Abstract
Dramatic decreases in the extent of Arctic multiyear ice (MYI) suggest this environment may disappear as early as 2100, replaced by ecologically different first-year ice. To better understand the implications of this loss on microbial biodiversity, we undertook a detailed census of the microbial community in MYI at two sites near the geographic North Pole using parallel tag sequencing of the 16S rRNA gene. Although the composition of the MYI microbial community has been characterized by previous studies, microbial community structure has not been. Although richness was lower in MYI than in underlying surface water, we found diversity to be comparable using the Simpson and Shannon's indices (for Simpson t=0.65, P=0.56; for Shannon t=0.25, P=0.84 for a Student's t-test of mean values). Cyanobacteria, comprising 6.8% of reads obtained from MYI, were observed for the first time in Arctic sea ice. In addition, several low-abundance clades not previously reported in sea ice were present, including the phylum TM7 and the classes Spartobacteria and Opitutae. Members of Coraliomargarita, a recently described genus of the class Opitutae, were present in sufficient numbers to suggest niche occupation within MYI.
Keywords: multiyear sea ice, 454 pyrosequencing, sea ice bacteria
Introduction
The sea ice environment is currently extensive on Earth, covering some 27–36 km2 × 106 km2 in the high latitudes of both hemispheres (Cavalieri et al., 1997; Stroeve et al., 2007), an area approximately the size of Canada. This environment is composed primarily of two ice types; first-year ice (FYI) which forms in the fall and melts in the summer, and multiyear ice (MYI), perennial sea ice that has persisted through at least one complete year. In parallel with observed global climate trends, the MYI environment is in a state of sharp decline (Nghiem et al., 2007; Comiso et al., 2008; Barber et al., 2009; Boe et al., 2009). A rapid decrease in minimal sea ice extent suggests that the MYI environment could be non-existent by 2100 (Boe et al., 2009). This dramatic reduction in MYI is expected to have a significant impact on the polar microbial community as ocean surface temperature, salinity, nutrients and availability of the unique sea ice environment change (Kirchman et al., 2009; Vincent, 2010). These changes will in turn impact the way carbon and other elements are cycled through the Arctic marine food web, most readily through the coupling of primary and bacterial production (Kirchman et al., 2009).
In 2006 the project DNA of the Polar Seas was launched as part of the Danish expedition Galathea III, to more fully catalog microbial diversity within the climatically vulnerable Arctic and Southern oceans. As participants in a 2009 extension of DNA of the Polar Seas onboard the Swedish icebreaker Oden, we sampled MYI and surface seawater near the geographic North Pole with the aim of achieving a high-resolution analysis of microbial diversity in these proximate but disparate environments. Previous microbiological investigations of sea ice, dating back to at least 1893 (McLean, 1918), have identified a specialized sea ice microbial community (Bowman et al., 1997; Brown and Bowman, 2001; Junge et al., 2002). Because of methodological limitations, however, these studies have reported mainly on community composition rather than structure. They have focused almost exclusively on FYI. The only previous microbial investigation of Arctic MYI, using fluorescence in situ hybridization and 16S rRNA clone libraries, reported a summer community dominated by Gammaproteobacteria, followed by Alphaproteobacteria and Cytophaga–Flavobacterium–Bacteriodes (Brinkmeyer et al., 2003), results similar to those obtained for a summer sampling of Antarctic MYI (Brown and Bowman, 2001). The dynamical processes that control microbial community structure in sea ice are not well understood, however, a growing body of literature suggests that microbial communities in MYI differ substantially from those in FYI (Brown and Bowman, 2001; Brinkmeyer et al., 2003; Deming, 2007).
Sea ice is considered selective for psychrophilic microbes (Junge et al., 2002), providing the ‘seed population' for the microbial community reported to dominate carbon cycling in Arctic coastal waters (Connelly et al., 2006). MYI in particular has been observed to host a strongly cold-active microbial enzyme assemblage compared with Arctic seawater and FYI (Deming, 2007). Loss of this seed population can be expected to impact the pelagic fate and downwards flux of carbon from sea surface to seafloor (Kellogg et al., 2010). If the microbial populations present in MYI are more complex than previously anticipated, then the predicted loss of MYI may result in a greater loss of biodiversity and an as yet unquantified impact on elemental cycling. In this study, we employed 454 pyrosequencing of the 16S rRNA gene to obtain a snapshot of microbial community structure within MYI at a greater resolution than previous studies, comparing it with that of surface seawater to test for a greater overlap of phylogenetic groups. This approach, in confirming the unique nature of the sea ice microbial community, also revealed an unexpectedly high degree of diversity in MYI and several new clades not previously found in sea ice.
Methods
Sample collection and preparation
Sea ice and seawater samples were collected during the LOMROG 2009 expedition on the Swedish icebreaker Oden. Two MYI samples (designated I13 and I14) and three surface seawater samples (S15, S16 and S17) were collected within 150 km of the geographic North Pole (Supplementary Figure S1, station coordinates given in Supplementary Table S1). Although the precise age of the sampled MYI floes could not be determined, ice thickness (301 cm and 310 cm), temperature and salinity conditions (Supplementary Figure S1), location (50–120 km from the geographic North Pole) and time of sampling suggest the ice had persisted through more than one summer. Detailed methods for sample collection and processing shipboard and for subsequent DNA extraction, purification, and sequencing and other analyses are given in Supplementary Information. For each MYI sample two cores spanning the depth of the MYI floe were sectioned and melted together as one sample. Microorganisms in the melted samples were captured on a 0.2-μm filter after a 2.0-μm pre-filtration. The decision to melt an entire core length precluded a study of vertical community structure within the ice, but ensured that enough DNA was recovered for this exploratory effort.
