Abstract
This study describes reconstruction of two highly unusual archaeal genomes by de novo metagenomic assembly of multiple, deeply sequenced libraries from surface waters of Lake Tyrrell (LT), a hypersaline lake in NW Victoria, Australia. Lineage-specific probes were designed using the assembled genomes to visualize these novel archaea, which were highly abundant in the 0.1–0.8 μm size fraction of lake water samples. Gene content and inferred metabolic capabilities were highly dissimilar to all previously identified hypersaline microbial species. Distinctive characteristics included unique amino acid composition, absence of Gvp gas vesicle proteins, atypical archaeal metabolic pathways and unusually small cell size (approximately 0.6 μm diameter). Multi-locus phylogenetic analyses demonstrated that these organisms belong to a new major euryarchaeal lineage, distantly related to halophilic archaea of class Halobacteria. Consistent with these findings, we propose creation of a new archaeal class, provisionally named ‘Nanohaloarchaea'. In addition to their high abundance in LT surface waters, we report the prevalence of Nanohaloarchaea in other hypersaline environments worldwide. The simultaneous discovery and genome sequencing of a novel yet ubiquitous lineage of uncultivated microorganisms demonstrates that even historically well-characterized environments can reveal unexpected diversity when analyzed by metagenomics, and advances our understanding of the ecology of hypersaline environments and the evolutionary history of the archaea.
Keywords: assembly, halophile, hypersaline, metagenome, Nanohaloarchaea
Introduction
Cultivation-independent molecular ecology techniques currently used to survey environmental microbiota include analysis of phylogenetic marker genes, targeted functional gene inventories and direct sequencing of DNA recovered from environmental samples (reviewed in Hugenholtz and Tyson, 2008; Wooley et al., 2010). Direct metagenomic sequencing is an appealing route for investigating microbial community composition because it provides simultaneous insight into phylogenetic composition and metabolic capabilities of uncultivated populations (Allen and Banfield, 2005; Wilmes et al., 2009). Gene fragments from individual sequencing reads and small assembled contigs can be annotated and assigned to approximate phylogenetic bins based on comparison with databases of known reference genomes (Mavromatis et al., 2007). However, cultivation biases limit the phylogenetic and physiological breadth of available reference genomes (Wu et al., 2009). Single cell genomics can potentially broaden genomic databases, but often provides highly fragmented data because of amplification biases (Lasken, 2007; Woyke et al., 2009). As a result of skewed genomic representations in reference data sets, metagenome analysis methods that rely on previously described sequence examples (for example, fragment recruitment approaches) share an inherent potential bias against novel findings. This anti-novelty bias can be overcome by de novo sequence assembly, which does not rely on external reference sequences, and can facilitate resolution of phylogeny-to-function linkages for individual community members. Yet de novo sequence assembly techniques are rarely applied to metagenomic sequences because of sampling deficiencies and/or computational challenges (Allen and Banfield, 2005; Baker et al., 2010).
Habitats characterized by low diversity microbial communities have proven useful for validating molecular (eco-)systems biology approaches to examine the genetic and functional organization of native microbial consortia (Tyson et al., 2004; Allen and Banfield, 2005; Ram et al., 2005; Lo et al., 2007; Raes and Bork, 2008; Wilmes et al., 2009). High salt-impacted habitats are distributed globally in the form of hypersaline lakes, salt ponds and solar (marine) salterns, where evaporative processes result in salt concentrations close to and exceeding saturation. These environments contain microbial communities of intermediate complexity (Oren, 2008), providing excellent model systems for developing scalable analytical techniques applicable to environments with greater species richness and evenness.
The biochemical and physiological challenges faced by extremely halophilic organisms have resulted in unique adaptations to maintain osmotic balance, overcome reduced water activity because of the hygroscopic effects of saturating salt concentrations, and deter DNA damage induced by intense solar irradiation (Bolhuis et al., 2006; Hallsworth et al., 2007). The most extreme halophiles maintain osmotic balance using a ‘high salt-in' strategy, which allows intracellular salt concentrations to reach levels approximately isosmotic with the external environment (Oren, 2008). Microorganisms using the salt-in strategy not only endure extreme ionic strength, they require it for growth. Although salt-in adaptation can be energetically more favorable than transporting salt out and the accumulation of compatible solutes (Oren, 1999), it requires significant modifications to the intracellular machinery, including specialized protein amino acid compositions to maintain solubility, structural flexibility, and water availability necessary for enzyme function (Fukuchi et al., 2003; Bolhuis et al., 2008; Paul et al., 2008; Rhodes et al., 2010).
