Fig. 4A). Between factors, there was weak, but positive correlation between CTCF and either MYC or RNAPII targets (Pearson correlation coefficient r = ∼0.2), and moderate correlation between MYC and RNAPII (r = ∼0.4), consistent with a functional relationship among the three factors (Fig. 4A). We further investigated single or combinatorial occupancy of these factors at their target sites and genes. The largest proportion of binding sites was occupied by only one factor, but a significant proportion was co-occupied by at least two factors (Fig. 4B; Supplemental Fig. S12). We saw similar relationships between target genes occupied singly or in combination by these three factors (Fig. 4C; Supplemental Fig. S13). Associations between these factors were further supported by the fact that both CTCF and MYC were co-enriched at RNAPII sites (Supplemental Fig. S14). These results suggest that a substantial set of genes may be regulated by combinatorial binding of these three factors, in particular MYC and RNAPII. In general, co-occupied sites were overrepresented in promoters as compared to sites occupied by single factors, particularly when a combination included RNAPII. Seventy percent of the MYC–RNAPII and CTCF–MYC–RNAPII combinatorial sites were in promoters, which was an enrichment over the RNAPII-only sites seen in promoters (Fig. 4D).
Figure 4.
CTCF, MYC, and RNAPII can regulate their target genes in a combinatorial manner. (A) Heat map of correlations between the target genes of each combination of factor and cell type. The Pearson correlation coefficient was calculated between the set of target genes of each factor and those of other factors in a binary mode (target or non-target). TFs and cell type combinations are arranged in the same order along both axes, as listed on the vertical axis. (B) Proportion of single and combinatorial binding sites of CTCF, MYC, and RNAPII in K562. (C) Proportion of single and combinatorial target genes of the three factors in each cell type. The percentage of target genes in each category is shown on the vertical axis, for each of the cell types shown below. (D) The distribution of single or combinatorial binding sites of the three factors in five different genomic regions. The average percentage of binding sites in each region across all cell lines is shown on the vertical axis, for each of the combinations shown below. (E) Heat map showing gene-wise patterns of TF occupancy in a 20-kb window around the TSS. Data were clustered using K-means clustering. Functional enrichment of genes in indicated functional categories for the groups indicated by a brown vertical bar is shown as the negative log of the P-value in brackets on the right.
Clustering of global occupancy patterns of the three factors revealed a few distinct clusters of binding patterns, with genes implicated in a wide range of functions (Fig. 4E). Genes bound by the combination of MYC–RNAPII or CTCF–MYC–RNAPII showed an enrichment for genes involved in translation, RNA processing, splicing, and ribosome biogenesis across all cell types, which suggests combinatorial control of genes implicated in general biological processes (Supplemental Table S4).
CTCF, MYC, or RNAPII binding positively correlates with target gene expression
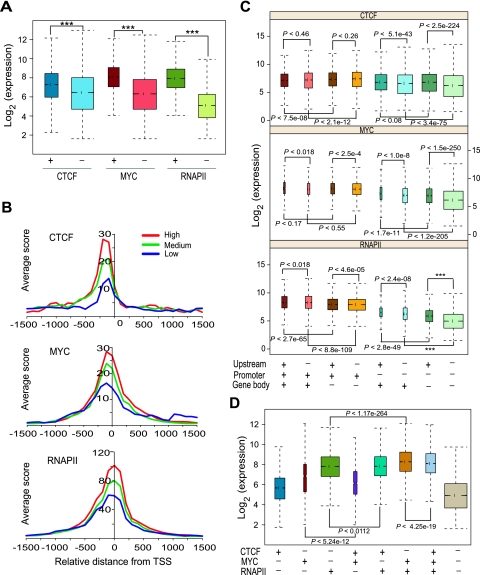
We investigated the relationship between CTCF, MYC, and RNAPII binding and target gene expression by comparing their transcript levels. Genes whose promoters were occupied by any one of the three TFs showed significantly higher expression than genes whose promoters were not occupied by that TF, across all cell types (Fig. 5A). The positive relationship between binding and transcript levels was also evident in the average binding profiles in the high, medium, and low expression level groups (Fig. 5B; Supplemental Fig. S15).
Figure 5.
CTCF, MYC, or RNAPII binding is associated with activated expression of their target genes. (A) Boxplots show that genes downstream from promoters (within ±2 kb from TSS) bound by any one of the three factors have significantly higher expression than genes not occupied by them. Data shown are across all cell types. Groups along the x-axis indicate promoters either occupied (+) or not occupied (−) by the indicated TF. The y-axis shows transcript expression levels. P-values were calculated by Wilcoxon rank sum test. Three asterisks (***) indicate a P-value of zero. (B) TSS profiles of CTCF, MYC, and RNAPII binding in K562 cells over genes with different expression levels. Genes were divided into three different expression groups based on their expression level in K562 cells as top 33% (High), middle 33% (Medium), bottom 33% (Low). The y-axis indicates average ChIP-seq score. (C) Boxplots show distribution of gene expression levels of genes bound by TFs in three different genomic regions: promoters, upstream, and gene body. Occupancy of each factor in the indicated genomic location is shown as + (presence) and − (absence) at the bottom. The y-axis shows the log-transformed expression level of genes. Data shown are across all cell types. P-values were calculated by Wilcoxon rank sum test. (D) Boxplots show expression of genes with single and combinatorial binding of the three TFs across all cell types. Combinatorial binding of MYC and RNAPII enhances their target gene expression. The x-axis represents single-factor-bound or multi-factor-bound gene groups. Occupancy of each factor was shown as + (presence) and − (absence) at the bottom. The y-axis shows the log-transformed expression level of genes.
