Abstract
Highly acidic (pH 0–1) biofilms, known as ‘snottites', form on the walls and ceilings of hydrogen sulfide-rich caves. We investigated the population structure, physiology and biogeochemistry of these biofilms using metagenomics, rRNA methods and lipid geochemistry. Snottites from the Frasassi cave system (Italy) are dominated (>70% of cells) by Acidithiobacillus thiooxidans, with smaller populations including an archaeon in the uncultivated ‘G-plasma' clade of Thermoplasmatales (>15%) and a bacterium in the Acidimicrobiaceae family (>5%). Based on metagenomic evidence, the Acidithiobacillus population is autotrophic (ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), carboxysomes) and oxidizes sulfur by the sulfide–quinone reductase and sox pathways. No reads matching nitrogen fixation genes were detected in the metagenome, whereas multiple matches to nitrogen assimilation functions are present, consistent with geochemical evidence, that fixed nitrogen is available in the snottite environment to support autotrophic growth. Evidence for adaptations to extreme acidity include Acidithiobacillus sequences for cation transporters and hopanoid synthesis, and direct measurements of hopanoid membrane lipids. Based on combined metagenomic, molecular and geochemical evidence, we suggest that Acidithiobacillus is the snottite architect and main primary producer, and that snottite morphology and distributions in the cave environment are directly related to the supply of C, N and energy substrates from the cave atmosphere.
Keywords: Acidimicrobium, acidophile, Acidithiobacillus, metagenomics, pyrosequencing, Thermoplasmatales
Introduction
Acidophilic microorganisms control biogeochemical cycling in a variety of natural and anthropogenically influenced environments. Acidophiles from high-temperature environments such as hot springs and fumaroles (Schleper et al., 1995; Inskeep et al., 2010), and iron-rich systems such as bioleaching operations and acid mine drainage (AMD) (Johnson and Hallberg, 2003; Tyson et al., 2004) have been the target of numerous studies linking microbial activity with geochemistry. Extremely acidophilic communities from low-temperature, sulfur-rich environments are common but less well studied, and include concrete-corroding subaerial biofilms in sewers (Okabe et al., 2007) and subaerial ‘snottites' from sulfide-rich caves (Macalady et al., 2007).
Snottites are extremely acidic (pH 0–1) microbial biofilms that form on the walls and ceilings of caves where sulfide-rich springs degas H2S into the cave air (Figure 1; Hose et al., 2000; Macalady et al., 2007; Jones et al., 2008). Sulfide oxidation produces sulfuric acid, which dissolves the limestone walls of the cave. Microcrystalline gypsum precipitates as a corrosion residue that eventually limits pH buffering by the underlying limestone and enables the development of extremely acidic wall surfaces. Previous research using rRNA methods showed that snottites have very low biodiversity and are dominated by Acidithiobacillus spp., sometimes with other less abundant populations of bacteria and archaea (Hose et al., 2000; Vlasceanu et al., 2000; Macalady et al, 2007).
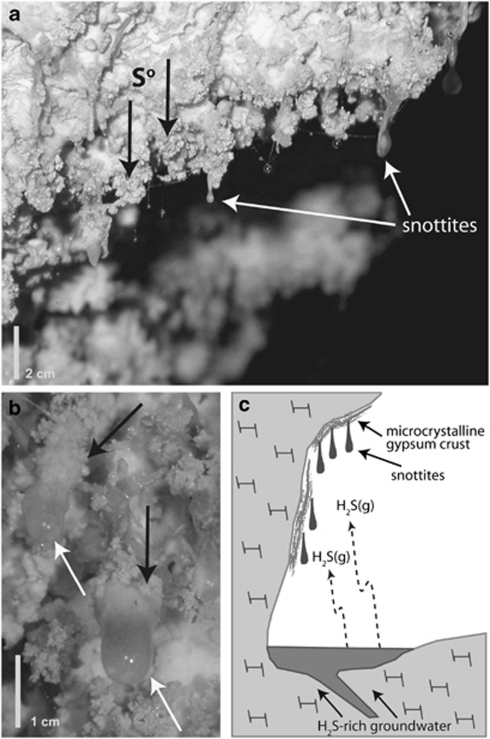
Figure 1.
(a, b) Field photographs of the collection site for snottite sample RS24. Note elemental sulfur (black arrows) occurring in close association with biofilm surfaces. (c) Schematic diagram depicting the formation of sulfidic cave snottites. Snottites form on subaerial cave surfaces in areas exposed to H2S(g) degassing from circumneutral cave streams. Snottites have extremely acidic pH values (0–1), because they are isolated from limestone cave walls and gypsum corrosion residues that would otherwise buffer the pH >2.
Snottites in the Frasassi cave system are components of a thriving, sulfur-based chemosynthetic ecosystem isolated from surface-derived organic matter (Galdenzi and Maruoka, 2003; Macalady et al., 2006). Biofilms are a prominant feature of the cave environment, both above and below the water table. Ongoing work at Frasassi, including measurements of water table sulfide degassing rates, indicates that subaerial sulfuric acid production is significant and possibly more important than acid production below the water table. Because sulfuric acid dissolves limestone and enlarges the cave, there may be important feedbacks between the physiology and activity of snottite microorganisms and meter- to kilometer-scale geochemical processes. However, little is known about the metabolic potential of snottite populations. For example, the nearest relatives of several Frasassi snottite organisms, including Thermoplasmatales-group archaea and Acidimicrobiaceae (Actinobacteria), originate from iron-dominated environments (Clark and Norris, 1996; Tyson et al., 2004). Therefore, their physiologies and ecological roles in the iron-poor, H2S-fed snottite communities are unclear. The sulfur-dependent energy metabolism of snottite Acidithiobacillus can be inferred from 16S rRNA sequence phylogenies, but many other important aspects of their physiology are unknown.
Metagenomics—genomic DNA sequencing directly from a mixed community gene pool—is an important source of genetic information from environmental samples (Allen and Banfield, 2005). In this study, we used metagenomics in combination with rRNA methods and lipid analyses to probe the metabolic potential and ecological roles of snottite microorganisms. The objectives were: (1) to resolve the composition and structure of the snottite community, including populations overlooked by rRNA methods due to primer and probe biases; (2) to investigate the metabolic potential and ecological role(s) of snottite Acidithiobacillus and other populations, including their pathways for carbon fixation, nitrogen fixation, sulfur oxidation and heterotrophy; and (3) to propose adaptations for survival in the extreme acidity (pH 0–1) of the biofilm matrix. These objectives were met using a relatively small metagenomic dataset, in which the genomic coverage of the dominant Acidithiobacillus population was estimated to be 2–3 × .
Materials and methods
Sample collection, DNA extraction and rRNA analyses
We collected roughly 3 g of biofilm (sample RS24) from 1 m2 of cave wall at site RS2 in the Frasassi cave system, Italy (Supplementary Figure S1). Biofilm pH was measured in the field with pH paper (range 0–2.5). Environmental DNA was extracted from RS24 as described in Bond et al. (2000), after first diluting the RNAlater (Ambion/Applied Biosystems, Foster City, CA, USA) preserved sample with three parts phosphate-buffered saline to one part sample. To remove excess polysaccharides from the final extract, we reprecipitated the DNA under high salt concentrations as follows: the pellet was resuspended in 200 μl Tris (200 m, pH 8.0), 100 μl NaCl (5 ) and 600 μl ethanol (100%), incubated at −20 °C for 30 min and pelleted for 20 min at 4 °C. Near-full-length 16S rRNA gene sequences were cloned from sample RS24 using archaeal specific primers 344f (ACGGGGYGCAGCAGGCGCGA) (Raskin et al., 1994) and deg1392r (ACRGGCGGTGTGTRC) (modified from 1392r, Lane, 1991), using the cloning procedure described previously (Macalady et al. 2008). Fluorescent in situ hybridization (FISH) was performed using probes THIO1, ACM732, EUBMIX, ARCH915 and FER656 as described in Macalady et al. (2007).