Extraction, purification and sequencing were conducted at the University of Copenhagen. Following lysis and phenol-chloroform DNA extraction, amplification of a section of the 16S RNA gene including the V3 hypervariable region was conducted with the universal primers PRK341F (5′-CCTAYGGGRBGCAACAG-3′) and PRK806R (5′-GGACTACNNGGGTATCTAAT-3′) (Yu et al., 2005). Sequencing was performed on a GS FLX pyrosequencer (454 Life Sciences, Branford, CT, USA). All sequences used in this study are available from the NCBI sequence read archive under run accession number SRR254219.2.
Community structure
Analysis was conducted using the microbial ecology community software program Mothur (http://www.mothur.org/wiki/Download_mothur) (Schloss et al., 2009). The reads were processed by removing tags and primer, only accepting reads with an average quality score above 20 and read lengths between 200 nt and 300 nt. Identical sequences were grouped and a representative aligned against the Greengenes reference set (DeSantis et al., 2006) using the Needleman–Wunsch algorithm (Needleman and Wunsch, 1970). A further screening step (pre-cluster) was applied to reduce sequencing noise by clustering reads differing by only 1 bp (Huse et al., 2010). Chimeric sequences were detected and removed using the implementation of ChimeraSlayer developed by the Broad Institute. The remaining high-quality reads were used to generate a distance matrix and clustering with the furthest-neighbor algorithm. Representative sequences for shared operational taxonomic units (OTUs) as defined by 100% (unique), 97%, 93% and 90% similarity were obtained.
Sequence coverage was calculated as the percentage of unique reads observed against the Chao1 estimate of richness (Chao, 1984), with coverage ranging 30.8–35.8%. Rarefaction curves were generated for each sample at the 100%, 97%, 90% and 80% levels of similarity. To describe community structure, diversity was calculated using Simpson's index (Simpson, 1949), which estimates the probability that two randomly selected reads will belong to the same OTU. Because coverage was less than 100%, diversity was also calculated using a non-parametric instance of the Shannon's index (Chao and Shen, 2003). The Pearson's correlation coefficient was calculated to determine the level of agreement between these two methods.
To analyze community breadth, the distance between all clades present in each community, a distance matrix of all sequences contained within the ice and seawater communities was created using ClustalX (Larkin et al., 2007), correcting for multiple substitutions (Kimura, 1983) and ignoring sites with gaps. Central tendencies were used to describe the distribution of distances within each community.
Community composition
Community taxonomy at the unique sequence level was determined using Mothur's classification tool with 1000 iterations and a bootstrap cutoff score of 60. The taxonomic database consisted of 14 956 bacterial and 2297 archaeal sequences from the SILVA database (Pruesse et al., 2007) classified with the Ribosomal Database Project (Cole et al., 2007). Samples were grouped by their respective environment (MYI or seawater) and classified against this database. Maximum-likelihood trees were constructed of all genera identified as common to both the MYI and seawater samples (present above 0.035% of the total community in both environments) to evaluate whether lineages within these genera were exclusive to either MYI or seawater. Reference trees were constructed from all high-quality, >1200-bp sequences available on the Ribosomal Database Project for each genus, with the addition of all type strains available for the relevant family, using the General Time Reversible model and rapid bootstrapping (100 bootstraps) option in the parallelized version of RaxML vs 7.0.4 (http://wwwkramer.in.tum.de/exelixis/software.html) (Stamatakis, 2006). The online version of placer (Matsen et al, 2010) was used to place the reads attributed to each genus on the appropriate reference tree.
Community similarity
Differences between communities were analyzed by first constructing a maximum-likelihood tree of all unique sequences aligned with Mothur using RAxML as previously described. Differences in tree topology were then assessed using the unweighted and weighted UniFrac algorithms (Lozupone and Knight, 2005). Abundance information was not embedded into the tree files necessary for these algorithms beyond the taxonomic level of unique. Therefore, Sørensen's abundance and richness-based indices of similarity were used to compare the communities at different taxonomic levels (unique, 97%, 93% and 90% similarity). All indices were calculated with Mothur. For compatibility with pplacer, trees were constructed in RAxML using the GTRGAMMA option, precluding calculating of UniFrac significance scores from predicted branch length. To calculate significance, a separate UniFrac analysis was conducted on a neighbor-joining tree constructed using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/#) (Larkin et al., 2007) and bootstrapped 1000 times.
16S rRNA divergence
Divergence within the 16S rRNA gene was assessed using both taxonomy and taxonomy-independent approaches. In the taxonomy approach, the number of member reads and number of unique members was tallied for each clade at each taxonomic level. The slope of the best-fit line for both seawater and sea ice communities was determined using a least-squares analysis. Slopes for the two communities were compared using a Student's t-test. To assess 16S rRNA gene divergence independent of taxonomy, and thus avoid bias due to unequal characterizations of the MYI and seawater communities, the number of OTUs was estimated for various definitions of OTU, from 0 to 20% divergence, for each community using the Chao1 estimate of richness (Chao, 1984). The number of OTUs calculated for seawater was normalized to that of sea ice by reducing the number estimated at each definition to 36%, the fraction of maximum seawater richness found within the ice. The relationship between the number of OTUs for both communities was described using a line of best fit. The difference between the slope of the best-fit line and unity was assessed with a Student's t-test.