The study of microbial populations in extreme hypersaline environments is well established; the first cultivated halophilic microorganism appeared in Bergey's manual over a century ago (Oren, 2002a). Despite the extreme conditions in salt-saturated habitats, microbial cell densities often exceed 107 cells ml–1 (Oren, 2002b). Although salt-adapted organisms derive from all three domains of life, most extreme hypersaline habitats are dominated by halophilic archaea belonging to the monophyletic class Halobacteria (phylum Euryarchaeota), including members of the genera Haloquadratum, Halobacterium, Halorubrum and Haloarcula (Oren, 2008). Pure isolates of halophilic archaea currently include >96 species distributed among 27 genera, with genome sequence information available for more than a dozen species (Oren et al., 2009). Numerous cultivation-independent biodiversity surveys have been performed in hypersaline environments using PCR amplification of archaeal and bacterial 16S ribosomal RNA (rRNA) genes, as well as direct metagenomic sequencing of community DNA (Grant et al., 1999; Benlloch et al., 2001; Ochsenreiter et al., 2002; Burns et al., 2004; Demergasso et al., 2004; Jiang et al., 2006; Maturrano et al., 2006; Mutlu et al., 2008; Pagaling et al., 2009; Sabet et al., 2009; Oh et al., 2010; Rodriguez-Brito et al., 2010). These studies confirm high abundance of a few dominant species with widespread geographical distribution, but the intermittent recovery of atypical, unconfirmed sequence fragments hints at additional, unrecognized diversity among halophilic archaea (Grant et al., 1999; Lopez-Garcia et al., 2001; Pagaling et al., 2009; Oh et al., 2010; Sime-Ngando et al., 2010).
The lure of uncovering biological novelty is a major incentive driving metagenomic investigations in many habitats worldwide. This study demonstrates that even historically well-characterized habitats like extreme hypersaline lakes and solar salterns can reveal unexpected genes, metabolic features and entire lineages overlooked previously. The ‘assembly-driven' community metagenomic approach applied in the current study has led to the discovery and reconstruction of near-complete genomes for two new archaeal genera representing the first members of a previously undescribed taxonomic class of halophilic archaea. We demonstrate that members of this new archaeal class are present in high abundance and broadly distributed in other hypersaline habitats worldwide.
Materials and methods
Sample collection
Surface water samples (0.3 m depth) were collected from Lake Tyrrell (LT), Victoria, Australia and a high salinity crystallizer pond at South Bay Salt Works, Chula Vista (CV) California. Detailed locations, sampling dates, and physical characteristics of the collection sites are provided in Supplementary Figure S1.
Water samples of 20l each were passed through a 20 μm Nytex prefilter, followed by sequential filtration through a series of polyethersulfone, 142 mm diameter membrane filters (Pall Corporation, Port Washington, NY, USA) of decreasing porosities (3 μm>0.8 μm>0.1 μm) using a peristaltic pump. After each stage of filtration, filters were frozen for future DNA extraction, 16S rRNA gene analysis and metagenomic sequencing. Aliquots of filtered water were fixed with formaldehyde (7% final concentration) overnight at 4 °C. Fixed water samples were collected on 0.2 μm polycarbonate GTTP filters (Millipore, Billerica, MA, USA) for fluorescence in situ hybridization (FISH) and direct count microscopy.
Library construction and assembly
Genomic DNA was extracted from individual, bar-coded 0.8 and 0.1 μm filters. Filter-specific DNA libraries were constructed with insert sizes of 8–10 kbp and/or 40 kb (fosmids) at the J Craig Venter Institute, as described previously (Goldberg et al., 2006). Details of genomic DNA sequence libraries are provided in Supplementary Table S1.
16S rRNA gene clone libraries were constructed by amplification of LT metagenomic DNA using universal archaeal primer sequences Arc21F and Arc529R (Table 1), as previously described (Bik et al., 2010). A group-specific primer for Nanohaloarchaea (LT_1215R) was designed using the NCBI primer design tool, and used together with universal archaeal primer Arc21F to amplify both LT and CV community DNA. Amplification products were cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA) and sequenced bi-directionally with M13F and M13R primers.
Table 1. Primers and probes for detecting 16S rRNA sequences.
Sanger and pyrosequencing read libraries were assembled both individually and in various combinations, using Celera Assembler software version 5.4 (Myers et al., 2000), in a series of iterative assemblies guided by phylogenetic binning. Detailed genome assembly procedures are provided in Supplementary Information.
Genome annotation
J07AB43 and J07AB56 draft genomes were annotated using the Integrated Microbial Genome Expert Review service of the Joint Genome Institute (Markowitz et al., 2009b). Genome completeness was estimated for the J07AB56 and J07AB43 scaffold groups by comparing genes involved in transcription, translation and replication to those identified as highly conserved in previously sequenced archaeal genomes (Ciccarelli et al., 2006; Wu and Eisen, 2008; Puigbo et al., 2009). Orthologs shared between the J07AB43 and J07AB56 proteomes were detected using the reciprocal smallest distance algorithm (threshold e-value=1e-05; sequence divergence=0.4) (Wall and Deluca, 2007).