We also investigated the effects of TF binding upstream (between 2 and 20 kb) of a TSS or within the gene body (within exons and introns of the gene). We first assigned binding sites to the nearest gene, then evaluated the expression levels of genes in each of the eight groups formed by the combination of upstream, promoter, and gene-body binding by the TF (Fig. 5C). The effects of upstream or gene-body binding often depended on the status of promoter binding. For instance, CTCF upstream binding was associated with an increase in its target gene expression only when it did not also bind at the promoter. CTCF gene-body binding was associated with reduced target gene expression when it also bound to the promoter, but associated with increased expression when it was depleted at promoters (Fig. 5C). Upstream binding of both MYC and RNAPII was associated with a positive effect on target gene expression, but the effect was stronger in the absence of their promoter binding. Gene-body binding of MYC corresponded to increased target gene expression only when MYC did not also bind to promoters, whereas RNAPII gene-body binding showed a positive relationship to expression regardless of its binding at the other locations (Fig. 5C). Interestingly, while there was a modest association of upstream binding with increased expression levels compared to no binding, there was no significant drop-off with increasing distance of binding ranging from 2 to 20 kb (Supplemental Fig. S16).
We also examined the expression level of genes whose promoters were bound by different combinations of the three factors. Genes whose promoters were bound by a single TF exhibited the highest expression levels for RNAPII binding and lowest expression levels for CTCF, with MYC being intermediate (Fig. 5D). Genes occupied by both MYC and RNAPII showed higher expression levels than the genes bound by either MYC or RNAPII alone whereas genes with combinatorial binding of MYC and CTCF exhibited lower expression than genes occupied by MYC without CTCF. These results suggest that promoters occupied by a single TF of the three we examined are generally active, but promoters showing combinatorial occupancy of TFs can either be more or less active depending on the combination of the three factors.
RNAPII regulates gene expression in four distinctive binding patterns across a gene
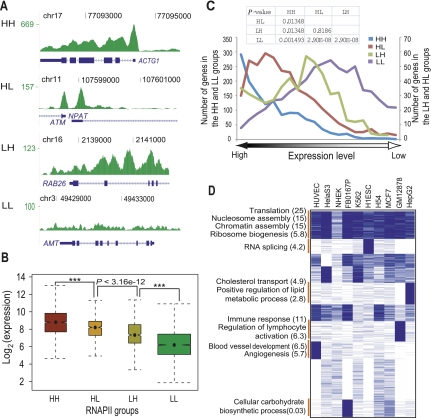
To identify distinct modes of RNAPII association with genes and examine their relationship to transcription, we first classified genes into four groups based on how RNAPII was bound to different gene regions: HH, showing high occupancy in both the promoter and the gene body; HL, high occupancy in the promoter and low occupancy in gene body; LH, low occupancy in promoter and high occupancy in the gene body; LL, low occupancy in both promoter and gene body (Fig. 6A; Methods), and then examined the expression level of each group of genes. Genes showing the HH pattern of RNAPII binding had the highest expression levels whereas genes in the LL group showed the lowest expression (Fig. 6B). Compared to the HH group, the HL group containing paused RNAPII at proximal promoters but little signal in the gene body showed significantly lower gene expression (Fig. 6B; Supplemental Fig. S17), which is consistent with results from RNAPII profiling in Drosophila melanogaster (Zeitlinger et al. 2007). We then ranked genes by their expression values and plotted the distribution of genes in each of the four modes of RNAPII occupancy as a function of expression. The proportion of genes in the HH group decreased with decreasing expression levels while those in the LL group gradually increased (Fig. 6C). Genes in the HL group were more biased toward highly expressed genes than genes in the LH category (Fig. 6C; Supplemental Fig. S18).
Figure 6.
RNAPII binding regulates gene expression in four distinct binding modes. (A) Wiggle track images show examples of four RNAPII binding groups classified based on its occupancy signal in the promoter and the body of a gene. (HH) High occupancy in both the promoter and the body of a gene; (HL) high occupancy only in the promoter of a gene; (LH) high occupancy only in the body of a gene; (LL) low occupancy signal in the promoter as well as the body of a gene. (B) Notched boxplot shows the distribution of expression level among four classes of RNAPII binding sites in K562 cells. The x-axis represents four different RNAPII binding groups. The y-axis shows log-transformed expression level of corresponding genes. Data for other cell types are shown in Supplemental Figure S17. (C) Distribution of genes of four RNAPII groups in the expression rank map. The x-axis indicates expression ranking from highest (left) to lowest (right). The y-axis represents number of genes in four groups. Two y-axes were used to represent the range of genes in each of the four groups. (D) Gene functional enrichment in cell-type specific as well as ubiquitous RNAPII targets in the HH group. Cell types are shown on top. Binding data were rendered in blue (occupied) and white (not-occupied) and clustered by K-means clustering. The functional annotation program, DAVID (Huang et al. 2007), was used to analyze functional enrichment. Annotated functional groups of each cluster are shown with the negative log of the P-values (Bonferroni corrected) in brackets.
We clustered genes in the four RNAPII binding groups to ascertain whether these distinct occupancy patterns corresponded to functional outcomes. Only genes in the HH group showed strong functional enrichment, with housekeeping functions enriched in the ubiquitous clusters where genes showed RNAPII occupancy constitutively in all cell types (Fig. 6D). Genes in the HH group where RNAPII occupancy was observed in a cell-type specific manner showed functional enrichment for cell-type specific functions, such as angiogenesis for HUVECs and lymphocyte activation in lymphoblastoid cells (Fig. 6D). The other three RNAPII occupancy groups including HL, LH, and LL showed little functional enrichment (data not shown).