Results
Environmental
Averaged vertically, the sampled MYI was slightly warmer than the surface seawater (−1.1 °C and −1.2 °C vs −1.48 °C and −1.51 °C, respectively) and lower in bulk salinity (1.9 and 0.7 compared with 30.92 and 32.57) (Supplementary Table S1). These values are typical for MYI (Eicken et al., 1995). Ice thickness, at 3.0 m for I13 and 3.3 m for I14, was also consistent with values expected for MYI (Eicken et al., 1995; Haas, 2004). At the time of sampling, the ice was observed to be drifting rapidly (5.19 m min–1 at 318.97° and 4.84 m min–1 at 198.73° for I13 and I14, respectively). These variations in drift are common for ice in the central Arctic Ocean and consistent with general transport from the Siberian Sea toward Fram Strait (Pfirman et al., 1997). Models from 2002 suggest that ice at the sampled locations should be between 4- and 8-years old, though the same study points to a dramatic decrease in the age of ice across most of the Arctic Ocean (Rigor and Wallace, 2004). As a result, the sampled ice could be younger than 4 years.
At 1.68 cells ml−1 and 8.40 cells ml−1 × 104 cells ml−1 melted sea ice, total bacterial and archaeal abundance (averaged vertically throughout the entire ice thickness) was lower in MYI than in the surface seawater (1.84 cells ml−1 × 105 cells ml−1 for sample S17), though concentrations of chlorophyll a were similar (sea ice: 0.025 mg m–3 and 0.055 mg m–3 melted ice averaged vertically, seawater: 0.037 mg m−3). Concentrations of particulate exopolymers were low in both ice samples and undetectable in seawater (sample S17; Supplementary Table S1). Calculating the concentration factor for the sea ice brine as the inverse of the brine volume fraction equation of Cox and Weeks (Cox and Weeks, 1983; Eicken, 2009) suggests that the abundance of microbes, salt, particulate exopolymers, chlorophyll a and other constituents partitioned into the liquid brine fraction would be an order of magnitude higher than these values (Supplementary Table S1).
Community structure
Richness was higher in seawater than sea ice for all definitions of OTU, reaching average values at the unique sequence level of 1552 for MYI and 3042 for seawater with a confidence level of 89% (P=0.11; Table 1). The limited degrees of freedom (df=3) precluded tighter confidence levels. Even though a pre-clustering step was performed to reduce pyrosequencing noise, the absolute values for richness should be treated cautiously because pyrosequencing noise can artificially inflate sample richness (Quince et al., 2009), as can sequence analysis of the 16S rRNA gene by other techniques (Acinas et al., 2004).
Table 1. Mean values of richness and diversity for multiple OTU definitions for the MYI and seawater communities.
|
Chao1 |
Np-Shannon's index |
Simpsons's index |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OTU | Sea ice | Seawater | t | P | Sea ice | Seawater | t | P | Sea ice | Seawater | t | P |
| Unique | 1552 | 3042 | 5.57 | 0.11 | 3.66 | 3.93 | 0.25 | 0.84 | 0.15 | 0.11 | 0.65 | 0.56 |
| 97% | 614 | 1309 | 2.34 | 0.26 | 3.38 | 3.72 | 0.34 | 0.79 | 0.16 | 0.11 | 0.65 | 0.56 |
| 93% | 305 | 768 | 2.38 | 0.25 | 3.08 | 3.51 | 0.47 | 0.72 | 0.17 | 0.12 | 0.75 | 0.51 |
| 90% | 230 | 600 | 2.15 | 0.28 | 2.84 | 3.35 | 0.53 | 0.69 | 0.20 | 0.12 | 0.87 | 0.45 |
Abbreviations: MYI, multi-year ice; OTU, operational taxonomic unit.
Student's t-test results and significance between MYI and seawater samples are given as t and P, respectively.
Diversity was similar for all definitions of OTU between ice and seawater samples (Table 1). Although the MYI community appeared less diverse by both Shannon's index of diversity and Simpson's index, the differences were not statistically significant by a Student's t-test (Table 1). Good agreement was observed between the parametric Simpson's index and the non-parametric Shannon's index (df=18, P<10−4; Supplementary Figure S2), suggesting that despite low coverage using Simpson's definition of abundance when describing these samples is appropriate.
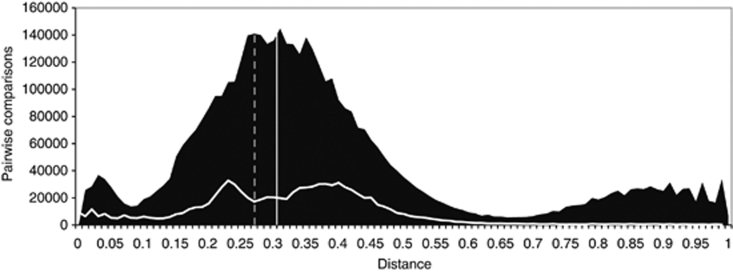
Breadth was significantly different for the ice and seawater communities. A histogram of distances (Figure 1) shows a multimodal distribution for the seawater community (mean=0.43, s.d.=0.30, n=4 941 729), compared with a more simplified (bimodal) distribution within the ice (mean=0.33, s.d.=0.16, n=912 025). A reanalysis of the seawater distribution using only data from the domain Bacteria revealed a narrower breadth (mean=0.30, s.d.=0.12, n=3 988 009). The difference between this mean and the mean calculated for the MYI community by a Student's t-test was significant (P<10−4, df=4 900 036).