Amino acid composition analysis
Amino acid frequencies in predicted proteins from J07AB56, J07AB43 and 1455 archaeal and bacterial genomes were compared using the Primer 6 software program (Clarke and Gorley, 2006) to perform Non-Metric Multidimensional Scaling (NM-MDS) analysis (Ramette, 2007). For each genome, the frequency of each amino acid for all predicted proteins was calculated using a custom perl script. These values were standardized by Z-score, then used to calculate a Euclidean distance similarity matrix. NM-MDS analysis was performed using default program parameters (25 random starts, Krustal fit scheme of 1 and a minimum stress value of 0.01). In addition to NM-MDS analysis, a cluster analysis was performed to define groups within the NM-MDS plot using a multidimensional distance parameter of 4%.
Phylogenetic analysis
16S rRNA sequences and ribosomal proteins from euryarchaeal genomes in the JGI-IMG database (Markowitz et al., 2009a) and GenBank were compared with metagenomic gene sequences obtained by (i) extraction from assembled scaffolds and (ii) amplification and sequencing of 16S rRNA genes from LT and CV clone libraries. Maximum likelihood trees were constructed using TreeFinder v.10.08 (Jobb et al., 2004) and PhyML v.3.0 (Guindon and Gascuel, 2003). The robustness of each maximum likelihood tree was estimated using non-parametric bootstrap analysis. Details of alignment curation and tree construction are provided in Supplementary Information.
Predicted proteins in assembled genomes were evaluated for phylogenetic relatedness to known sequences in NCBI GenBank nr using the DarkHorse program, version 1.3, with a threshold filter setting of 0.05 (Podell and Gaasterland, 2007; Podell et al., 2008). Minimum quality criteria for match inclusion in the DarkHorse analysis were that BLASTP alignments to GenBank nr sequences cover at least 70% of total query length and have e-value scores of 1e-5 or better.
Fluorescence in situ hybridization
Fluorophore-conjugated custom 16S rRNA probes (Table 1) were designed using ARB (Ludwig et al., 2004), screened for specificity in silico using ProbeCheck (Loy et al., 2008) and synthesized by Integrated DNA Technologies Inc. (San Diego, CA, USA). FISH was performed on CV and LT water samples collected on 0.2 μm polycarbonate GTTP filters (Millipore) at every stage of filtration (post 20 μm, post 3 μm and post 0.8 μm). The Nanohaloarchaea-specific probe Narc_1214 conjugated with Cy3 along with unlabeled helper probes LT_1198h1, LT_976h2 and LT_127h3 (Fuchs et al., 2000) were used for FISH analysis. Universal probes Arc915 (archaeal) and EubMix (a bacterial probe consisting of an equimolar mixture of Eub338 and Eub338plus) were also used for the purpose of cell counts. Hybridization conditions were optimized at 46 °C for 2 h, as previously described (Pernthaler et al., 2001). Filters were mounted with Vectashield medium (Vector Laboratories, Burlingame, CA, USA), and imaged at 1000 × with a Nikon Eclipse TE-2000U inverted microscope (Nikon Instruments Inc., Irvine, CA, USA). Cell counts were performed on multiple fields per slide, normalizing 16S rRNA-specific probe counts to total number of cells stained with the DNA-binding dye 4′,6-diamidino-2-phenylindole.
Accession numbers
16S rRNA gene sequences have been deposited to DDBJ/EMBL/GenBank under accession numbers HQ197754 to HQ197794. Assembled genomes with annotations have been deposited as Whole Genome Shotgun projects under accession numbers AEIY01000000 (J07AB43) and AEIX01000000 (J07AB56).
Results
Metagenomic assembly
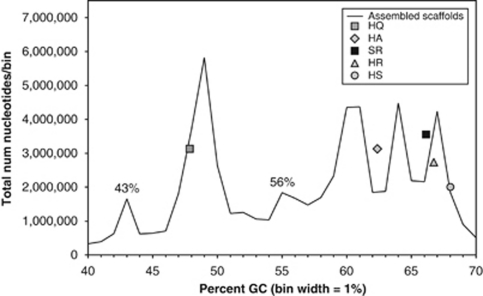
Seven independent DNA sequencing libraries were constructed from size-fractionated surface water samples collected at LT, Australia (Supplementary Figure S1 and Supplementary Table S1). Initial assembly of the combined 632 903 Sanger sequencing reads produced 15 008 scaffolds (maximum length=2 764 168 bp; scaffold N50=29 346 bp). These scaffolds included at least six different relatively abundant microbial populations, each with a distinct nucleotide percent G+C composition. A length-weighted histogram of percent G+C versus total assembled scaffold nucleotides showed peaks corresponding to these populations (Figure 1). The largest peak in this histogram, at 48% G+C, included scaffolds containing 16S rRNA sequences from multiple strains of Haloquadratum walsbyi, consistent with previous observations noting the dominance of this species in similar hypersaline environments (Cuadros-Orellana et al., 2007; Oh et al., 2010). Three additional peaks at 60% G+C or higher included scaffolds containing 16S rRNA genes with 89–99% identity to clone sequences annotated as uncharacterized halophilic archaea (class Halobacteria). Microbial populations associated with these peaks are currently under investigation, but fall outside the scope of the present report.