Novel promoters and alternative promoter usage of RNAPII
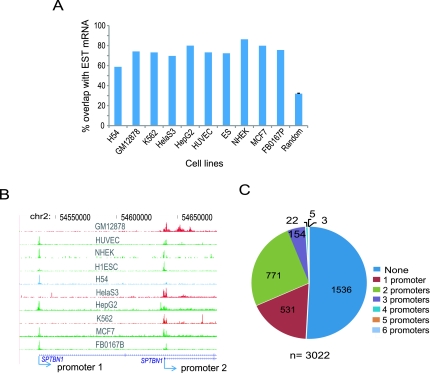
Recent studies have revealed that RNAPII can bind outside of genes, for example, to enhancers (Koch et al. 2008; De Santa et al. 2010; Kim et al. 2010). Indeed, even after mapping RNAPII sites relative to genes using combined gene annotations from RefSeq, UCSC, Ensembl, Vega, and SIB genes downloaded from the UCSC Genome Browser, we found a considerable proportion of RNAPII binding sites (7%–20%) in distal (upstream and intergenic) regions. These RNAPII sites could either represent unannotated novel promoters or enhancers. A large majority (93%) of these distal RNAPII sites contained core promoter motifs such as initiator (INR), TATA box, TFIIB recognition element (BRE), downstream core promoter element (DPE), or motif 10 element (MTE) (Jin et al. 2006), but this representation was not different from background (not shown). We therefore compared these distal RNAPII binding sites with expressed sequence tag (EST) data to see whether they could be associated with transcription. We considered a RNAPII site to be associated with a transcript and potentially a novel promoter if it lay from 2 kb upstream to 300 bp downstream from the 5′ end of the EST. On average, 74% of the distal RNAPII sites across all cell types corresponded in this manner to EST tag mRNA, which was a ∼2.3-fold enrichment for EST overlap compared to background (Fig. 7A). This suggests that, while the majority of distal RNAPII sites could be promoters for novel uncharacterized transcripts, up to ∼26% could be enhancers marked by RNAPII occupancy.
Figure 7.
Novel promoters and cell-type specific alterative promoter usage. (A) Percent overlap of RNAPII distal sites with expressed sequence tags (ESTs). The random background control was generated from the average overlap between 10,000 random intergenic loci and ESTs, repeated 10 times and averaged. The small error bar shows the standard deviation for the randomized control. (B) An example of cell-type specific alternative promoter usage is shown in genome browser tracks. Chromosomal coordinates are shown on top and transcripts at bottom. Arrows indicate two TSS locations as well as direction of transcription. Blue, red, and green indicate cell lines using promoter 1, promoter 2, or both promoters, respectively. (C) Pie chart showing the number of genes utilizing different numbers of alternative promoters in K562. “n” indicates the total number of genes having at least two promoters.
To evaluate the extent to which alternative promoters are used in diverse cell types, we first identified alternative promoters for RefSeq annotated genes by flagging genes that had the same gene symbol but different TSS annotations in the RefFlat file (http://genome.ucsc.edu/), then mapped RNAPII binding sites to these alternative promoters. Figure 7B shows one example of cell-type specific alternative promoter usage. We found that a considerable number of genes (∼4.3%) were transcribed utilizing at least two alternative promoters (Fig. 7C; Supplemental Fig. S19).
RNAPII shows higher occupancy at exons than adjacent introns
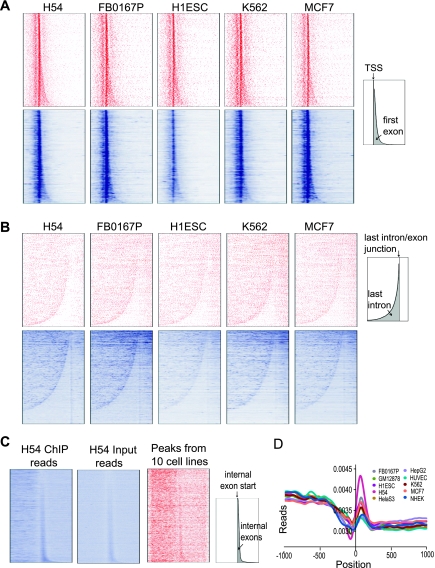
Recent studies indicate that chromatin structure differs at exons and introns, likely reflecting an effect of co-transcriptional splicing (Schwartz et al. 2009; Spies et al. 2009; Huff et al. 2010). We examined whether co-transcriptional splicing might be more directly reflected in RNAPII occupancy over exons and introns. We first examined RNAPII occupancy around the initial and terminal exon/intron junctions. Strong RNAPII binding around the TSS, combined with the highly variable lengths of the first exon and intron, makes it difficult to reliably quantify specific differences in occupancy between the first exon and intron. We visualized RNAPII occupancy over the first exon/intron junction by generating gene-wise heat maps where genes were aligned at their TSS and sorted by the length of their first exons. This analysis showed that, in addition to the high occupancy at the TSS, RNAPII also binds preferentially to the first exon compared with its downstream intron. This enrichment was seen in all 10 cell types when either input-corrected RNAPII ChIP peaks or raw RNAPII ChIP-seq reads were plotted (Fig. 8A; Supplemental Fig. S20). Similarly, when we aligned genes by the 5′ end of their last exon and sorted them by the length of their last intron, RNAPII occupancy was lower within the last intron relative to upstream and downstream exons (Fig. 8B; Supplemental Fig. S20). We obtained the same results if we considered only constitutively spliced exons, which are always included in all splice isoforms and therefore unlikely to contain alternative transcription start sites, indicating that the higher occupancy of RNAPII over exons is not entirely due to internal initiation sites (not shown).