Lipid analyses
Total lipid extracts were prepared from the RS24 biofilm using a modified Bligh–Dyer extraction as described by Talbot et al. (2003), with dichloromethane substituted for chloroform. Analyses of ether lipids and bacteriohopanepolyols (‘hopanoids') were performed with an Agilent 6310 high pressure liquid chromatograph/mass spectrometrometer (Agilent Technologies, Santa Clara, CA, USA) following the procedures of Hopmans et al. (2000) and Talbot et al. (2003), with minor modifications (see Supplementary Material).
Metagenomics
DNA from sample RS24 was pyrosequenced at the Pennsylvania State University Center for Genomic Analysis with a GS20 platform (454 Life Sciences, Branford, CT, USA; Margulies et al., 2005). All metagenome reads were compared via BLASTX (Altschul et al., 1997) to the NCBI non-redundant (nr) database, after first removing identical read copies and putative rRNA gene sequences. Before BLASTX analyses, protein sequences from ‘Thermoplasmatales archaeon Gpl' in the AMD microbiome were added to the nr database (Tyson et al., 2004; Markowitz et al., 2008; available at http://img.jgi.doe.gov/m). Reads matching nr with a bit score <40 were removed before subsequent analyses. As we were unable to assemble more than 5% of reads into non-chimeric contigs >500 bp, we analyzed only unassembled metagenomic sequences. We annotated reads to functional categories identified in the clusters of orthologous groups of proteins (COGs) classification system (Tatusov et al., 2003), using reverse position specific BLAST with a bit score cutoff of 35. We binned RS24 metagenome reads to taxonomic groups using MEGAN v.3.2.1 (Huson et al., 2007), using a bit score threshold of 40, min support of 1 and top percent of 10%. We identified P-type ATPase transporters using links between TC family and COGs provided at TransportDB (Ren et al., 2004). We calculated functions that were overrepresented in the snottite metagenome by comparing COG assignments from eight other publicly available metagenomes also generated with GS20 pyrosequencing technology.
In order to ensure that function and taxonomy were accurately assigned for metagenomic reads, we determined appropriate parameters for BLAST analyses, COG assignments and taxonomic binning with MEGAN, using simulated datasets constructed from full-length protein and genome sequences. For example, appropriate MEGAN parameters were determined by BLAST analysis of simulated datasets against two versions of the nr database: the original nr database with query sequences present and a modified database from which the query organisms were removed. Complete details for all methods are provided in the Supplementary Material. Metagenomic sequences are deposited in the National Center for Biotechnology Information Sequence Read Archive under accession number SRA026550. Genbank accession numbers are HM754546-HM754573 for 16S rRNA and HM852513-HM852515 for squalene–hopene cyclase (SHC) sequences.
Results
Field observations and geochemistry
Snottites at site RS2 are viscous biofilms attached to cave walls at their base and hang down as far as 3 cm into the cave atmosphere (Figure 1). Snottites collected for sample RS24 were attached exclusively to microcrystalline gypsum wall crusts and were associated with elemental sulfur precipitates on biofilm and adjacent gypsum surfaces. Temperature (13 °C) and humidity (close to 100%) are virtually constant throughout the year. Individual snottites had pH values between 0 and 1. H2S(g) concentration at site RS2 was 8–24 ppm by volume (ppmv), CO2(g) was 3300 ppmv, CH4(g) was 1.9–2.2 ppmv, and NH3(g), NO2(g) and SO2(g) were below detection (0.25, 0.1 and 0.1 ppmv, respectively).
Pyrosequencing of RS24 snottites
We obtained 12.9 Mb of quality metagenomic sequence from the GS20 pyrosequencer. After screening to remove duplicates that arise as an artifact of pyrosequencing, the dataset reduced to 11.9 Mb and 118 624 total reads. These sequences had an average length of 100.3 bp, an average G+C content of 51.5% and a mode G+C of 54%. Using criteria described in the methods, 40.5% of reads had significant matches to the nr database, 39.4% of these (17.1% of total reads) were assigned to COG categories (Supplementary Figure S2) and 0.23% matched rRNA genes (0.08% to 16S rRNA genes).
Snottite community composition
In earlier work that included FISH and 16S rRNA cloning (Macalady et al., 2007), we found that the most abundant bacterial 16S rRNA phylotypes (>98% 16S rRNA similarity) in snottites collected throughout the Frasassi cave system are relatives of Acidithiobacillus thiooxidans and the genera Acidimicrobium and Ferrimicrobium (Acidimicrobiaceae family, Actinobacteria). Consistent with this earlier work, FISH analyses of snottite sample RS24 indicated that Acidithiobacillus and Acidimicrobiaceae are the most abundant bacterial populations (Figure 2, Supplementary S3 and S4). Detailed phylogenetic analyses of Acidimicrobiaceae 16S rRNA sequences from Macalady et al. (2007) showed that they belong to a monophyletic sister group to the genus Ferrimicrobium (Supplementary Figure S5).
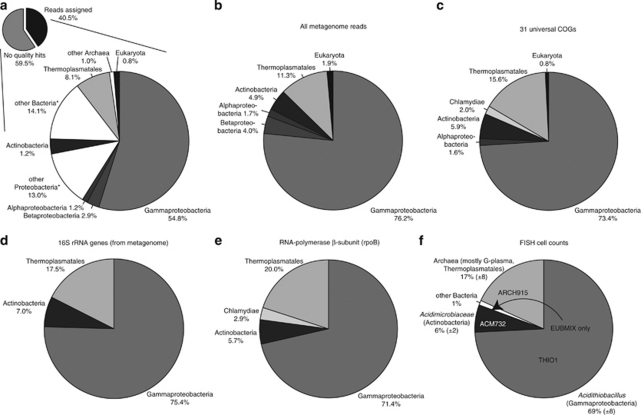
Figure 2.
Comparison of RS24 community composition based on FISH and metagenomic data. (a) Taxonomic classification and binning of all metagenomic reads. Using the criteria described in the methods, 40.5% of total metagenome reads were assigned to taxa. *groups include reads that cannot be assigned to a more specific taxonomic group. Taxa that make up <0.5% of all matches are omitted from the figure. (b) Community composition based on taxonomic classification of all metagenomic reads, after removing reads assigned to non-specific groups (for example, ‘other bacteria'). (c–e) Community composition based on phylogenetic markers from the metagenome: (c) 31 universal genes (Ciccarelli et al., 2006), (d) 16S rRNA genes and (e) RNA polymerase-β subunit sequences. (f) Community composition determined from FISH cell counts. Numbers in parentheses represent one s.d. FISH probes used to generate the data were THIO1, genus Acidithiobacillus, ACM732, Acidimicrobiaceae family, EUBMIX, bacteria and ARCH915, archaea.
FISH analyses with the archaeal domain probe ARCH915 indicated that archaea make up 17% of the RS24 community (Figure 2f and Supplementary Figure S3). Ferroplasma populations identified previously in Frasassi snottites using FISH (Macalady et al., 2007) were not detected in sample RS24. Analysis of metagenome sequences matching 16S rRNA genes revealed that RS24 archaea are most closely related to members of the ‘G-plasma' group in the Thermoplasmatales. From the G-plasma metagenomic sequences, we learned that universal primer 1392r (Lane, 1991) has a 1-bp mismatch to 16S rRNA sequences from G-plasma. Other primers 1492r (Lane, 1991) and archaeal-specific 21f (DeLong, 1992) also have mismatches with the G-plasma clade. Therefore, we added a degenerate nucleotide to the sequence of primer 1392r (new primer deg1392r) and subsequently used it to amplify 16S rRNA sequences from RS24 environmental DNA. In the resulting library of 28 clones, 26 sequences belonged to a single G-plasma phylotype (Supplementary Figure S6). On the basis of the new knowledge that most sequenced RS24 archaea are G-plasma, we added metagenomic sequences from the Iron Mountain AMD G-plasma (Tyson et al., 2004) to the nr database, which increased the number of reads in the snottite G-plasma bin by 172%.