Figure 1.
Distribution of genetic distances for the MYI and seawater communities. The shaded region indicates the distribution of distances for the seawater community. The multi-modal distribution is a result of the dominance of a single genus (Pelagibacter) and the two-domain structure of the community. The white lateral line indicates the distribution of distances for the MYI community and the solid vertical line, the mean distance. The dashed vertical line shows the mean distance for the seawater community, excluding the domain Archaea.
Community composition
Within the seawater samples, 2927 reads (represented by 106 unique reads) could not be classified at the domain level using the reference taxonomy (Supplementary Figure S3). NCBI BLASTN (Altschul et al., 1997) was used to find similar sequences. All scored as highly similar (E⩽10−24) to sequences classified as belonging to isolates or environmental sequences of the domain Archaea. A much smaller number of reads in MYI (13, six unique) could not be classified at the domain level, but they showed a high degree of similarity with sequences classified as Archaea (E⩽10−37). Few archaeal reads were detected in MYI (16, 0.13% of all reads) compared with seawater (5485, 15.9%). Within the seawater a large number of archaeal reads were Euryarchaeota (21.9%), with the remainder unclassified below the level of domain. In silico analysis of the universal prokaryotic primer set indicated a low-binding efficiency between primer and marine Crenarchaeota (Supplementary Figure S4). As a result, Crenarchaeota are likely underrepresented in this data set.
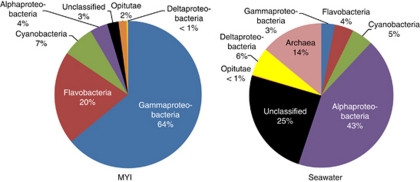
Although the bacterial phylum Proteobacteria was numerically dominant in both the MYI and seawater communities, representing 66.3% and 62.3% of total reads, respectively, substantial differences in community composition between sample types were observed below the level of phylum (Figure 2). Gammaproteobacteria and Flavobacteria (62.3% and 19.9% of total reads) dominated the MYI community, while Alphaproteobacteria dominated the seawater community (48.9% of total reads). The majority (64.6%) of seawater Alphaproteobacteria were classified as Pelagibacter. At the family level the MYI community was dominated by Moraxellaceae, accounting for 42.8% of all ice reads. Large numbers of reads were also assigned to the Flavobacteriaceae family (19.6%). Although not numerically dominant in either case, cyanobacteria were identified in both the ice and seawater samples, accounting for 6.8% of total ice reads and 5.4% of total seawater reads. Cyanobacterial abundance was likely underestimated, as the primer was predicted to have low-binding efficiency with the cyanobacterial 16S rRNA gene (Supplementary Figure S4).
Figure 2.
Numerically dominant clades in the MYI and seawater communities. Gammaproteobacteria and Flavobacteria dominated the ice community; Alphaproteobacteria and Archaea, the seawater community. Deltaproteobacteria and Archaea comprised the smallest fractions of the MYI community; Opitutae, of the seawater community.
At the level of genus (the lowest level assigned), the reads represented 41 identifiable genera for MYI and 37 for seawater. At this level 32.1% of ice reads and 64.3% of seawater reads could not be classified (Supplementary Figure S3). A complete list of read assignments by clade is given in Supplementary Table S3 (MYI) and Supplementary Table S4 (seawater). Of the clades listed for MYI, 54 have not been reported previously in sea ice. Because the bottom, porous horizons of the MYI cores were included in this analysis we cannot rule out that some of these organisms were contained within entrained seawater. Excluding all clades observed within the two seawater samples leaves 38 clades not previously described in either FYI or MYI.
Four genera were identified as common to both environments; Polaribacter, Rubritalea, Psychrobacter and Pelagibacter. Visual inspection of maximum-likelihood trees constructed of these genera (not shown) suggests that only Polaribacter has lineages that have evolved to occupy separate niches within MYI and seawater.
16S rRNA gene divergence
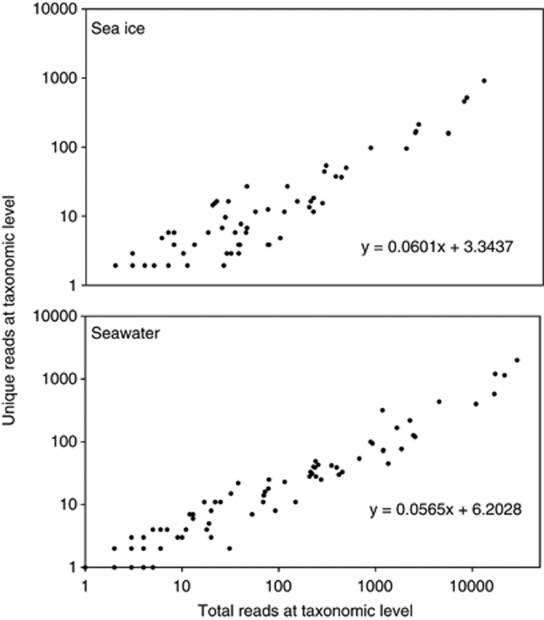
To assess the level of divergence within the MYI and seawater communities, all taxonomies as determined by our classification were tested for a correlation between membership and the number of unique members (Figure 3). For a given taxonomy more members (greater sampling) should produce more unique members. To account for greater coverage of the ice samples, (Table 2) the number of unique members for ice was reduced by 3.50% (the difference in mean coverage between the ice and seawater communities). If the unique MYI and seawater communities were subject to similar evolutionary pressures and timescales, they should display a similar relationship between taxon membership and number of unique members. Strong correlations were observed for both the MYI and seawater communities (P<10−4, df=117 and P<10−4, df=130, respectively), but the relationships differed: slopes of the regression lines fit to the data (m=0.0601 for ice, 0.0565 for seawater) were significantly different with a Student's t-test (df=247, P<10−4). This difference suggests that MYI taxa may accumulate more unique reads than seawater for the same amount of sampling.