Figure 8.
RNAPII is enriched in exons. (A) Heat maps of gene-wise RNAPII occupancy. RNAPII signals in the form of input-corrected peak scores (red) or read counts (blue) were assigned to 10-bp bins across the 4-kb region shown in these plots (1 kb upstream of and 3 kb downstream from the TSS). Only genes with at least one peak or read occurrences within 4 kb of their TSS are plotted. As a result, the plotted gene sets in the top row (red) are different from that in the bottom row (blue). Five representative cell lines are shown and data for additional cell lines are in Supplemental Figure S20. The schematic at the right shows the overall gene map, with the first exon area shaded in gray. (B) RNAPII occupancy is higher in the last exon compared to the last intron. Heat maps similar to A, but genes were aligned by the start of their last exons and sorted by the length of their last introns. RNAPII signal across the 9-kb region (7 kb upstream of and 2 kb downstream from the start of the last exon) is represented similarly as in A. Most of the long genes at the bottom of the read plots (blue) do not have significant RNAPII binding and therefore are not part of the peak plots (red), a phenomenon contributing to the pattern difference between read plots and peak plots. (C) RNAPII is enriched on internal exons. Internal constitutively spliced exons were aligned by their start and sorted by exon length. RNAPII occupancy 1.5 kb upstream of and downstream from the exon start sites is plotted. In the representative H54 cell type shown, reads for both RNAPII ChIP and input showed stronger intensity within internal exons (first two panels, blue). However, the combined RNAPII peaks (input-corrected and normalized) from all 10 cell types showed modest enrichment at internal exons (third panel, red). (D) Read profiles for RNAPII ChIP-seq, after correcting for input signal using our binomial correction method. Zero indicates the start (5′ end) of constitutive internal exons.
To evaluate RNAPII occupancy at internal exons, we generated heat maps of its occupancy by aligning all constitutive internal exons by their 5′ ends and sorting by exon length. The small exon sizes and relatively sparse RNAPII peaks over this region made it difficult to visualize a significant difference in occupancy between exons and introns when we considered peaks in individual cell types (Supplemental Fig. S21). Heat maps of raw RNAPII ChIP-seq reads revealed higher signal within internal exons compared with adjacent regions, but a similar enrichment over internal exons was also seen in input samples (Fig. 8C, left and middle panels; Supplemental Fig. S22). This enrichment is partly, but not entirely, due to more unique sequence content within exons which results in higher alignability (Supplemental Fig. S22). We did not see similar high signals in input samples at the first or last exons (Supplemental Fig. S23). Combining the data for RNAPII ChIP peaks, which was corrected for input signal, across all 10 cell types confirmed the higher RNAPII occupancy within constitutive internal exons (Fig. 8C, right panel). The average read count for RNAPII around constitutive internal exons, corrected for input signal using our binomial correction method also showed higher ChIP signal over internal exons (Fig. 8D). Thus, RNA RNAPII occupancy tends to be higher at exons than at adjacent introns.
Motif analysis
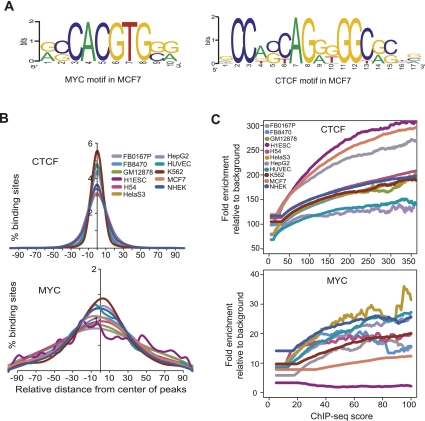
To identify sequence motifs in the binding sites of the sequence specific TFs CTCF and MYC, we used the Discriminating Matrix Enumerator (DME) algorithm (Smith et al. 2005). We divided binding sites into strong, moderate, and weak groups based on their ChIP-seq scores, and searched for motifs de novo in these three groups. The algorithm identified only the previously known canonical motif for both CTCF (Kim et al. 2007) and MYC (Blackwell et al. 1993) in all three groups in all cell types except in the case of MYC in ES cells (Fig. 9A; Supplemental Fig. S24). While an alternative motif was discovered in the strongest MYC binding sites in ES cells (Supplemental Fig. S24), we noted that many of these strong sites occurred in clusters near regions of low complexity; the significance of this alternative motif is therefore unclear. Next, we examined the location of the putative binding motifs relative to the position of the transcription factor binding peak. In virtually all cases, the most likely position for the motif was the position of the binding peak for both CTCF and MYC (Fig. 9B). Motif enrichment relative to background gradually increased with ChIP-seq score for both CTCF and MYC in all 11 cell types (Fig. 9C).
Figure 9.
Motif enrichment in binding sites. (A) Motifs discovered de novo from strong binding sites in MCF7 for CTCF and MYC, using the DME algorithm. (B) Distribution of motifs for CTCF and MYC around the center of individual binding sites for each factor. The plots show the distribution of the canonical motif, discovered de novo from our ChIP-seq data. However, for MYC in H1ES cells, where the canonical motif was not discovered de novo, the plot shows the distribution of the known canonical motif, CACGTG. (C) Relationship of motif enrichment relative to background and the ChIP-seq occupancy score. The plots show the behavior of the canonical motif, discovered de novo from our ChIP-seq data.