Taxonomic classification of phylogenetic markers in the metagenome confirms the RS24 community composition determined from FISH population counts (Figure 2). We used metagenomic 16S rRNA genes, RNA polymerase-β subunit (Case et al., 2007) and a set of 31 universal genes identified by Ciccarelli et al. (2006) to construct independent estimates of community composition (Figures 2c–e). Together, these phylogenetic markers show that RS24 is dominated by Gammaproteobacteria (71.4–75.4%), Thermoplasmatales (15.6–20.0%) and Actinobacteria (5.7–7.0%), with the addition of several low-abundance taxa (Figure 2). These include Chlamydiae identified with both RNA polymerase-β subunit and the universal marker gene set (Figures 2c and d). The Chlamydiae sequences are most closely related to protist endosymbionts, including Candidatus Protochlamydia amoebophila UWE25 (Collingro et al., 2005). Eukaryotic sequences were rare but detectable in the metagenome (Figure 2), and we observed large nucleated cells resembling protists and fungi in FISH analyses.
Gene and genome coverage of snottite organisms
We estimated the expected genome coverage for each snottite population (defined as total nucleotides sequenced/genome size) in sample RS24 based on FISH population counts, the average genome size of related organisms and the size of the metagenomic dataset (Supplementary Figure S7; Whitaker and Banfield, 2006). Expected sequence coverages for Acidithiobacillus, G-plasma and Acidimicrobiaceae are 3.0 × , 0.72 × and 0.24 × , respectively. Assuming a Poisson distribution of DNA sequencing, 95% of the Acidithiobacillus genome is present in the metagenome, and 51% and 22% of the G-plasma and Acidimicrobiaceae are present, respectively. Based on metagenome reads that we could assign to 18 single copy genes from Acidithiobacillus, the average coverage of that population is 2.1 × (ranging from 0.8 to 4.3 × , see Supplementary Material).
To evaluate how well our metagenome represents the gene content of the different populations, we simulated random shotgun sequencing by a 454 GS20 platform and calculated the proportion of total genes represented by at least one read (Supplementary Figure S8). From this simulation, we found that for a coverage of 3 × , 100% of the genes and 95% of the total nucleotide positions are represented. For coverages of 0.72 × and 0.24 × , although only 51% and 21% of the total nucleotide positions are represented, 98% and 82% of the genes are represented by at least one read (Supplementary Figure S8). Accordingly, the Acidithiobacillus and G-plasma bins have all amino-acyl tRNA-synthases represented by at least one read, with the exception of asparaginyl–tRNA synthetase in Acidithiobacillus (Table 1). As for other Acidithiobacilli, snottite Acidithiobacillus populations may synthesize Asn–tRNA by transamidation of mischarged Asp–tRNAAsn (Salazar et al., 2001). The Acidimicrobiaceae bin contains reads for nine AA–tRNA synthetases.
Table 1. AA–tRNA synthetase COGs present in each taxonomic bin.
| tRNA synthetase | COG ID |
Bin |
||
|---|---|---|---|---|
| Acidithiobacillus | G-plasma | Acidimicrobium | ||
| Ala–tRNA synthetase | COG0013 | 24 | 3 | — |
| Arg–tRNA synthetase | COG0018 | 14 | 1 | — |
| Asp/Asn–tRNA synthetases | COG0017 | —a | 7 | 1 |
| Asp–tRNA synthetase | COG0173 | 22 | — | 1 |
| Cys–tRNA synthetase | COG0215 | 9 | 5 | — |
| Glu– and Gln–tRNA synthetases | COG0008 | 20 | 3 | 1 |
| Gly-tRNA synthetase | ||||
| α-Subunit | COG0752 | 7 | — | 1 |
| β-Subunit | COG0751 | 16 | — | — |
| Class II | COG0423 | — | 4 | — |
| His–tRNA synthetase | COG0124 | 18 | 2 | — |
| Ile–tRNA synthetase | COG0060 | 19 | 4 | 3 |
| Leu–tRNA synthetase | COG0495 | 18 | 4 | 3 |
| Lys-tRNA synthetase | ||||
| Class I | COG1384 | — | 4 | — |
| Class II | COG1190 | 17 | — | — |
| Met–tRNA synthetase | COG0143 | 19 | 4 | — |
| Phe-tRNA synthetase | ||||
| α-subunit | COG0016 | 6 | 2 | — |
| β-subunit | COG0072 | 8 | — | — |
| Pro–tRNA synthetase | COG0442 | 10 | 1 | 1 |
| Se–tRNA synthetase | COG0172 | 6 | 3 | — |
| Thr–tRNA synthetase | COG0441 | 14 | 12 | 1 |
| Trp–tRNA synthetase | COG0180 | 5 | 1 | — |
| Ty–tRNA synthetase | COG0162 | 3 | 3 | — |
| Val–tRNA synthetase | COG0525 | 17 | 6 | 3 |
Abbreviation: COGs, clusters of orthologous groups of proteins.
Asn–tRNA likely formed via transamination pathway in Acidithiobacillus.
Sulfur oxidation pathways
We identified reads matching sulfur oxidation genes in the snottite Acidithiobacillus bin (Table 2), including sulfide–quinone reductase (SQR), tetrathionate hydrolase, thiosulfate:quinone oxidoreductase and three of four central components of the sox system (soxAX, soxB and soxYZ). There is no evidence for soxCD, or for flavocytochrome C or dissimilatory sulfite reductase (DSR). We did not retrieve any sequences matching the recently identified bacterial sulfur oxygenase-reductase genes (Chen et al., 2007), including the predicted sulfur oxygenase-reductase in Acidithiobacillus caldus (Valdés et al., 2009).
Table 2. Summary of metabolic capabilities of snottite microorganisms based on metagenomic data.
| Acidithiobacillus | G-plasma | Acidimicrobium | |
|---|---|---|---|
| CO2 fixation | Yes | No evidence | No evidence |
| Reductive pentose–phosphate pathway | Yes | — | — |
| S-oxidation | Yes | No evidence | Probable |
| SQR | Yes | — | Yes |
| sox system | Yes (soxAB, X, YZ) | — | No evidence |
| soxCD | No evidence | — | — |
| TQO | Yes | — | No evidence |
| TTH | Yes | — | No evidence |
| SOR | No evidence | — | No evidence |
| fccC | No evidence | — | No evidence |
| dsrAB | No evidence | — | No evidence |
| N-metabolism | |||
| N2-fixation (nifH, D, K) | No evidence | No evidence | No evidence |
| Ammonia assimilation | Yes | Yes | Yes |
| Ammonia permease | Yes | Yes | No evidence |
| Glutamine synthetase | Yes | Yes | Yes |
| Nitrate assimilation | Yes | No evidence | Probable |
Abbreviations: dsrAB, dissimilatory sulfite reductase ; fccC, flavocytochrome C ; SOR, sulfur oxygenase reductase; THH, tetrathionate hydrolase; SQR, sulfide–quinone reductase; TQO, thiosulfate:quinol reductase.
We found no evidence for sulfur oxidation in the G-plasma bin. No evidence was found for archaeal SQR, thiosulfate:quinone oxidoreductase or sulfur oxygenase-reductase, even though homologs of SQR and sulfur oxygenase-reductase are known from several Thermoplasmatales organisms (Kletzin, 2007; Chan et al., 2009).
The Acidimicrobiaceae bin contains three reads matching SQR. We found no other evidence for sulfur oxidation enzymes in the Acidimicrobiaceae bin.