Figure 1.
Length-weighted histogram of percent G+C for all scaffolds assembled from the LT community, binned in 1% GC increments. Symbols represent reference control points, indicating where five previously sequenced halophile genomes would have fallen, if they had been present in this data set. Data points are plotted based on total number of nucleotides in each scaffold (y axis) versus average percent GC for the entire scaffold (x axis). HA, Haloarcula marismortui; HQ, Haloquadratum walsbyi; HR, Halorubrum lacusprofundi; HS, Halobacterium salinarum R1; SR, Salinibacter ruber. Peaks labeled at 43% and 56% GC are the focus of this study.
Two groups of scaffolds, with peaks at 43% and 56% G+C, shared an intriguing pattern of unusual characteristics. In addition to distinctive G+C content, >90% of the reads that co-assembled in these scaffolds were obtained from microorganisms that had passed through a 0.8 μm filter, but were retained on a 0.1 μm filter, suggesting small cell size. The 16S rRNA gene sequences contained in these scaffolds were <78% identical to any previously known cultured isolate, although they did resemble 16S rRNA gene fragments periodically recovered in culture-independent surveys of microbial diversity in hypersaline waters (Grant et al., 1999; Oh et al., 2010; Sime-Ngando et al., 2010).
To optimize assembly efficiency for these unusual populations, the full set of metagenomic reads were subjected to a series of iterative assemblies guided by phylogenetic binning. The 43% G+C peak was thereby consolidated into seven major scaffolds (J07AB43) and the 56% G+C peak into three major scaffolds (J07AB56) (Supplementary Table S2). The J07AB43 and J07AB56 scaffold groups were subsequently analyzed as draft genomes, each representing the consensus sequence of an individual microbial population. Overall properties of these draft genomes are summarized in Table 2. These properties differ substantially from previously sequenced extreme halophiles in both nucleotide composition, expressed as percent G+C, and total genome size (Markowitz et al., 2009a). With the exception of H. walsbyi, at 48% G+C, all other previously described halophilic archaea, as well as the halophilic bacterium Salinibacter ruber, have nucleotide compositions of 60% or greater G+C, compared with 43% and 56% for these new organisms. Estimated total genome size and predicted number of coding sequences for J07AB43 and J07AB56 (Table 2) were also considerably smaller than other known extreme halophiles, which currently range from 2.7 to 5.4 Mbp.
Table 2. General features of the J07AB43 and J07AB56 draft genomes.
| J07AB43 | J07AB56 | |
|---|---|---|
| Genome size, bp | 1 227 157 | 1 215 802 |
| G+C percentage | 44% | 56% |
| Scaffold number | 7 | 3 |
| rRNA operons | 1 | 1 |
| tRNAs | 59 | 38 |
| Predicted CDSs | 1678 | 1411 |
| CDSs w/func. Pred. | 773 | 719 |
Abbreviations: CDS, coding sequence; rRNA, ribosomal RNA; tRNA, transfer RNA.
Genome completeness
To estimate the extent of genome completeness of J07AB43 and J07AB56, functional annotations for all predicted proteins were searched against a set of 53 housekeeping genes, previously identified as universally present in all archaeal genomes sequenced as of 2009 (Puigbo et al., 2009). These highly conserved genes are physically dispersed throughout the genome (non-clustered) and include ribosomal proteins, amino acid tRNA synthetases, translation initiation and elongation factors, molecular chaperones and proteins essential for DNA replication and repair. All 53 of the universal archaeal housekeeping proteins were identified in J07AB56 while 44/53 (83%) were found in the J07AB43 draft genome (Supplementary Table S3). The presence of these core proteins, a single rRNA operon and transfer RNAs enabling translation of all 20 amino acids, suggests that both draft genomes are nearly complete.
Community abundance
Community abundance of J07AB43 and J07AB56 was initially assessed by sequencing 16S rRNA gene clone libraries, constructed by amplifying LT community DNA with universal archaeal primers Arc21F and Arc529R (Table 1). Amplified sequences with >91% identity to the J07AB43 and J07AB56 draft genomes were found in 124/315 (39%) of archaeal clones obtained from organisms retained on 0.1 μm pore filters, but only 24/254 (9%) of clones retained on 0.8 μm pore filters. These results are consistent with the observed enrichment of J07AB43 and J07AB56 reads specifically derived from 0.1 μm filter fractions in the assembled data set.