Discussion
Combinatorial regulation of expression
The higher occupancy of TFs at ubiquitous sites relative to cell-type specific sites is consistent with the idea that cell-type specific regulation is likely to involve binding by a combination of several TFs. While occupancy of putative target genes by any one of these diverse TFs was associated with increased transcription, combinatorial occupancy gave varied effects. Combinations of MYC and RNAPII occupancy were associated with additional increases in transcription, but CTCF occupancy in conjunction with MYC and/or RNAPII was associated with modest repression (Fig. 5D). The strongest effect on transcription was when regions close to the TSS were occupied. Although binding at a distance was still associated with increased transcription, the effect was weaker and not distance dependent (Supplemental Fig. S16). The positive effect of CTCF, MYC, and RNAPII binding in upstream regions is consistent with many of these upstream regions acting as enhancers (De Santa et al. 2010; Kim et al. 2010). Our data suggest that a prominent means of modulating the transcriptional activity of genes is by altering the cohort of transcription factors that bind near the core promoter, rather than altering the activity of any given transcription factor. Interestingly, while action at a distance is a well-established mode for transcription factor function, we found that binding sites were positively correlated with gene density. Binding sites for all three factors were much rarer in gene deserts, suggesting there are limits both on the distance over which transcription factors act upon genes as well as the extent to which novel active transcriptional units remain to be discovered in gene-poor regions.
Different classes of RNAPII occupancy patterns
Categorizing RNAPII binding over genes into distinct patterns revealed how these binding patterns relate to gene expression. The fact that the HL and LH clusters did not show enrichment for cell-type specific functions (while HH did) suggests that the HL and LH classes contain paused RNAPII while HH is the most transcriptionally active. However, even though the HL group having promoter-paused RNAPII showed lower expression of its target genes compared to the HH group, genes in the HL group were still highly expressed. This suggests that many actively transcribed genes also possess paused RNAPII at their proximal promoters, but the majority of RNAPII pausing is transient rather than long-lasting, which would prevent genes from being expressed. Our observation is consistent with recent studies in D. melanogaster showing that RNAPII proximal pausing is prevalent even in actively transcribed genes (Gilchrist et al. 2010). The fact that the majority of RNAPII sites were ubiquitous was surprising and might reflect the likelihood that a major mode of transcriptional regulation is not at the level of recruitment of RNAPII but a post-recruitment step such as relief of pausing or elongation.
RNAPII exon enrichment and co-transcriptional splicing
Since splicing occurs while the pre-mRNA is being actively transcribed (Allemand et al. 2008), it is possible that the transcription and splicing machineries interact. RNAPII interacts with the splicing initiator SR proteins and this interaction is essential for both pre-mRNA splicing and RNAPII elongation (Zhong et al. 2009). Recent studies also reveal a possible relationship between chromatin modifications and splicing by demonstrating that several histone modifications, especially H3K36me3 are enriched at exons (Andersson et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Huff et al. 2010). While this may reflect a slowing down of RNAPII elongation which in turn can favor exon inclusion (de la Mata et al. 2003; Schwartz et al. 2009; Ip et al. 2011), there is little direct evidence regarding whether RNAPII slows down and shows higher occupancy at exons. Conflicting results were obtained when RNAPII elongation rates were assayed on reporter genes containing varying number and length of introns (Alexander et al. 2010; Brody et al. 2011). Possibly because the difference of RNAPII occupancy over exons and introns is small, and because mammalian exons are on average much smaller than introns, studies at individual loci may give conflicting results. ChIP-ChIP studies in plants have shown that RNAPII preferentially binds to exons rather than introns (Chodavarapu et al. 2010). We found that RNAPII occupancy is consistently higher within exons in vivo across 10 different human cell types, reflecting a relationship between transcriptional elongation by RNAPII and exon–intron boundaries, which has not been directly observed before on a genome-wide scale in human genes. Further studies examining the relationship of the splicing status of genes with the extent of exon bias of RNAPII binding will clarify whether the RNAPII exon bias is directly related to co-transcriptional splicing or if it reflects some other mode of demarcating exons and introns.
Our study shows how genome-wide binding of diverse transcription factors is related to gene regulation in a multitude of human cell types. The data we have generated and made publicly available will be a useful reference data set both for investigators studying these or related transcription factors as well as those studying similar or related cell or tissue types. Future analysis of these data in relation to other genome-wide data sets being generated by the ENCODE Consortium and other groups such as the Epigenomics Initiative, on histone modifications, DNA methylation, high-resolution RNA analyses using RNA-seq, phylogenetic conservation, and genetic variation, will illuminate how gene regulation across the human genome is orchestrated.
Methods
Cell lines and culture
The ENCODE Consortium has designated GM12878, K562, HeLaS3, HepG2, HUVEC, NHEK, and H1ESC cells as Tier 1 and Tier 2 cell lines. The source and cell growth conditions for these cells are described at the ENCODE website (http://genome.ucsc.edu/ENCODE/cellTypes.html). Additional cell types analyzed in this study listed in Table 1 were cultured under standard culture conditions. Cells were grown to appropriate numbers and processed for chromatin immunoprecipitation.