Carbon metabolisms
We identified matches to ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) and phosphoribulokinase in the Acidithiobacillus bin (Table 2). These enzymes are diagnostic of carbon fixation by the reductive pentose phosphate pathway (Shively et al., 1998). The Acidithiobacillus bin also contains several reads matching carbonic anhydrase (COG0288) and carboxysome shell proteins (COG4577). We found no evidence for any of the known carbon fixation pathways in the Acidimicrobiaceae and G-plasma bins (see Supplementary Material).
The metagenome contains 40 reads matching RuBisCO large and small subunits. Although some of these sequences binned as Acidithiobacillus, because RuBisCO is a highly conserved protein, the majority of these reads are not assigned to specific taxonomic bins (Figure 2a) by our MEGAN criteria. Therefore, to better identify C fixing organisms, we aligned metagenome reads matching RuBisCO against reference RuBisCO sequences. From this alignment, we identified at least two divergent, overlapping RuBisCO large subunit sequences, both most similar to RuBisCO from Acidithiobacillus spp. by nucleotide sequence. Therefore, we suggest that the snottite Acidithiobacillus has multiple copies of RuBisCO, at least one of which is type I. The metagenome also contains two reads most similar to environmental RuBisCO sequences amplified from grassland soil by Videmšek et al. (2009). These two reads are only distantly related to RuBisCO from Acidithiobacillus spp., Acidimicrobium ferrooxidans and archaea.
The Acidimicrobiaceae bin contains multiple reads matching genes for the breakdown of complex organics (Supplementary Table S1). These include four hits to enoyl-CoA hydratase, two hits to chitinase, two hits to extradiol ring-cleavage dioxygenases and matches to glycosidases and galactosidases.
Nitrogen metabolisms
We found no evidence for nitrogen fixation (nifH, nifD or nifK) in the metagenome, whereas several COGs for ammonia and nitrate assimilation are strongly overrepresented (Supplementary Figure S9). Reads matching ammonia permeases are found in both the Acidithiobacillus and G-plasma bins, and all three bins contain hits to glutamine synthetase, indicating the potential for ammonium assimilation. The Acidithiobacillus bin contains reads matching nitrate reductase narB and 45 hits to nitrite reductase nirB from other Acidithiobacilli including Acidithiobacillus thiooxidans (Levicán et al., 2008). We also identified one read similar to nitrate reductase-α subunit narG (COG5013), which is part of a membrane bound nitrate reductase (narGHJI) used for either respiratory or assimilatory functions (Richardson et al., 2001; Malm et al., 2009). Although this read is not assigned to the Acidimicrobiaceae bin using our MEGAN criteria, the top 10 blast matches are assigned to members of the Actinobacteria. We found no COGs related to nitrate or nitrite assimilation in the G-plasma bin, nor any reads matching archaeal nitrate and nitrite reductases listed by Cabello et al. (2004). We found no evidence for dissimilatory ammonia oxidation by ammonia monooxygenase in the metagenome.
Biofilm formation
The Acidithiobacillus bin contains reads matching flagella and type IV pilus biosynthesis genes, which are important in early biofilm formation (Davey and O'toole, 2000). It also has reads matching genes for the biosynthesis of expolymeric substance precursors UDP-glucose, UDP-galactose (GalEU) and dTDP-rhamnose (rfbABCD) (Barreto et al., 2005).
Membrane lipids
Membrane lipid structures and other membrane modifications are known to contribute to the maintenance of pH homeostasis in acidophiles. We identified multiple metagenomic reads matching SHC, an essential enzyme in bacteriohopanepolyol (‘hopanoid') biosynthesis (Pearson et al., 2007). SHC is present in both the Acidithiobacillus and Acidimicrobiaceae bins, in addition to a sequence affiliated with the Betaproteobacteria (Supplementary Figure S10a). Direct amplification and cloning of SHC genes from the RS24 DNA extract retrieved three unique, partial SHC sequences (see Supplementary Material). Sequence SDPr_DirF shares 94% identity at the amino acid level with Acidithiobacillus ferrooxidans ATCC 53993 and the other two sequences are from the Proteobacteria (clone SDPr_Cl121) and an unknown clade (clone SDCy21B) (Supplementary Figure S10b; Pearson et al., 2009). Sequence SDPr_DirF was obtained by direct sequencing of the PCR product without cloning. Notably, the ability to obtain any amount of clean sequence in the absence of cloning is consistent with the expectation that a close relative of Acidithiobacillus ferrooxidans is the dominant bacterium in the biofilm and that it is the major source of the hopanoid lipids detected in this sample.
We confirmed that snottite organisms are actively producing hopanoids in the environment by high pressure liquid chromatograph/mass spectrometrometer analysis of intact polar lipids. We detected four bacteriohopanepolyol structures in the RS24 biofilm total lipid extract, including adenosylhopane, bacteriohopanetetrol, aminotriol and cyclitol ether (Supplementary Figure S11). We also analyzed the RS24 lipid extract for tetraether-linked membrane lipids that have previously been shown to be critical for archaeal life in acidic environments (Macalady et al. 2004). The sample contained five isoprenoid glycerol dialkyl glycerol tetraether lipid structures (Supplementary Figure S12). No archaeal or bacterial diethers were detected. Glycerol dialkyl glycerol tetraether lipids from RS24 contain between 0 and 4 cyclopentane rings (no cyclohexane rings) and have structures previously identified in Thermoplasmatales-group isolates.
Discussion
Carbon and energy metabolisms of snottite populations
Our results indicate that Acidithiobacillus are the main primary producers in the snottite community (Figure 3). Acidithiobacillus make up approximately 70% of cells and appear to be lithoautotrophs, consistent with the observation that snottites do not occur on cave walls where sulfide gas concentrations are less than 0.2 ppm (Macalady et al., 2007). Carbon fixation occurs by the reductive pentose-phosphate pathway, facilitated by multiple copies of RuBisCO, carbonic anhydrase and carboxysomes.
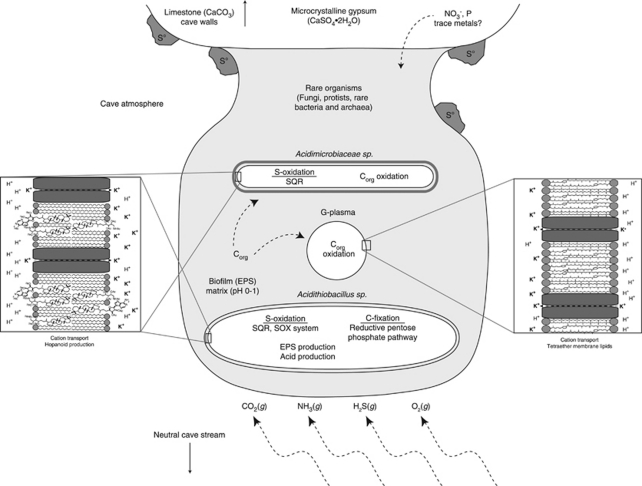
Figure 3.
Conceptual model of the biogeochemistry of Frasassi snottite biofilms, based on evidence from metagenomic, rRNA, lipid and geochemical analyses. Carbon fixation, expolymeric substance (EPS) production and acid generation occur largely due to the metabolism of lithoautotrophic, sulfide-oxidizing Acidithiobacillus. Energy substrates (H2S, O2) and C and N for primary production (CO2, NH3) are from gasses in the cave atmosphere. Trace metals and other non-volatile nutrients such as phosphorous ultimately derive from limestone cave walls, corrosion residues or downward percolating groundwater solutions.