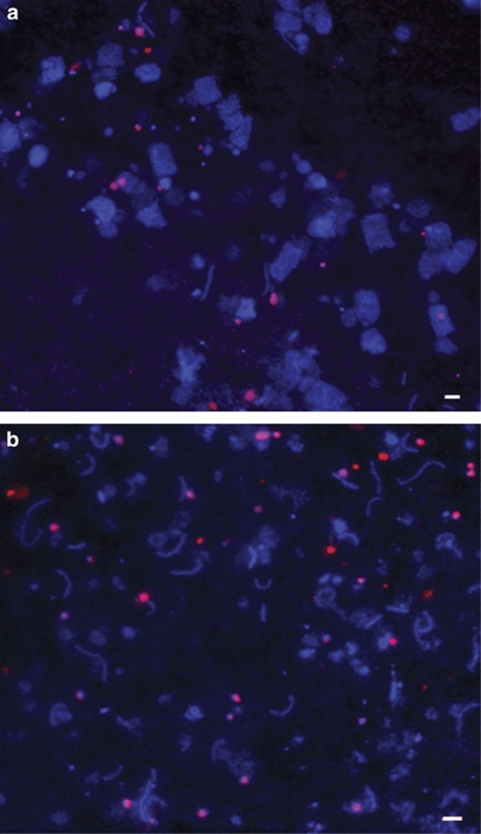
As a second, independent test of community abundance, new lineage-specific 16S rRNA probes were designed to visualize J07AB43 and J07AB56 cells in environmental samples by FISH (Table 1). These probes were used in combination with the DNA-binding dye 4′,6-diamidino-2-phenylindole and universal bacterial and archaeal probes to obtain direct cell counts in LT and CV water samples (Figure 2). Cells approximately 0.6 μm in diameter were labeled with lineage-specific probe NArc_1214 in samples from both locations. These results are consistent with size estimates of <0.8 μm but >0.1 μm based on filter-specific composition for both amplified 16S rRNA clones and metagenomic reads. Direct counts of fluorescently labeled cells indicated that the combined abundance of strains matching the new, lineage-specific probes was approximately 14% of all 4′,6-diamidino-2-phenylindole-labeled cells in water samples from LT, and 8–11% in samples from CV (Supplementary Table S4).
Figure 2.
FISH micrographs. (a) LT (0.1 to 3 μm filter fraction), (b) CV South Bay Salt Works (0.1 to 0.8 μm filter fraction). All cells are stained with 4′,6-diamidino-2-phenylindole (blue). Nanohaloarchaea cells shown are stained with lineage-specific Cy3 probe Narc_1214 (red). Scale bar=2 μm.
Community abundance of the organisms responsible for the J07AB43 and J07AB56 draft genomes was further examined using statistical properties of the assembled metagenomic sequence data. The number of reads that co-assembled to create each composite population scaffold group was divided by the total number of reads available and normalized for estimated genome size. Assuming the two new genomes are approximately 1.2 Mbp each, and other microbial species sampled from LT have an average genome size of 3 Mbp, J07AB43 was estimated to represent at least 6.7% of the LT sampled community (17 066 reads) and J07AB56 at least 3.4% (8652 reads), totaling approximately 10% for the two populations combined (3.0/1.2*25 718/632 903). Calculations based on metagenomic assembly most likely underestimate true population abundance, because they may exclude closely related polymorphic strains containing DNA sequence variations that were not incorporated into the consensus population assembly.
Taxonomic position of J07AB43 and J07AB56
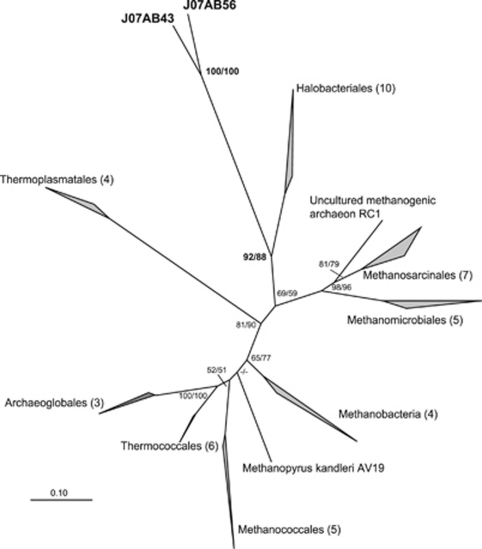
J07AB43 and J07AB56 16S rRNA shared sequence identities of 68% to 75% with previously sequenced, cultured representatives of class Halobacteria (Supplementary Table S5). An unrooted maximum likelihood phylogenetic tree of euryarchaeotal 16S rRNA gene sequences placed J07AB43 and J07AB56 as a deep sister group of class Halobacteria (Figure 3), with significant bootstrap support.
Figure 3.
Unrooted maximum-likelihood 16S rRNA gene phylogenetic tree of the Euryarchaeota. Tree is based on 48 sequences, 1275 positions. Numbers of sequences in each collapsed node are indicated in parentheses. Numbers at nodes represent bootstrap values inferred by TreeFinder/PhyML. Bootstrap values <50% are indicated by a ‘–' sign. Scale bar represents 0.1 substitutions per site. A full, uncollapsed version of this tree is presented in Supplementary Figure S2a.