Chromatin immunoprecipitation and deep sequencing (ChIP-seq)
ChIP assays were performed as described previously (Lee et al. 2011). Briefly, cells were cross-linked with 1% formaldehyde for 10 min at room temperature. The cross-linked cells were sheared by sonication until the average fragmented DNA size reached 500 bp, then TF–DNA complexes were pulled down with a specific antibody for either CTCF (07-729, Millipore), MYC (SC-764X, Santa Cruz Biotech), or the large subunit of RNAPII (POLR2A, MMS-126R, Covance Inc.). An input sample was processed in parallel but leaving out the immunoprecipitation step. After reversal of cross links, DNA was purified, quantitated, and analyzed by deep sequencing primarily using single-end Illumina sequencing technology, with the exception of one out of three replicates that was sequenced using the Applied Biosystems SOLiD platform (Life Technologies).
Peak calling and statistical correction
Thirty-two to thirty-six base pair reads from the ends of ChIP-enriched DNA fragments and corresponding control DNA (input) were mapped back to the human genome (hg18) using the Maq aligner (Li et al. 2008). We combined reads from the biological replicates for each unique factor–cell line combination. The number of mapped reads obtained from each cell type is listed in Supplemental Table S1. To identify discrete regions (binding sites) occupied by a TF from this high-throughput sequencing data, we used a Parzen window density estimation algorithm as described previously with some modifications (Shivaswamy et al. 2008; Lee et al. 2011). Each aligned read was assigned a value representing the frequency of observing that read in the sequenced library; however, to avoid distortions from sequencing artifacts the maximum number of reads at a given location was capped at five for each replicate. After extending reads in the 3′ direction by half the library fragment length (67 bp), a Gaussian kernel with a defined bandwidth was applied to weight the occupancy scores based on the proximity of neighboring nucleotides. These density profiles yielded the initial set of binding locations, with local maxima defining the center of binding, and the interquartile range (IQR) of each peak along the chromosome defining the binding site. The chromosomal coordinates and total number of reads were recorded for each binding site. The resulting set of candidate binding sites was then subjected to input correction, filtering for additional copy number artifacts, and determination of statistical significance.
First, to normalize for regions of high signal in the input control for each cell line, each binding site was paired with the corresponding input site (within 200 bp) with the highest read count. A binomial P-value was computed for each binding site under the null hypothesis that ChIP and input reads were equally likely. The ratio of total ChIP to input reads was used to normalize for differences in overall sequencing depth before calculating the binomial P-value. The binomial P-value was then used to adjust the binding site's read count by calculating the number of ChIP reads there would be if no input reads were present, solving (using R terminology) pbinom(input, chip+input, 0.5) = pbinom(0, corChip, 0.5) for corChip. This “binomial P-value corrected number of reads” (binCorRd) score was recorded for each binding site and was used in all further occupancy score-based analyses.
Only sites where the sequencing-depth-scaled ChIP read count exceeded input were retained. Binding sites falling in previously defined genomic regions with aberrantly high signal due to copy number differences were also discarded (Boyle et al. 2011). High-confidence sites were identified based on the empirical cumulative distribution function of the filtered binding site scores. Wiggle tracks showing the aligned reads and putative peak regions for each data set were generated and loaded into a local mirror of the UCSC Genome Browser (see Fig. 1 for an example). We referred to these data tracks while determining appropriate score thresholds for each data set. Differences in the nature of binding of the three factors and/or the quality of the three antibodies for ChIP, coupled with differences between cell types make it difficult to set a single uniform threshold across all data sets. Qualitatively, CTCF binding sites tend to be narrow and sharp while the dynamic RNAPII transcription machinery produces broad, dense binding signals. MYC sites appeared to be more variable and fewer in number, with lower signal/noise than either CTCF or RNAPII (Supplemental Fig. S25). We therefore initially chose a different top percentage level of highest-scoring binding sites for each TF: 4% for CTCF, 2% for RNAPII, and 0.5% for MYC, and identified a corresponding threshold score. These initial thresholds were slightly adjusted for a few data sets, based on minimum and maximum score considerations, to account for experiment-specific quality differences. Data sets with initial target score thresholds below that corresponding to a binomial P-value of 0.0005 had their thresholds adjusted upwards (removing the lowest scoring sites) and data sets with scores corresponding to a binomial P-value of 1−10 had thresholds adjusted downward (capturing additional high-scoring sites). The final count of significant binding sites identified for each data set, along with the corresponding score thresholds and percentage of top-scoring sites represented, is shown in Supplemental Table S2.
RNA expression profiling
RNA expression profiling was carried out independently as part of the cell line phenotyping component of the ENCODE project. For GM12878, K562, HeLaS3, HepG2, and H1ES, RNA was generated from the same culture of cells used for ChIP; for HUVEC, NHEK, MCF7, FB8470, FB0167P, and H54 cells, different cultures were used for RNA and for ChIP. Details of RNA expression have been previously described, and these data were deposited to the Gene Expression Omnibus (GSE15805) (McDaniell et al. 2010).