Figure 3.
Divergence of the 16S rRNA gene using a taxonomy-based method. Divergence of the 16S rRNA gene (unique reads) with abundance (total reads) for seawater (top): r=0.96, P<10–4, df=130 and for MYI (bottom): r=0.96, P<10–4, df=117. The MYI community appears more divergent than the seawater community, with more new clades formed from increasing membership: the slopes of the lines are significantly different (Student's t-test, t=15.06, P<10–4, df=247).
Table 2. Estimates of richness and diversity for operational taxonomic unit definition of 100% similarity for each multi-year ice and seawater sample.
| Sample | Unique reads | Chao1 | 95% confidence interval | Estimated % coverage | Shannon's index (non-parametric) | Simpson's index | 95% confidence interval |
|---|---|---|---|---|---|---|---|
| I13 | 543 | 1516 | 1242–1896 | 35.8 | 2.96 | 0.237 | 0.23–0.25 |
| I14 | 552 | 1588 | 1298–1990 | 34.8 | 4.36 | 0.071 | 0.065–0.076 |
| S15 | 1057 | 3214 | 2769–3776 | 32.9 | 4.19 | 0.079 | 0.076–0.082 |
| S16 | 853 | 2770 | 2320–3358 | 30.8 | 3.64 | 0.137 | 0.137–0.148 |
| S17 | 996 | 3143 | 2675–3742 | 31.7 | 3.95 | 0.111 | 0.106–0.115 |
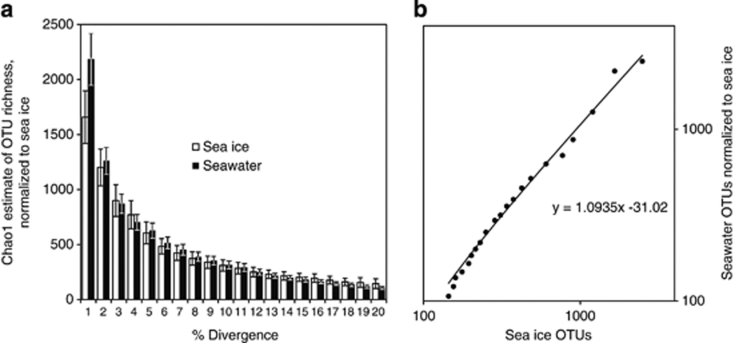
The non-taxonomic approach to divergence yielded a different result. For a given definition of OTU (% divergence), the seawater samples hosted a greater number of OTUs (Figure 4a) when corrected for greater overall richness. A best-fit line for the number of OTUs present in ice for each definition of OTU plotted against the number in seawater yielded a slope significantly different from unity (df=18, P<10−4; Figure 4b) suggesting that seawater hosts more OTUs for an increasingly narrow definition of OTU than does MYI.
Figure 4.
Divergence of the 16S rRNA gene using a taxonomy-independent analysis. The number of OTUs in the seawater community for varying definitions of OTU (0–20% difference in read sequence) was normalized to the lower richness observed in the MYI community. The accumulation of new OTUs at low percentage divergence appears faster for the seawater than sea ice community (a). Error bars indicate 95% confidence. The slope of a best-fit line (shown with log scale on both axis) for values from both environments (b) is significantly different than one (df=18, P=10−4).
Community similarity
Pairwise differences in community composition and structure were assessed for all samples using two richness-based approaches (Sørensen's richness based, unweighted UniFrac) and two abundance-based approaches (Sørensen's abundance based and weighted UniFrac) (Table 3 and Supplementary Table S2). The abundance-based approaches showed substantially lower (more similar) scores between the two sea ice and the three seawater samples than did the richness-based approaches. Using a separate, neighbor-joining tree to calculate the significance of dissimilarity, only seawater samples S16/S17 were not significantly different (P=0.06) for unweighted UniFrac, while S16/S17 and I13/I14 were not significantly different using weighted UniFrac (P=0.19 and P=0.32, respectively).
Table 3. Pairwise comparison of sample similarity for operational taxonomic unit definition of 100% similarity.
| Sample | Pair | Richness- based Sørensen | Unweighted UniFrac | Abundance- based Sørensen | Weighted UniFrac |
|---|---|---|---|---|---|
| I13 | I14 | 0.281 | 0.592 | 0.914 | 0.195 |
| I13 | S15 | 0.013 | 0.841 | 0.544 | 0.684 |
| I13 | S16 | 0.012 | 0.835 | 0.303 | 0.635 |
| I13 | S17 | 0.014 | 0.813 | 0.548 | 0.694 |
| I14 | S15 | 0.015 | 0.813 | 0.391 | 0.651 |
| I14 | S16 | 0.027 | 0.539 | 0.317 | 0.226 |
| I14 | S17 | 0.022 | 0.836 | 0.362 | 0.698 |
| S15 | S16 | 0.239 | 0.824 | 0.961 | 0.642 |
| S15 | S17 | 0.212 | 0.515 | 0.942 | 0.164 |
| S16 | S17 | 0.199 | 0.469 | 0.953 | 0.098 |
High values for Sørensen's index and UniFrac, indicate a high level of similarity. Value in bold were shown to be significantly similar using a separate weighted UniFrac analysis on a neighbor-joining tree bootstrapped 1000 times.