Concatenated ribosomal protein data sets have been shown to be particularly useful for resolving deep evolutionary relationships (Brochier et al., 2002; Matte-Tailliez et al., 2002; Rokas et al., 2003; Rannala and Yang, 2008). Phylogenetic analysis of 57 ribosomal proteins from the J07AB43 and J07AB56 draft genomes showed, like the 16S rRNA tree, robust placement of these genomes as a deeply branching sister group of class Halobacteria, with bootstrap values of 98% (PhyML) and 74% (TreeFinder). This relationship was corroborated using Dayhoff04 recoding of ribosomal protein alignments (Hrdy et al., 2004; Susko and Roger, 2007), to rule out possible artifacts of biased amino acid composition or fast-evolving lineages (Supplementary Figure S2b). The long branch lengths separating J07AB43 and J07AB56 from members of class Halobacteria indicate that these two sister-lineages are only distantly related, consistent with the average divergence of 35% observed between Halobacteria and J07AB43 and J07AB56 16S rRNA gene sequences (Supplementary Table S5). By contrast, 16S rRNA variability within the Halobacteria is <16%.
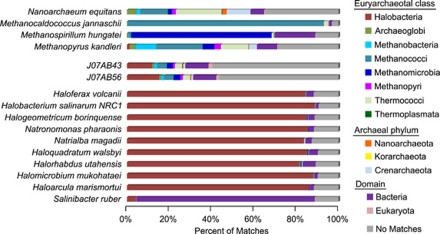
Nearly 60% of predicted proteins in J07AB43 and J07AB56 had no GenBank database matches close enough to enable confident phylogenetic assignment. Of those that could be assigned, fewer than 20% matched proteins from members of class Halobacteria (Figure 4). In contrast, >80% of predicted proteins in the genomes of previously sequenced Halobacteria had closest non-self matches to other members of their own class, leaving fewer than 20% unmatched. Similar patterns of protein sequence conservation were observed in organisms with many sequenced database relatives, including Methanocaldococcus janaschii, Methanospirillum hungatei and Salinibacter ruber, but not in sparsely sampled species that are only distantly related to other known lineages, such as Nanoarchaeum equitans and Methanopyrus kandleri (Branciamore et al., 2008).
Figure 4.
Phylogenetic distribution of non-self-protein BLAST matches for euryarchaeotal genomes. Searches against the GenBank nr database were classified by euryarchaeotal class, archaeal phylum, domain or no match using the DarkHorse algorithm, as described in Materials and methods section.
Genome characteristics of J07AB43 and J07AB56
Although the J07AB43 and J07AB56 genomes are more closely related to each other than to any previously sequenced organisms, gene content analysis identified only 480 (30%) shared protein ortholog pairs between them. Of these, 143 (approximately 10% of each genome) were not found in other halophilic archaea. The majority of these shared lineage-specific sequences were too dissimilar to previously characterized proteins to assign a functional annotation. The remainder was dominated by housekeeping proteins involved in translation and ribosomal structure. Each genome included only one rhodopsin-like gene, compared with multiple paralogs present in the genomes of other extreme halophilic archaea (Ihara et al., 1999), and the extremely halophilic bacterium Salinibacter ruber (Mongodin et al., 2005). Notably absent from both genomes were homologs to the highly conserved Gvp family of gas vesicle proteins found in most halophilic archaea, Cyanobacteria and purple photosynthetic bacteria (Walsby, 1994).
Both J07AB43 and J07AB56 have highly unusual amino acid compositions compared with previously sequenced archaeal and bacterial genomes. These unusual compositions appear to support a ‘salt-in' strategy of maintaining osmotic balance, as evidenced by the over-representation of amino acids with negatively charged side chains (aspartic and glutamic acid) and the under-representation of residues with bulky hydrophobic side chains (tryptophan, phenylalanine and isoleucine), to enhance protein structural flexibility and solubility under intracellular conditions of high ionic strength and low water availability. Although a similar salt-in strategy is employed by other extreme halophiles, J07AB43 and J07AB56 use their own, distinct combination of amino acids to achieve this end, preferring glutamic to aspartic acid, serine to threonine, and reduced frequencies of alanine, proline and histidine (Supplementary Table S6). The large number of proteins annotated with ‘hypothetical' functions in the J07AB43 and J07AB56 genomes may be at least partially because of their unusual amino acid compositions, which can hinder recognition of database homologs in sequence-based similarity searches.
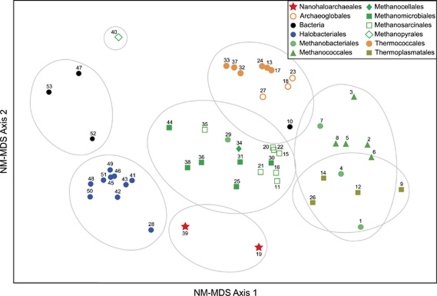
The peculiar amino acid compositions of J07AB43 and J07AB56 compared with other halophilic archaea are highlighted in a NM-MDS plot of intergenomic distances based on frequencies for all 20 standard amino acids (Figure 5). The data used to construct this matrix included all protein sequences from euryarchaeal genomes used to build the phylogenetic tree in Figure 3, supplemented with four bacterial species found in high salt environments: Salinibacter ruber (Bacteroidetes), Halorhodospira halophila (Gammaproteobacteria), Chromohalobacter salexigens (Gammaproteobacteria) and Halothermothrix orenii (Firmicutes).