Mapping binding sites to gene features and CpG islands
Binding sites were mapped to within ±2 kb from the TSS of all annotated genes generated by combining gene lists from RefSeq (November 2010), UCSC, Ensemble, Vega, and SIB downloaded from UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgTables?command=start), resulting in a total of 348,592 TSS. We used a more comprehensive TSS annotation for mapping binding sites relative to gene features because we did not want to overestimate the proportion of intergenic binding sites. Not every true TSS might be included in the RefSeq annotation, so using a larger, more comprehensive annotation is more conservative. For the other analyses such as TSS/TTS profiles and exon/intron binding, we preferred to use a standard and well-defined gene annotation set such as in the RefSeq set. However, using a larger gene annotation data set does not change the overall distribution pattern of TF binding sites in the TSS/TTS and exon/intron plots. We defined a distinct gene as a unique combination of chromosome number, start, and number of exons across annotation files. We defined a promoter as 2 kb up- or downstream from the TSS, and upstream as between 2 and 20 kb upstream of the TSS. Binding sites that could not be mapped to within 20 kb upstream of any TSS, or to any exon or intron, were termed intergenic. To assess the number of binding sites mapping to different genomic features, we assigned each site to only one gene feature using the hierarchy: promoter > upstream > intron > exon > intergenic, since TFs in general have a preference for binding near the TSS. Genes that had TF binding sites within the promoter were defined as TF target genes. For classification in terms of CpG islands, binding sites were assigned to CpG islands when the peak was located within the boundaries of CpG islands in the CpG island annotation file downloaded from UCSC.
Mapping binding sites of bidirectional promoters
We defined a bidirectional promoter as a genomic region that is upstream of the TSS of a gene and also located between the TSS of two genes which could be separated by a maximum of 2 kb and divergently transcribed from opposite DNA strands. Based on this criterion, we identified 1233 bidirectional promoters corresponding to 2466 genes (11.06%) among the 22,279 genes annotated in RefSeq. To evaluate binding within bidirectional promoters, we first assigned target genes for the transcription factors by mapping their binding sites to within 2 kb upstream of all annotated TSS in RefSeq. Among those target genes, we determined the number of genes which were regulated by bidirectional promoters. We calculated the significance of the enrichment of binding to bidirectional promoters using the hypergeometric probability distribution function.
TSS/TTS binding profiles and heat maps
RefSeq annotations (hg18, November 2010) were used to define genomic regions upstream of and downstream from each distinct TSS or TTS, which were then binned into 50-bp segments. Occupancy scores for CTCF, MYC, or RNAPII were added to each bin so that the peak area overlapped by at least 25 bp. Heat maps of binding around TSS were generated by K-means clustering of the resulting data matrix. Average profiles of peak scores around the TSS or TTS were generated by averaging corresponding bin scores across all genes.
Overlap analysis
Binding site overlap among different ChIP-seq data sets was evaluated by scanning for overlapping binding sites across the genome whose centers resided within 300 bp of each other. In this evaluation, we compared each binding site of one binding data set, using its ChIP-seq score, with the corresponding binding site of another data set at a relaxed score threshold obtained by multiplying the first score by 0.5. This was done to avoid erroneous designation of the site as a non-overlapping binding site just because one of the two sites had a slightly weaker binding site with a lower score.
Analysis of RNAPII binding patterns
We classified RNAPII binding patterns into four groups (HH, high occupancy in both the promoter and the body of a gene; HL, high occupancy only in the promoter; LH, high occupancy only in the gene body; LL, low occupancy signal in both promoter and gene body) based on the ratio of the ChIP-seq score at proximal promoters (ranging from 2 kb upstream to 300 bp downstream from the TSS) to the ChIP-seq score over the gene body. We first assigned ChIP-seq occupancy scores to promoters and gene bodies by mapping RNAPII binding to them. Using the empirical cumulative distribution function (ECDF), we then ranked both the promoter and gene body scores from highest to lowest occupancy. To distinguish the four RNAPII binding patterns, we used a high occupancy threshold of 0.7 and low occupancy threshold as 0.3. We considered the HL group as genes having paused RNAPII at their promoters. We also determined the significance of enrichment of CTCF and MYC around RNAPII sites in promoters as well as gene bodies using the hypergeometric distribution function.
RNAPII exon/intron occupancy analysis
November 2010 RefSeq annotations for hg18 were used to define distinct genomic regions upstream of and downstream from each annotation type (TSS, last exon, or all middle exons), which were then binned into 10-bp segments. For analysis of all middle exons, we considered only genes with at least two exons, and only exons that appeared in all isoforms of a gene. For peak analyses, occupancy scores for RNAPII binding sites were added to the bin containing the maximum peak position; for read analysis, all 3′-extended reads in each bin were aggregated. For uniqueness analysis, a uniqueness score was assigned to each genomic locus based on the UCSC Genome Browser “Mapability—ENCODE Duke Uniqueness of 20-bp sequences” track (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/encodeDCC/wgEncodeMapability/wgEncodeDukeUniqueness20bp.wig.gz, described at http://genome.ucsc.edu/cgi-bin/hgTrackUi?g=wgEncodeMapability). These uniqueness scores were summed for each bin, at either 10- or 1-bp bin resolution. Heat maps of binding around exons and introns (Fig. 8) were generated by sorting the per-gene or per-exon matrix by the length of the appropriate segment. Average profiles were generated by summing corresponding bins across all genes and dividing by the matrix total.
Motif analysis
Motif analysis of sequence-specific TF (CTCF and MYC) binding sites was carried out using the DME algorithm (Smith et al. 2005). First, we divided the binding sites of a TF into three groups, strong (top 25%), moderate (middle 50%), and weak (bottom 25%), based on their ChIP score, and considered the top 500 sites from each group for motif searches. A 200-bp DNA sequence (100 bp in each direction) around the binding peak was extracted from the human genome assembly hg18, and used for motif discovery. A background set was generated by sampling 200-bp sized 100,000 random sequences from the genome. Since ∼15% of CTCF sites and 50% of MYC sites across the cell types occurred in promoters ±2 kb of a TSS, this proportion was maintained in the random samples corresponding to each factor.
Data access
All primary data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE32883.