Discussion
Numerous investigations have characterized the composition and structure of the microbial community within Arctic surface seawater, including some analysis by the 454 method (Galand et al., 2009; Kirchman et al., 2010). Although a further characterization of this environment is not the purpose of this paper, a brief comparison is in order due to spatial and seasonal variation of the sea surface microbial community. As observed in this study, Alphaproteobacteria typically dominate Arctic surface water (Bano and Hollibaugh, 2002; Alonso-Sáez et al., 2008; Galand et al., 2009), though in one coastal study a greater abundance of Gammaproteobacteria was observed (Kirchman et al., 2010). Gammaproteobacteria comprised only a small fraction of the seawater reads in this study (<2%), which may reflect a regional difference given the mid-ocean location of this study. None of these studies reported cyanobacteria among the major clades present, though they have been found as allochthonous forms in river-impacted, coastal Arctic waters (Waleron et al., 2007). Further differences were restricted to variations in the relative abundance of the major clades. Deltaproteobacteria, Bacteriodetes, Verrucomicrobiae and Actinobacteria are widely recognized as abundant Arctic seawater clades within the domain Bacteria.
Previous phylogenetic studies of the microbial communities in spring and summer sea ice (FYI and MYI) have indicated dominance by the clades Gammaproteobacteria, Alphaproteobacteria and Cytophaga–Flavobacterium–Bacteriodes (Bowman et al., 1997; Petri and Imholff, 2001; Junge et al., 2002; Brinkmeyer et al., 2003), followed by Chlamydiales and Verrucomicobiales (Brown and Bowman, 2001). With the exception of the order Chlamydiales, which has no representative reads in this data set, our findings confirm the dominance of these clades. Reports of Archaea in sea ice were inconclusive (Deming, 2009) until Collins et al. (2010) confirmed their presence in winter FYI. They have not been detected in FYI of other seasons or in MYI using less sensitive techniques than 454 pyrosequencing. Although our data are biased by the performance of the universal primer set, our findings of limited archaeal reads in the MYI community suggest that marine archaea, along with many pelagic bacterial clades, are not competitive against the spring/summer sea ice bacterial community. Some Archaea may persist interannually in MYI, or the archaeal reads observed in our MYI samples may be the result of recent seawater infiltration of the lower ice horizons. Similarly, cyanobacteria have not previously been reported in either FYI or MYI, though they are common to freshwater polar environments (Waleron et al., 2007; Jungblut et al., 2009). Their presence within MYI may be the result of colonization of surface melt ponds, with subsequent incorporation into MYI through infiltration or melt pond burial. Further study is required to determine whether cyanobacteria may be indigenous to MYI, perhaps occupying upper ice horizons where light and nutrient conditions may provide an open niche.
Several clades not previously reported in sea ice were observed exclusively within MYI, including the Gram-positive phylum TM-7 and family Micrococcineae. These and several other new clades, including the Spartobacteria and Verrucomicrobiales, are commonly found in soil environments, suggesting that they may have reached the MYI environment through dust deposition or inclusion of riverine sediments (Brinkmeyer et al., 2003). Other newly observed clades are likely to be of marine origin, including the genera Persicivirga, Ulvibacter, Granulosicoccus and Lewinella. Representatives from the first three clades have characteristics that suggest an association with marine algae (Nedashkovskaya et al., 2004; Barbeyron et al., 2010; Kurilenko et al., 2010). The possibility that these clades were present in MYI because of sea ice diatoms warrants further study. Other newly observed clades of biogeochemical interest include Methylmicrobium, a dimethylsulfide-oxidizing genus of the Gammaproteobacteria, Cycloclasticus, a polycyclic aromatic hydrocarbon-degrading clade also of this class and Jannaschia, an anoxygenic phototroph of the Roseobacter clade. Although all of these taxa were present at low levels within MYI (below 0.36% of all MYI reads), they may represent an important seed population for bloom conditions within the surrounding seawater or sea ice, a hypothesis further supported by their absence from the seawater samples.
A final newly observed genus of note is Coraliomargarita, a recently described genus of the class Opituae (Yoon et al., 2007). Coraliomargarita accounted for 197 reads in this data set (1.6% of all MYI reads), split among 14 lineages. This genus has no psychrophilic members in culture and, despite a sequenced genome for C. akajimensis (Mavromatis et al., 2010), there is little in the natural history of this genus to suggest why it should be associated with MYI. Like many sea ice-associated microbes C. akajimensis is copiotrophic, a feature that might favor its persistence within organic-rich horizons of MYI. The frequency of Coraliomargarita reads in MYI also raises questions about the role of psychrotolerant bacteria in sea ice.