There is evidence that the snottite Acidithiobacillus uses multiple sulfide oxidation pathways (Table 2). SQR catalyzes the two-electron oxidation of sulfide to zero-valent sulfur (Griesbeck et al., 2002; Wakai et al., 2007), tetrathionate hydrolase disproportionates tetrathionate to thiosulfate, sulfate and elemental sulfur (Kanao et al., 2007), and thiosulfate:quinone oxidoreductase oxidizes thiosulfate to tetrathionate (Müller et al., 2004). In sulfur-oxidizing acidophiles including At. ferrooxidans, these components are proposed to operate together in a pathway known as the ‘SQR-system', in which sulfate is an end product (Rohwerder and Sand, 2007; Valdés et al., 2008a; Valdés et al., 2008b). Acidithiobacillus also has components of a partial SOX system, including soxAX, soxB and soxYZ, but no evidence for soxCD. In microorganisms with a complete SOX system (for example, Friedrich et al., 2001; Sauvé et al., 2007), soxCD catalyzes the six-electron oxidation of a soxYZ-bound sulfane to a sulfone, which is later released as sulfate. Organisms known to use a partial SOX system without soxCD produce zero-valent sulfur as an intermediate or end product (Hensen et al., 2006). In these organisms, DSR is thought to be necessary for further oxidation of the elemental sulfur produced (Pott and Dahl, 1998, 2005; Hensen et al., 2006), but DSR was not detected in the snottite metagenome.
The presence of both an SQR and a partial SOX system could provide flexibility in the sulfur metabolism of the Acidithiobacillus population, and the occurrence of S° deposits on snottite surfaces might result from multiple end products of biological sulfide oxidation. Specifically, the absence of DSR in the metagenome indicates that S° could be produced when the partial SOX pathway is expressed. The absence of DSR also indicates that sulfate reduction has a minor role, if any, in the snottite community. This is in sharp contrast to sulfur-oxidizing communities below the cave water table, which host substantial populations of sulfate-reducing bacteria that supply sulfide-oxidizing lithoautotrophs with an alternative sulfide source when concentrations in the bulk water are low (Macalady et al. 2006, 2008).
We found no evidence for lithotrophy or carbon fixation by the snottite G-plasma population, so we suggest that they are heterotrophic. Although coverage of the G-plasma is low, simulated sequencing (Supplementary Figure S8) and matches to AA–tRNA synthetases (Table 1) indicate that our metagenome contains a nearly complete representation of the gene content of this population. Our results are consistent with other work showing that heterotrophy is the most common carbon metabolism among known members of the Thermoplasmatales (Schleper et al., 1995; Tyson et al., 2004). However, more work will be required to obtain a detailed understanding of the role of G-plasma in snottite biogeochemistry.
Based on metagenomic evidence, it is likely that the snottite Acidimicrobiaceae population is capable of organotrophic or mixotrophic growth (Supplementary Table S1). The Acidimicrobiaceae bin contains three SQR fragments, which could indicate either lithotrophic sulfur oxidation or sulfide detoxification (Griesbeck et al., 2000). The nearest relatives of the snottite Acidimicrobiaceae are from acidic, iron-rich environments and include obligate heterotrophs Ferrimicrobium acidophilum and Ferrithrix thermotolerans (Johnson et al., 2009) and mixotrophic Acidimicrobium spp. (Clark and Norris, 1996; Cleaver et al., 2007). The ability to oxidize both sulfide and organic carbon represents a distinct niche and could allow Acidimicrobiaceae to avoid direct competition with lithoautotrophic Acidithiobacillus populations in the biofilm.
Nitrogen cycling
Possible sources of fixed nitrogen in the cave system are nitrate in downward-percolating groundwater and ammonia degassing from the water table. Dripping groundwater collected near Frasassi cave entrances can have up to 50 μ NO3− (unpublished data). Although the RS2 sample site is not near a cave entrance and we have not observed active dripping at the site, diffuse seepage of downward-percolating water could theoretically deliver nitrate–nitrogen to snottite microorganisms. The potential for ammonia degassing in sulfidic caves was first recognized by Stern et al. (2003), based on low nitrogen isotopic values measured on the walls of Lower Kane Cave, Wyoming (δ15N below −15‰). Extremely low δ15N values (below −20‰) have also been measured on Frasassi cave walls (Vlasceanu et al., 2000; Jones et al., 2008) and are thought be the result of N isotopic fractionation during the ammonia degassing process. Streamwaters at site RS2 have pH 7.3 and 30–80 μ NH4+ (Macalady et al., 2006). Thus, field and laboratory observations point to ammonia degassing from the circumneutral water table and subsequent trapping in the acidic biofilm matrix as a plausible mechanism to supply fixed nitrogen to snottites.
No evidence for N2 fixation (nifH, D or K) was found in the RS24 metagenome. As the nif genes are highly conserved and genome coverage of the dominant snottite organism (Acidithiobacillus) is nearly complete, the metagenomic results indicate that Acidithiobacillus do not fix nitrogen in this snottite community. Nonetheless, we recognize that low abundance populations of N2 fixers may be present. We cannot rule out the potential for N2 fixation by Acidimicrobiaceae (∼6% of cells) or other even rarer populations. However, ammonia permeases and nitrate assimilation functions are overrepresented in the metagenome (Supplementary Figure S9). Thus, metagenomic data support the hypothesis that fixed sources of nitrogen in the snottite environment supply N for the lithoautotrophic growth of the main biofilm population.
Adaptations for survival at extremely low pH
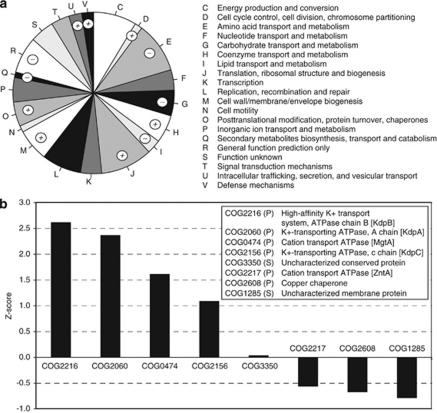
In order to thrive at extremely low pH values, snottite microorganisms must maintain a cytoplasmic pH that is several orders of magnitude higher than that of their external environment. Proton leakage in many acidophiles is offset by maintaining an inside-positive membrane potential as a charge barrier (Cox et al., 1979). This mechanism is thought to work by uptake of potassium and other cations (Baker-Austin and Dopson, 2007). Accordingly, reads matching ion transporters are abundant in the metagenome and, in particular, P-type ATPases for active potassium transport are overrepresented in the metagenome (Figure 4).
Figure 4.
(a) Proportion of reads assigned to COG categories. COG categories are listed to the right. Categories marked by a + or a − symbol are strongly over- or underrepresented in the snottite metagenome, respectively. (b) Standardized abundance scores of COGs classified as P-type ATPases according to the TransportDB (Ren et al., 2004). The COG id, category, description and gene name if available are provided in the legend.
There is growing evidence that hopanoids in bacteria (Welander et al., 2009) and tetraether-linked lipids in archaea (van de Vossenberg et al., 1998; Macalady et al., 2004) function to reduce membrane permeability to protons. Based on evidence from two independent methods (metagenomics and PCR), snottite Acidithiobacillus have the SHC gene essential for hopanoid production. Both methods also show that other bacterial populations in the snottite community including Acidimicrobiaceae, contain the SHC gene. Pearson et al. (2007) estimate that the fraction of hopanoid producing bacteria in nature is less than 10%. Thus, the presence of SHC in both major snottite bacterial populations is significant and supports the hypothesis that hopanoid production is an important bacterial adaptation to extreme acid. Ether-linked lipids in the snottite total lipid extract were exclusively tetraethers (Supplementary Figure S12), indicating that snottite G-plasma have membrane chemistry expected for extreme acidophiles. Interestingly, functions devoted to membrane/cell wall/envelope biogenesis are strongly overrepresented in the snottite metagenome (Figure 4), which had the highest proportion of COGs in this category among all the metagenomes we compared.