Figure 5.
NM-MDS comparison of amino acid compositions. Euryarchaeal genomes were supplemented with four halophilic bacteria genomes. Symbols denote taxonomic classifications. Numbers rank genomes in increasing order of G+C content (1–10: 29–38%, 11–20: 38–43%, 21–30: 43–50%, 31–40: 50–60%, 41–53: 60–67%). Grey circles indicate hierarchical clustering, based on a 4% distance setting to define groups. A complete list of these genomes and their amino acid compositions is presented in Supplementary Table S6.
Although genome percent G+C compositions were not explicitly included as one of the factors in this analysis, there is a trend for microorganisms with lower G+C (denoted with lower label numbers in Figure 5) to be located further to the right along the horizontal axis. This trend is consistent with the known influence of G+C composition on usage frequency for some amino acids because of codon bias (Liu et al., 2010). In contrast, position along the vertical axis of Figure 5 was unrelated to percent G+C. Instead, amino acid composition differences captured along this axis appear to correlate more closely with common ancestry and/or shared environmental adaptations. The outlier positions of J07AB43 (#19) and J07AB56 (#39) along the vertical axis of Figure 5 clearly demonstrate their unusual amino acid compositions relative to other archaea. Similar outlier positions were observed for these two genomes when analyzed in the context of a much larger microbial genomic data set, including 1382 bacterial and 73 archaeal species (data not shown).
Inferred metabolic capabilities of the J07AB43 and J07AB56 genomes are consistent with a predominately aerobic, heterotrophic lifestyle. The absence of identifiable anaerobic terminal reductases suggests they are incapable of anaerobic respiration although the presence of lactate dehydrogenases suggests possible fermentative metabolism under microaerophilic conditions. Both genomes contain enzymes necessary to support glycolysis, as well as operons encoding key enzymes for glycogen synthesis and catabolism. Several of these enzymes, including a glycogen debranching enzyme and predicted alpha-1,6-glucosidase activity, are not present in any other known members of class Halobacteria. However, these enzyme activities are frequently found in archaea from classes Methanococci and Thermoplasmata that utilize starch as an internal storage molecule (König et al., 1985, 1982). This suggests a possible common ancestral origin, with subsequent gene loss in the Halobacteria lineage.
In addition to the Embden–Meyerhoff pathway, genes supporting the entire pentose phosphate pathway were observed in both genomes, including both oxidative and non-oxidative branches. The presence of a complete pentose phosphate pathway has not been demonstrated previously in any other archaea, by either biochemical or bioinformatic methods (Verhees et al., 2003). The key, rate-limiting enzyme for this pathway is glucose-6-phosphate dehydrogenase, which converts glucose-6-phosphate into 6-phosphoglucono-δ-lactone. Although both J07AB43 and J07AB56 appear to have complete genomic copies of this gene, the closest database relatives to their sequences are all bacterial, suggesting this functionality may have been acquired by ancient horizontal gene transfer. The nearest homolog of the glucose-6-phosphate dehydrogenases in J07AB43 and J07AB56 is from the genome of Salinibacter ruber, a common bacterial inhabitant of hypersaline environments believed to have experienced frequent horizontal gene exchange with archaea (Mongodin et al., 2005).
Geographical distribution and diversity
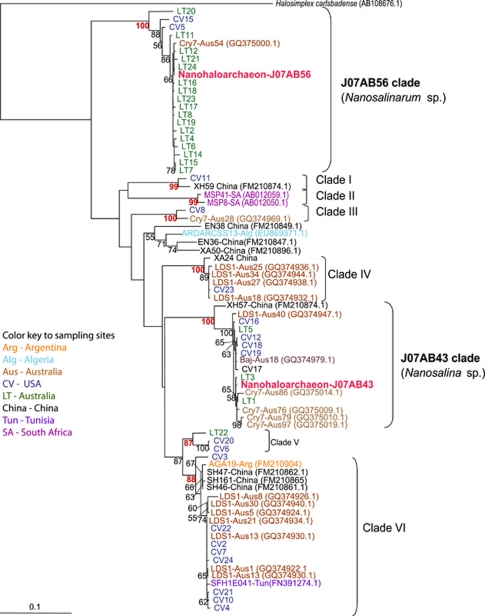
Lineage-specific PCR primer, LT_1215R (Table 1) and general archaeal primer Arc21F were used to construct clone libraries from environmental DNA samples collected from both LT and CV, yielding 43 new 16S rRNA gene sequences. Additional 16S rRNA gene sequences, with >85% identity to J07AB43 and J07AB56, were identified in public databases. These published sequences originated in environmental samples from Africa, Asia and South America, as well as Australia and North America (Supplementary Table S7). The phylogenetic analysis of these 16S rRNA gene sequences reveals at least eight distinct clades with strong bootstrap support (bootstrap values >87%, Figure 6). Based on degree of sequence divergence, each clade most likely represents a new genus or higher taxonomic level. Classification of J07AB43 and J07AB56 into separate genera is strongly supported by tree topology, 16% sequence divergence in the 16S rRNA gene (Supplementary Table S5) and a 13% difference in genomic G+C content.