Past efforts to quantify microbial community structure within sea ice likely underestimated both richness and diversity. Brown and Bowman (2001) were able to measure these parameters on a single MYI sample from the Southern Ocean, identifying it as the most diverse of several sampled ice types. From a clone library they estimated richness at 24±12 and Shannon's index at 1.050, with coverage at 53.8%. Brinkmeyer et al. (2003) conducted a similar clone-library-based analysis on several MYI cores recovered from Fram Strait. This study reports similar coverage (53.3–76%), richness (15–19 OTUs) and diversity (Shannon's index between 1.065±0.002 and 1.291±0.002). By comparison with 454 sequencing and only 34.8% and 35.8% coverage we observed richness an order of magnitude higher, with the Chao1 estimate two orders of magnitude higher. Shannon's index of diversity was calculated to be threefold higher than these earlier estimates.
The low coverage obtained even with the application of 454 sequencing, the identification of new clades not previously found in sea ice and continuing questions regarding the dynamical processes of recruitment, succession and persistence highlight the need for a greater sequencing effort across the sea ice environment. Given the high degree of temporal and spatial heterogeneity inherent to the sea ice environment, and resulting differences in microbial community structure, a three-dimensional sequencing effort across time, space and sequence depth is needed. Such an effort could provide not only the missing characterization of a vast and changing environment, but also identify where analyses targeting specific metabolic genes and gene expression could be applied to our best advantage.
Acknowledgments
We thank Christian Marcussen (GEUS) for the opportunity to participate in LOMROG 2009, Matthias Wietz and Jens Blom for assistance with sampling, Shelly Carpenter, Pia Friis, and Peter Holmsgaard for assistance with laboratory analysis, and John Hopper for calculating the ice drift velocities. This work was funded by the Villum Kann Rasmussen Foundation and Lundbeckfonden, the Danish Agency for Science, Technology, and Innovation (SR), an award from the Gordon and Betty Moore Foundation (JSB), an NSF IGERT fellowship through the University of Washington Astrobiology Program (JSB), and NSF OPP award 0908724 (JWD).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol. 2004;186:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Sáez L, Sánchez O, Gasol JM, Balagué V, Pedrós-Alio C. Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ Microbiol. 2008;10:2444–2454. doi: 10.1111/j.1462-2920.2008.01674.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nuc Acids Res. 1997;25:2289–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano N, Hollibaugh JT. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl Environ Microbiol. 2002;68:505–518. doi: 10.1128/AEM.68.2.505-518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DG, Galley R, Asplin MG, De Abreu R, Warner K-A, Pu, et al. Perennial pack ice in the southern Beaufort Sea was not as it appeared in the summer of 2009. Geophys Res Lett. 2009;36:L24501. [Google Scholar]
- Barbeyron T, Lerat Y, Sassi J-F, Le Panse S, Helbert W, Nyvall Collen P.2010Persicivirga ulvanivorans sp. nov., a marine Flavobacteriaceae degrading ulvan from green algae Int J Syst Evol Microbiole-pub ahead of print 10 September 2010; doi: 10.1099/ijs.0.024489-0 [DOI] [PubMed]
- Boe J, Hall A, Qu X. September sea-ice cover in the Arctic Ocean projected to vanish by 2100. Nature Geosci. 2009;2:341–343. [Google Scholar]
- Bowman JP, McCammon SA, Brown MV, Nichols DS, McMeekin TA. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl Environ Microbiol. 1997;63:3068–3078. doi: 10.1128/aem.63.8.3068-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmeyer R, Knittel K, Jurgens J, Weyland H, Amann R, Helmke E. Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl Environ Microbiol. 2003;69:6610–6619. doi: 10.1128/AEM.69.11.6610-6619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MV, Bowman JP. A molecular phylogenetic survey of sea-ice microbial communities. FEMS Microbiol Ecol. 2001;35:267–275. doi: 10.1111/j.1574-6941.2001.tb00812.x. [DOI] [PubMed] [Google Scholar]
- Cavalieri DJ, Gloersen P, Parkinson CL, Comiso JC, Zwally HJ. Observed hemispheric asymmetry in global sea ice changes. Science. 1997;278:1104–1106. [Google Scholar]
- Chao A. Nonparametric estimation of the number of classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
- Chao A, Shen T-J. Nonparametric estimation of Shannon's index of diversity when there are unseen species in sample. Environ Ecolog Stat. 2003;10:429–443. [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nuc Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Rocap G, Deming JW. Persistence of bacterial and archaeal communities in sea ice through an Arctic winter. Environ Microbiol. 2010;12:1828–1841. doi: 10.1111/j.1462-2920.2010.02179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comiso JC, Parkinson CL, Gersten R, Stock L. Accelerated decline in the Arctic sea ice cover. Geophys Res Lett. 2008;35:L01703. [Google Scholar]
- Connelly TL, Tilburg CM, Yager PL. Evidence for psychrophiles outnumbering psychrotolerant marine bacteria in the springtime coastal Arctic. Limnol Ocean. 2006;51:1205–1210. [Google Scholar]
- Cox GFN, Weeks WF. Equations for determining the gas and brine volumes in sea-ice samples. J Glaciol. 1983;29:306–316. [Google Scholar]