Acknowledgments
We thank Tonya Severson, Fangfei Ye, and Lisa Bukovnik at the Duke IGSP Sequencing Core Facility, Keith Moon at Illumina, and Scott Hunicke-Smith at UT Austin for sequencing, Scott Tenenbaum and Sridar Chittur at the University at Albany–SUNY for expression data, the Progeria Research Foundation, Matt Wortham and Darell Bigner for cell lines, and Yungki Park for assistance with analysis. This work was supported by an NIH/NHGRI ENCODE Consortium grant U54 HG004563 and by R01 CA130075.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.127597.111.
References
- Adhikary S, Eilers M 2005. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 [DOI] [PubMed] [Google Scholar]
- Alexander RD, Innocente SA, Barrass JD, Beggs JD 2010. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell 40: 582–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand E, Batsche E, Muchardt C 2008. Splicing, transcription, and chromatin: A menage a trois. Curr Opin Genet Dev 18: 145–151 [DOI] [PubMed] [Google Scholar]
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J 2009. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res 19: 1732–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Huang J, Ma A, Kretzner L, Alt FW, Eisenman RN, Weintraub H 1993. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol 13: 5216–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M, et al. 2001. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J 20: 1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Song L, Lee BK, London D, Keefe D, Birney E, Iyer VR, Crawford GE, Furey TS 2011. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res 21: 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y 2011. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol 9: e1000573 doi: 10.1371/journal.pbio.1000573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova GN, Lobanenkov VV, Renkawitz R 1997. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol 17: 1281–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernukhin I, Shamsuddin S, Kang SY, Bergstrom R, Kwon YW, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D, et al. 2007. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol 27: 1631–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. 2010. Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling VH, Cole MD 2007. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol 27: 2059–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Lu H 2008. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J Cell Biochem 105: 670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol 19: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F 2006. The c-Myc target gene network. Semin Cancer Biol 16: 253–264 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G 2010. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384 doi: 10.1371/journal.pbio.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046 doi: 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, Zhong G, Rozowsky J, Bhardwaj N, Gerstein MB, Snyder M 2011. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet 7: e1002008 doi: 10.1371/journal.pgen.1002008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol 16: 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143: 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 16: 653–699 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC et al. 2007. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 35: W169–W175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff JT, Plocik AM, Guthrie C, Yamamoto KR 2010. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol 17: 1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ 2011. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res 21: 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin VX, Singer GA, Agosto-Perez FJ, Liyanarachchi S, Davuluri RV 2006. Genome-wide analysis of core promoter elements from conserved human and mouse orthologous pairs. BMC Bioinformatics 7: 114 doi: 10.1186/1471-2105-7-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN 2006. Myc influences global chromatin structure. EMBO J 25: 2723–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Jourquin F, Ferrier P, Andrau JC 2008. Genome-wide RNA polymerase II: Not genes only! Trends Biochem Sci 33: 265–273 [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 doi: 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladomery M, Dellaire G 2002. Multifunctional zinc finger proteins in development and disease. Ann Hum Genet 66: 331–342 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lee BK, Bhinge AA, Iyer VR 2011. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Res 39: 3558–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Collins PJ, Trinklein ND, Fu Y, Xi H, Myers RM, Weng Z 2007. Transcription factor binding and modified histones in human bidirectional promoters. Genome Res 17: 818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniell R, Lee BK, Song L, Liu Z, Boyle AP, Erdos MR, Scott LJ, Morken MA, Kucera KS, Battenhouse A, et al. 2010. Heritable individual-specific and allele-specific chromatin signatures in humans. Science 328: 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Penn LZ 2008. Reflecting on 25 years with MYC. Nat Rev Cancer 8: 976–990 [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K 2010. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol 17: 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M 2010. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci 107: 3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD 2002. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev 16: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT 2010. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 20: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G 2009. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol 16: 990–995 [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR 2008. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol 6: e65 doi: 10.1371/journal.pbio.0060065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ III, Mandal SS, Reinberg D 2004. Recent highlights of RNA-polymerase-II-mediated transcription. Curr Opin Cell Biol 16: 263–271 [DOI] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Zhang MQ 2005. Identifying tissue-selective transcription factor binding sites in vertebrate promoters. Proc Natl Acad Sci 102: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB 2009. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, Brosh R, Buganim Y, Reiner A, Zuk O, Yitzhaky A, Koudritsky M, Rotter V, Domany E 2007. Wide-scale analysis of human functional transcription factor binding reveals a strong bias towards the transcription start site. PLoS ONE 2: e807 doi: 10.1371/journal.pone.0000807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L, Kamakaka RT 2006. Chromatin insulators. Annu Rev Genet 40: 107–138 [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A 2008. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods 5: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309 [DOI] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM 2009. A census of human transcription factors: Function, expression and evolution. Nat Rev Genet 10: 252–263 [DOI] [PubMed] [Google Scholar]
- Vostrov AA, Quitschke WW 1997. The zinc finger protein CTCF binds to the APBβ domain of the amyloid β-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem 272: 33353–33359 [DOI] [PubMed] [Google Scholar]
- Vostrov AA, Taheny MJ, Quitschke WW 2002. A region to the N-terminal side of the CTCF zinc finger domain is essential for activating transcription from the amyloid precursor protein promoter. J Biol Chem 277: 1619–1627 [DOI] [PubMed] [Google Scholar]
- Wierstra I, Alves J 2008. The c-myc promoter: Still MysterY and challenge. Adv Cancer Res 99: 113–333 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD 2009. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell 35: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]