Snottite ecology and biogeochemistry
Metagenomic data reveal important insights into the structure and function of Frasassi snottite communities, beyond those that we obtained using rRNA methods. Together with geochemical and lipid analyses, these data provide a conceptual model of snottite ecology that will guide future work at Frasassi and other sulfidic caves distributed globally (Figure 3). A role for Acidithiobacillus as the main primary producer, sulfuric acid generator and biofilm architect in the Frasassi snottite community is supported by metagenomic analysis. Other major populations (for example, G-plasma, Acidimicrobiaceae and eukaryotic organisms) are likely heterotrophs or mixotrophs that rely on organic carbon fixed by Acidithiobacillus. Nitrogen is provided to the community either by rare nitrogen-fixing populations or by NH3 degassing from the circumneutral water table, or both.
As the cave atmosphere supplies H2S and O2 that support energy generation, as well as CO2 and (potentially) NH3 required for autotrophic growth, the formation of hanging biofilms with high surface area to volume ratio is a crucial feature of snottite ecology. Hanging biofilms also have minimal contact with cave wall minerals such as calcite and gypsum that would buffer the pH >2. Other cave wall biofilms (pH 4–6) in the sulfidic zones at Frasassi have species richness comparable to soil (Jones et al., 2008), demonstrating that neither complete darkness nor dispersal limitations are the root cause of low species richness in snottites. Metagenomic data show that Acidithiobacillus have flagella, pilin and expolymeric substance biosynthesis genes required to form biofilms. Equipped with adaptations for life in acid (for example, hopanoid production, K+ transporters), Acidithiobacillus thrive in the extremely acidic snottite matrix, which excludes microorganisms that may otherwise compete for nutrients and the abundant thermodynamic energy available from sulfide oxidation in the cave air.
The Frasassi cave snottite community provides an interesting comparison to the extreme AMD community described by Tyson et al. (2004). The closest relatives of several snottite organisms derive from AMD environments (Supplementary Figures S5, S6), despite striking differences in the geochemistry of the two environments including metal ion concentrations (toxic levels in AMD, potentially growth-limiting levels in limestone caves) and available electron donors (Fe2+ in AMD, H2S in snottites). Despite these differences, both communities are dominated by a large population of lithoautotrophic bacteria (>70% of total cells), with smaller populations of heterotrophs. Both communities have very low species richness, consistent with exclusion of most species by extremely low pH (0–1). Nitrogen-fixers represent 10–12% of cells in the AMD community, but were not detected in the snottite community, reflecting either nitrogen fixation by rare community members and/or the availability of sufficient fixed nitrogen in the cave environment (Figure 3). Membrane adaptations (for example, hopanoid biosynthesis) to extremely acidic conditions are found in the major bacterial populations in both snottite and AMD communities.
Advantages and limitations of metagenomic analysis with short reads
The RS24 metagenomic dataset was obtained using first generation 454 Life Sciences pyrosequencing technology, which produced short reads averaging 100 bp. This technology has proven to be an effective tool for metagenomics (for example, Edwards et al., 2006; Dinsdale et al., 2008). Due to recent advances in sequencing technology, platforms such as Illumina/Solexa (San Diego, CA, USA) and ABI SOLiD (Applied Biosystems, Foster City, CA, USA) produce reads between 50–100 bp in length, and these platforms are becoming increasingly popular for metagenomic analyses because they combine low cost with extremely high throughput (for example, Lazarevic et al., 2009; Qin et al., 2010). Therefore, short-length sequence reads are likely to remain an important tool for metagenomics in the future and will be associated with both advantages and disadvantages in comparison with other metagenomic approaches.
There are significant challenges to using short sequences for metagenomic analysis. Shorter reads contain less information, with the result that it is more difficult to assign taxonomic affiliations and gene functions. In the snottite metagenome, reads assigned to Gammaproteobacteria could be reassigned to Acidithiobacillus, Actinobacteria to Acidimicrobiaceae and archaea to G-plasma. This binning approach was dependent on 16S rRNA cloning and FISH population counts indicating that a single species overwhelmingly dominated each of these higher-level clades. The use of FISH (an in situ technique) also allowed us to evaluate potential DNA extraction bias in the metagenome. In addition to constraints derived from FISH, we used stringent criteria for sequence annotations and tested our methods with a variety of simulated datasets to ensure that functional and taxonomic assignments were as accurate as possible (Supplementary Table S2). We could only assign 40% of all metagenomic reads using these conservative criteria, although this is high compared with other studies that used 454 GS20 technology for metagenomics (c.f. Biddle et al., 2008; Dinsdale et al., 2008). Analyses of simulated datasets with MEGAN indicate that unless a strain of the query organism is present in the nr database, binning rates using 454 GS20 data are expected to be similarly low (Supplementary Table S3). Biases due to the unequal representation of microbial taxonomic groups in genome databases are a major challenge for metagenomics in general (for example, Fuchsman and Rocap, 2006; Huson et al., 2007). Database biases are especially significant for short sequences, because we cannot use ab initio gene predictors and must determine function based on similarity to known sequences. The effects of database bias are evident in our analyses in several ways. First, the taxonomic classification of all reads is skewed towards Acidithiobacillus compared with other taxa (Figure 2b), because the genus Acidithiobacillus and its parent group Gammaproteobacteria are better represented in the nr database compared with G-plasma and Acidimicrobiaceae. Second, when we added Iron Mountain AMD G-plasma sequences (Tyson et al., 2004) to the nr database, the number of reads assigned to G-plasma increased by 174%. Likewise, the addition of the Acidimicrobium ferrooxidans genome to the nr database increased the number of reads and we could assign to Acidimicrobiaceae by 175%. Database biases will diminish with the rapidly increasing number of microbial genome and metagenome sequences, but remain an important challenge for current efforts.
However, as this study demonstrates that there are significant advantages to using short pyrosequencing reads for metagenomic analyses. The RS24 metagenome cost less than $3000 (United States dollars) and yet produced data that revealed important metabolic attributes of snottite organisms including carbon and nitrogen fixation, energy metabolisms and adaptations to life in extreme acid. These data will enhance future cultivation and sequencing efforts. Although still subject to DNA extraction biases that affect all DNA-based characterization technologies, pyrosequencing platforms do avoid biases associated with PCR and cloning. Notably, metagenomic data identified G-plasma as the major archaeal component of sample RS24, allowing us to address mismatches between G-plasma and published archaeal 16S rRNA primers. More generally, pyrosequencing provides a way to quantitatively assess archaeal contributions to microbial communities, despite long-known and significant biases against archaea in published PCR primers, probes and during cloning (Teske and Sørensen, 2007; Biddle et al., 2008). Finally, as microbial genomes continue to be added to public databases, metagenomes composed of short reads may become the most efficient path to metabolic profiling of entire communities, because representative fragments of a large number of genes can be detected with low overall sequencing effort.