- Deming JW.2007Life in ice formations at very low temperaturesIn: Gerday C and Glansdorff N (eds).Physiology and Biochemistry of Extremophiles ASM Press: Washington, DC; 135–145. [Google Scholar]
- Deming JW.2009Sea ice bacteria and virusesIn: Thomas DN and Dieckmann GS (eds).Sea ice—An Introduction to its Physics, Chemistry, Biology, and Geology2nd edn. Blackwell Science Ltd: Oxford; 247–282. [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicken H.2009Ice Sampling and Basic Sea Ice Core AnalysisIn: Eicken H, Gradinger R, Salganek M, Shirasawa K, Perovich D and Lepparanta M (eds).Field Techniques for Sea Ice Research University of Alaska Fairbanks Theatre Department: Fairbanks [Google Scholar]
- Eicken H, Lensu M, Leppäranta M, Tucker WB, III, Gow AJ, Salmela O. Thickness, structure, and properties of level summer multiyear ice in the Eurasian sector of the Arctic Ocean. J Geophys Res. 1995;100:22697–22710. [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. Ecology of the rare microbial biosphere of the Arctic Ocean. PNAS. 2009;106:22427–22432. doi: 10.1073/pnas.0908284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas C.2004Late-summer sea ice thickness variability in the Arctic transpolar drift 1991–2001 derived from ground-based electromagnetic sounding Geophys Res Lett 31L09402. [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungblut AD, Lovejoy C, Vincent WF. Global distribution of cyanobacterial ecotypes in the cold biosphere. ISME J. 2009;4:191–202. doi: 10.1038/ismej.2009.113. [DOI] [PubMed] [Google Scholar]
- Junge K, Imhoff F, Staley T, Deming W. Phylogenetic diversity of numerically important Arctic sea-ice bacteria cultured at subzero temperature. Microb Ecol. 2002;43:315–328. doi: 10.1007/s00248-001-1026-4. [DOI] [PubMed] [Google Scholar]
- Kellogg C, Carpenter S, Renfro A, Cochran J, Deming J.2010Evidence for microbial attenuation of particle flux in the Amundsen Gulf and Beaufort Sea: elevated activity on sinking aggregates Polar Biole-pub ahead print 5 may 2011.
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press: Cambridge; 1983. [Google Scholar]
- Kirchman DL, Cottrell MT, Lovejoy C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol. 2010;12:1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- Kirchman DL, Moran XAG, Ducklow H. Microbial growth in the polar oceans: role of temperature and potential impact of climate change. Nat Rev Micro. 2009;7:451–459. doi: 10.1038/nrmicro2115. [DOI] [PubMed] [Google Scholar]
- Kurilenko VV, Christen R, Zhukova NV, Kalinovskaya NI, Mikhailov VV, Crawford RJ, et al. Granulosicoccus coccoides sp. nov., isolated from leaves of seagrass (Zostera marina) Int J Syst Evol Microbiol. 2010;60:972–976. doi: 10.1099/ijs.0.013516-0. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsen FA, Kodner RB, Armbrust EV. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K, Abt B, Brambilla E, Lapidus A, Copeland A, Desphande S, et al. Complete genome sequence of Coraliomargarita akajimensis type strain (04OKA010-24T) SIGS. 2010;2:290–299. doi: 10.4056/sigs.952166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean AL. Bacteria of ice and snow in Antarctica. Nature. 1918;102:35–39. [Google Scholar]
- Nedashkovskaya OI, Kim SB, Han SK, Rhee MS, Lysenko AM, Falsen E, et al. Ulvibacter litoralis gen. nov., sp. nov., a novel member of the family Flavobacteriaceae isolated from the green alga Ulva fenestrata. Int J Syst Evol Microbiol. 2004;54:119–123. doi: 10.1099/ijs.0.02757-0. [DOI] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Nghiem SV, Rigor IG, Perovich DK, Clemente-Colón P, Weatherly JW, Neumann G. Rapid reduction of Arctic perennial sea ice. Geophys Res Lett. 2007;34:L19504. [Google Scholar]
- Petri R, Imholff JF. Genetic analysis of sea-ice bacterial communities of the western Baltic Sea using an improved double gradient method. Polar Biol. 2001;24:252–257. [Google Scholar]
- Pfirman SL, Colony R, Nürnberg D, Eicken H, Rigor I. Reconstructing the origin and trajectory of drifting Arctic sea ice. J Geophys Res. 1997;102:12575–12586. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Meth. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Rigor IG, Wallace JM.2004Variations in the age of Arctic sea-ice and summer sea-ice extent Geophys Res Lett 31L09401. [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson EH. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stroeve J, Holland MM, Meier W, Scambos T, Serreze M. Arctic sea ice decline: faster than forecast. Geophys Res Lett. 2007;34:L09501. [Google Scholar]
- Vincent WF. Microbial ecosystem responses to rapid climate change in the Arctic. ISME J. 2010;4:1087–1090. doi: 10.1038/ismej.2010.108. [DOI] [PubMed] [Google Scholar]
- Waleron M, Waleron K, Vincent WF, Wilmotte A. Allochthonous inputs of riverine picocyanobacteria to coastal waters in the Arctic Ocean. FEMS Microbiol Ecol. 2007;59:356–365. doi: 10.1111/j.1574-6941.2006.00236.x. [DOI] [PubMed] [Google Scholar]
- Yoon J, Yasumoto-Hirose M, Katsuta A, Sekiguchi H, Matsuda S, Kasai H, et al. Coraliomargarita akajimensis gen. nov., sp. nov., a novel member of the phylum ‘Verrucomicrobia' isolated from seawater in Japan. Int J Syst Evol Microbiol. 2007;57:959–963. doi: 10.1099/ijs.0.64755-0. [DOI] [PubMed] [Google Scholar]
- Yu Y, Lee C, Kim J, Hwang S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng. 2005;89:670–679. doi: 10.1002/bit.20347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.