Acknowledgments
We acknowledge the contributions of two anonymous reviewers whose comments improved the manuscript. This work was supported by grants to JLM from the National Science Foundation (EAR-0527046) and NASA NAI (NNA04CC06A). AP received support from NSF (EAR-0641899) and the David & Lucille Packard Foundation. We thank A Montanari for logistical support and the use of facilities and laboratory space at the Osservatorio Geologico di Coldigioco (Italy), and S Mariani, S Galdenzi and S Cerioni for expert advice and field assistance in Italy. We thank T Sowers for methane analyses, S Schuster, J Biddle, C House and M Rhodes for insightful discussions about metagenomic data analysis, and T Canich and D Futrick for computing support.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allen E, Banfield J. Community genomics in microbial ecology and evolution. Nat Rev Microbiol. 2005;3:489–498. doi: 10.1038/nrmicro1157. [DOI] [PubMed] [Google Scholar]
- Altschul S, Madden T, Schäffer A, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Austin C, Dopson M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007;15:165–171. doi: 10.1016/j.tim.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Barreto M, Jedlicki E, Holmes D. Identification of a gene cluster for the formation of extracellular polysaccharide precursors in the chemolithoautotroph Acidithiobacillus ferrooxidans. Appl Environ Microbiol. 2005;71:2902–2909. doi: 10.1128/AEM.71.6.2902-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle J, Fitz-Gibbon S, Schuster S, Brenchley J, House C. Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci USA. 2008;105:10583–10588. doi: 10.1073/pnas.0709942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond P, Smriga S, Banfield J. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl Environ Microbiol. 2000;66:3842–3849. doi: 10.1128/aem.66.9.3842-3849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello P, Roldán M, Moreno-Vivián C. Nitrate reduction and the nitrogen cycle in archaea. Microbiology. 2004;150:3527–3546. doi: 10.1099/mic.0.27303-0. [DOI] [PubMed] [Google Scholar]
- Case R, Boucher Y, Dahllöf I, Holmström C, Doolittle W, Kjelleberg S. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol. 2007;73:278–288. doi: 10.1128/AEM.01177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L, Morgan-Kiss R, Hanson T. Functional analysis of three sulfide: quinone oxidoreductase homologs in Chlorobaculum tepidum. J Bacteriol. 2009;191:1026–1034. doi: 10.1128/JB.01154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Liu Y, Wu J, She Q, Jiang C, Liu S. Novel bacterial sulfur oxygenase reductases from bioreactors treating gold-bearing concentrates. Appl Microbiol Biotechnol. 2007;74:688–698. doi: 10.1007/s00253-006-0691-0. [DOI] [PubMed] [Google Scholar]
- Ciccarelli F, Doerks T, Von Mering C, Creevey C, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Clark D, Norris P. Acidimicrobium ferrooxidans gen. nov., sp. nov.: mixed-culture ferrous iron oxidation with Sulfobacillus species. Microbiology. 1996;142:785–790. doi: 10.1099/00221287-142-4-785. [DOI] [PubMed] [Google Scholar]
- Cleaver A, Burton N, Norris P. A novel Acidimicrobium species in continuous cultures of moderately thermophilic, mineral-sulfide-oxidizing acidophiles. Appl Environ Microbiol. 2007;73:4294–4299. doi: 10.1128/AEM.02658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro A, Toenshoff E, Taylor M, Fritsche T, Wagner M, Horn M. andidatus Protochlamydia amoebophila', an endosymbiont of Acanthamoeba spp. Int J Syst Evol Microbiol. 2005;55:1863–1866. doi: 10.1099/ijs.0.63572-0. [DOI] [PubMed] [Google Scholar]
- Cox JC, Nicholls DG, Ingledew WJ. Transmembrane electrical potential and transmembrane pH gradient in the acidophile Thiobacillus ferro-oxidans. Biochem J. 1979;178:195–200. doi: 10.1042/bj1780195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M, O'toole G. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale E, Edwards R, Hall D, Angly F, Breitbart M, Brulc J, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- Edwards R, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson D, et al. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7:57. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C, Bardischewsky F, Rother D, Quentmeier A, Fischer J. Prokaryotic sulfur oxidation. Curr Opin Microbiol. 2005;8:253–259. doi: 10.1016/j.mib.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Friedrich C, Rother D, Bardischewsky F, Quentmeier A, Fischer J. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism. Appl Environ Microbiol. 2001;67:2873–2882. doi: 10.1128/AEM.67.7.2873-2882.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsman C, Rocap G. Whole-genome reciprocal BLAST analysis reveals that Planctomycetes do not share an unusually large number of genes with Eukarya and Archaea. Appl Environ Microbiol. 2006;72:6841–6884. doi: 10.1128/AEM.00429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdenzi S, Maruoka T. Gypsum deposits in the Frasassi Caves, central Italy. J Cave Karst Stud. 2003;65:111–125. [Google Scholar]
- Griesbeck C, Hauska G, Schütz M.2000Biological sulfide oxidation: sulfide-quinone reductase (SQR), the primary reactionIn: Pandalai SG (ed)Recent Research Developments in Microbiology Research Signpost: Trivadrum, India; 179–203. [Google Scholar]
- Griesbeck C, Schütz M, Schödl T, Bathe S, Nausch L, Mederer N, et al. Mechanism of sulfide-quinone reductase investigated using site-directed mutagenesis and sulfur analysis. Biochemistry. 2002;41:11552–11565. doi: 10.1021/bi026032b. [DOI] [PubMed] [Google Scholar]
- Hensen D, Sperling D, Trüper H, Brune D, Dahl C. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol Microbiol. 2006;62:794–810. doi: 10.1111/j.1365-2958.2006.05408.x. [DOI] [PubMed] [Google Scholar]
- Hopmans E, Schouten S, Pancost R, van der Meer M, Damstè J. Analysis of intact tetraether lipids in archaeal cell material and sediments by high performance liquid chromatography/atmospheric pressure chemical ionization mass spectrometry. Rapid Commun Mass Spec. 2000;14:585–589. doi: 10.1002/(SICI)1097-0231(20000415)14:7<585::AID-RCM913>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Hose LD, Palmer AN, Palmer MV, Northup DE, Boston PJ, DuChene HR. Microbiology and geochemistry in a hydrogen-sulphide-rich karst environment. Chem Geol. 2000;169:399–423. [Google Scholar]
- Huson D, Auch A, Qi J, Schuster S. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inskeep W, Rusch D, Jay Z, Herrgard M, Kozubal M, Richardson T, et al. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One. 2010;5:e9773. doi: 10.1371/journal.pone.0009773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, Bacelar-Nicolau P, Okibe N, Thomas A, Hallberg K. Ferrimicrobium acidiphilum gen. nov., sp. nov. and Ferrithrix thermotolerans gen. nov., sp. nov.: heterotrophic, iron-oxidizing, extremely acidophilic Actinobacteria. Int J Syst Evol Microbiol. 2009;59:1082. doi: 10.1099/ijs.0.65409-0. [DOI] [PubMed] [Google Scholar]
- Johnson D, Hallberg K. The microbiology of acidic mine waters. Res Microbiol. 2003;154:466–473. doi: 10.1016/S0923-2508(03)00114-1. [DOI] [PubMed] [Google Scholar]
- Jones D, Lyon E, Macalady J. Geomicrobiology of biovermiculations from the Frasassi cave system, Italy. J Cave Karst Stud. 2008;70:78–93. [Google Scholar]
- Kanao T, Kamimura K, Sugio T. Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans. J Biotechnol. 2007;132:16–22. doi: 10.1016/j.jbiotec.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Kletzin A.2007General characteristics and important model organismsIn: Cavicchioli R (ed). Archaea: Molecular and Cellular Biology ASM Press: Washington DC; 14–92. [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M (eds). Nucleic Acid Techniques in Bacterial Systematics Wiley: New York; 115–175. [Google Scholar]
- Lazarevic V, Whiteson K, Huse S, Hernandez D, Farinelli L, sterâs M, et al. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods. 2009;79:266–271. doi: 10.1016/j.mimet.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levicán G, Ugalde J, Ehrenfeld N, Maass A, Parada P. Comparative genomic analysis of carbon and nitrogen assimilation mechanisms in three indigenous bioleaching bacteria: predictions and validations. BMC Genomics. 2008;9:581–600. doi: 10.1186/1471-2164-9-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalady J, Vestling M, Baumler D, Boekelheide N, Kaspar C, Banfield J. Tetraether-linked membrane monolayers in Ferroplasma spp: a key to survival in acid. Extremophiles. 2004;8:411–419. doi: 10.1007/s00792-004-0404-5. [DOI] [PubMed] [Google Scholar]
- Macalady JL, Dattagupta S, Schaperdoth I, Jones DS, Druschel GK, Eastman D. Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. ISME J. 2008;2:590–601. doi: 10.1038/ismej.2008.25. [DOI] [PubMed] [Google Scholar]
- Macalady JL, Jones DS, Lyon EH. Extremely acidic, pendulous microbial biofilms from the Frasassi cave system, Italy. Environ Microbiol. 2007;9:1402–1414. doi: 10.1111/j.1462-2920.2007.01256.x. [DOI] [PubMed] [Google Scholar]
- Macalady JL, Lyon EH, Koffman B, Albertson LK, Meyer K, Galdenzi S, et al. Dominant microbial populations in limestone-corroding stream biofilms, Frasassi cave system, Italy. Appl Environ Microbiol. 2006;72:5596–5609. doi: 10.1128/AEM.00715-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malm S, Tiffert Y, Micklinghoff J, Schultze S, Joost I, Weber I, et al. The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology. 2009;155:1332–1339. doi: 10.1099/mic.0.023275-0. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman W, Attiya S, Bader J, Bemben L, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz V, Ivanova N, Szeto E, Palaniappan K, Chu K, Dalevi D, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F, Bandeiras T, Urich T, Teixeira M, Gomes C, Kletzin A. Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate: quinone oxidoreductase. Mol Microbiol. 2004;53:1147–1160. doi: 10.1111/j.1365-2958.2004.04193.x. [DOI] [PubMed] [Google Scholar]
- Okabe S, Odagiri M, Ito T, Satoh H. Succession of sulfur-oxidizing bacteria in the microbial community on corroding concrete in sewer systems. Appl Environ Microbiol. 2007;73:971–980. doi: 10.1128/AEM.02054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson A, Leavitt W, Sáenz J, Summons R, Tam M, Close H. Diversity of hopanoids and squalene-hopene cyclases across a tropical land-sea gradient. Environ Microbiol. 2009;11:1208–1223. doi: 10.1111/j.1462-2920.2008.01817.x. [DOI] [PubMed] [Google Scholar]
- Pearson A, Page S, Jorgenson T, Fischer W, Higgins M. Novel hopanoid cyclases from the environment. Environ Microbiol. 2007;9:2175–2188. doi: 10.1111/j.1462-2920.2007.01331.x. [DOI] [PubMed] [Google Scholar]
- Pott A, Dahl C. Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology. 1998;144:1881–1894. doi: 10.1099/00221287-144-7-1881. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf K, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin L, Stromley J, Rittmann B, Stahl D. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Kang K, Paulsen I. TransportDB: a relational database of cellular membrane transport systems. Nucleic Acids Res. 2004;32:D284–D288. doi: 10.1093/nar/gkh016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D, Berks B, Russell D, Spiro S, Taylor C. Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell Mol Life Sci. 2001;58:165–178. doi: 10.1007/PL00000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwerder T, Sand W. Oxidation of inorganic sulfur compounds in acidophilic prokaryotes. Eng Life Sci. 2007;7:301–309. [Google Scholar]
- Salazar J, Zúñiga R, Raczniak G, Becker H, Söll D, Orellana O. A dual-specific Glu-tRNAGln and Asp-tRNAAsn amidotransferase is involved in decoding glutamine and asparagine codons in Acidithiobacillus ferrooxidans. FEBS Lett. 2001;500:129–131. doi: 10.1016/s0014-5793(01)02600-x. [DOI] [PubMed] [Google Scholar]
- Sauvé V, Bruno S, Berks B, Hemmings A. The SoxYZ complex carries sulfur cycle intermediates on a peptide swinging arm. J Biol Chem. 2007;282:23194–23204. doi: 10.1074/jbc.M701602200. [DOI] [PubMed] [Google Scholar]
- Schleper C, Puehler G, Holz I, Gambacorta A, Janekovic D, Santarius U, et al. Picrophilus gen. nov., fam. nov.: a novel aerobic, heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around pH 0. J Bacteriol. 1995;177:7050–7059. doi: 10.1128/jb.177.24.7050-7059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively J, Van Keulen G, Meijer W. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu Rev Microbiol. 1998;52:191–230. doi: 10.1146/annurev.micro.52.1.191. [DOI] [PubMed] [Google Scholar]
- Stern L, Engel A, Bennett P. Nitrogen isotope evidence of ammonia vapor assimilation by cave wall microbial biofilms in a sulfidic cave, a novel mechanism of nutrient acquisition. AGU Fall Meeting Abstracts. 2003;84:B42E–B405. [Google Scholar]
- Talbot H, Squier A, Keely B, Farrimond P. Atmospheric pressure chemical ionisation reversed-phase liquid chromatography/ion trap mass spectrometry of intact bacteriohopanepolyols. Rapid Commun Mass Spec. 2003;17:728–737. doi: 10.1002/rcm.974. [DOI] [PubMed] [Google Scholar]
- Tatusov R, Fedorova N, Jackson J, Jacobs A, Kiryutin B, Koonin E, et al. The COG database: an updated version includes eukaryotes. BMC Bioinform. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske A, Sørensen K. Uncultured archaea in deep marine subsurface sediments: have we caught them all. ISME J. 2007;2:3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- Tyson G, Chapman J, Hugenholtz P, Allen E, Ram R, Richardson P, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Valdés J, Pedroso I, Quatrini R, Dodson R, Tettelin H, Blake R, et al. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics. 2008a;9:597. doi: 10.1186/1471-2164-9-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés J, Pedroso I, Quatrini R, Holmes D. Comparative genome analysis of Acidithiobacillusferrooxidans, A.thiooxidans and A. caldus: insights into their metabolism and ecophysiology. Hydrometallurgy. 2008b;94:180–184. [Google Scholar]
- Valdés J, Quatrini R, Hallberg K, Dopson M, Valenzuela P, Holmes D. Draft genome sequence of the extremely acidophilic bacterium Acidithiobacillus caldus ATCC 51756 reveals metabolic versatility in the genus Acidithiobacillus. J Bacteriol. 2009;191:5877–5878. doi: 10.1128/JB.00843-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vossenberg J, Driessen A, Konings W. The essence of being extremophilic: the role of the unique archaeal membrane lipids. Extremophiles. 1998;2:163–170. doi: 10.1007/s007920050056. [DOI] [PubMed] [Google Scholar]
- Videmšek U, Hagn A, Suhadolc M, Radl V, Knicker H, Schloter M, et al. Abundance and diversity of CO2-fixing bacteria in grassland soils close to natural carbon dioxide springs. Microb Ecol. 2009;58:1–9. doi: 10.1007/s00248-008-9442-3. [DOI] [PubMed] [Google Scholar]
- Vlasceanu L, Sarbu SM, Engel AS, Kinkle BK. Acidic cave wall biofilms located in the Frasassi Gorge, Italy. Geomicrobiol J. 2000;17:125–139. [Google Scholar]
- Wakai S, Tsujita M, Kikumoto M, Manchur M, Kanao T, Kamimura K. Purification and characterization of sulfide: quinone oxidoreductase from an acidophilic iron-oxidizing bacterium, Acidithiobacillusferrooxidans. Biosci, Biotechnol, Biochem. 2007;68:2519–2528. doi: 10.1271/bbb.70332. [DOI] [PubMed] [Google Scholar]
- Welander P, Hunter R, Zhang L, Sessions A, Summons R, Newman D. Hopanoids play a role in membrane integrity and pH homeostasis in Rhodopseudomonas palustris TIE-1. J Bacteriol. 2009;191:6145–6156. doi: 10.1128/JB.00460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker R, Banfield J. Population genomics in natural microbial communities. Trends Ecol Evol. 2006;21:508–516. doi: 10.1016/j.tree.2006.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.