Figure 6.
16S rRNA gene maximum likelihood tree of Nanohaloarchaea sequences recovered from worldwide hypersaline habitats. Tree is based on 709 nucleic acid positions in 77 sequences. Numbers at nodes represent bootstrap values (values <50% not shown). Scale bar shows average number of substitutions per site.
Discussion
This study has demonstrated that re-examination of a fairly simple, well studied environmental habitat using a combination of strategic environmental sampling, deep sequencing, and de novo metagenomic assembly can reveal significant new information. We have discovered and characterized nearly complete genomes representing a novel archaeal lineage prevalent in hypersaline systems worldwide, yet very different from all previously described members of class Halobacteria.
We propose the creation of a new class ‘Nanohaloarchaea' within phylum Euryarchaeota to accommodate this new lineage. We further propose partitioning class Nanohaloarchaea to place J07AB43 and J07AB56 into distinct genera, Candidatus ‘Nanosalina sp. J07AB43' and ‘Candidatus Nanosalinarum sp. J07AB56'. Evidence supporting these proposals includes: (i) comprehensive euryarchaeotal phylogenetic analyses based on 16S rRNA genes and ribosomal proteins; (ii) lineage-specific features, including numerous genes without previously described close relatives; and (iii) significant intra-lineage diversity and abundance within geographically distinct hypersaline habitats worldwide. Evolutionary distinctness of J07AB43 and J07AB56 from other halophilic archaea is reinforced by taxonomic patterns of BLASTP matches for their predicted proteomes against GenBank nr, as well as amino acid composition-based clustering. The sister-grouping of class Halobacteria and class Nanohaloarchaea reflects probable derivation from an ancient common halophilic ancestor with a ‘high salt-in' osmotic regulation strategy, followed by subsequent divergence along separate evolutionary paths.
Lineage-specific characteristics that distinguish ‘Candidatus Nanosalina sp.' and ‘Candidatus Nanosalinarum sp.' from most other extreme halophiles include their small physical size, compact genomes, single-copy rRNA operon, low G+C composition, unique proteome amino acid composition, absence of conserved gas vesicle genes and atypical predicted pathways associated with carbohydrate metabolism. Small compact genomes, as well as single-copy rRNA operons, have been proposed to minimize metabolic costs in habitats where neither broad metabolic repertoire nor high numbers of paralogous proteins are needed to accommodate rapid growth under fluctuating environmental conditions (Klappenbach et al., 2000). Small cell size, which increases surface to volume ratio, could be an adaptation for optimizing nutrient uptake capacity. Alternatively it is possible that small physical size allows Nanohaloarchaea to remain suspended in oxygenated surface waters to support aerobic metabolism, thus eliminating the need for gas vesicles to provide buoyancy.
The low G+C compositions of the two Nanohaloarchaea genomes, especially J07AB43 (43%), are surprising considering their prevalence in high light habitats. In the absence of compensatory mechanisms, lower G+C would be expected to increase susceptibility to ultraviolet-induced DNA damage. One possible explanation is that the low G+C composition of J07AB43 is related to ecological lifestyle. Low G+C composition and genomic streamlining have been associated with decreased nitrogen requirements and a slow-growing, energy-conservative lifestyle in marine bacteria (Giovannoni et al., 2005). However, the habitats from which these Nanohaloarchaea were isolated are not generally considered to be nutrient-limited (Oren, 2002b). Alternatively, it has been proposed that the low G+C composition of H. walsbyi (48%) compared with other halophiles is a specific adaptation to counteract the over-stabilizing effect of high magnesium concentrations on DNA structure (Bolhuis et al., 2006). If extremely high environmental magnesium cannot be adequately excluded from the cell, lower genomic G+C helps maintain DNA structural flexibility and avoids difficulties in strand separation caused by elevated melting temperatures. These same principles could apply to J07AB43, providing a possible selective advantage under high magnesium conditions expected in evaporative high salt environments.
Nanohaloarchaea are estimated to represent at least 10–25% of the total archaeal community in surface water samples from LT, Australia and CV, California, USA. We believe these values are robust, based on agreement of three independent analysis techniques: amplification of environmental 16S rRNA gene sequences; statistical analysis of metagenomic sequencing reads assembled into near-complete draft genomes; and quantitative FISH of cells from natural water samples labeled with lineage-specific probes. Microscopic counts reveal that Nanohaloarchaea are present at cell concentrations exceeding 106 cells ml–1 in hypersaline habitats of Australia and North America. The sporadic identification of Nanohaloarchaea in other surveys of hypersaline communities worldwide suggests that Nanohaloarchaea represent a significant yet neglected fraction of the biomass and diversity in these habitats.
The inability of earlier studies to recognize the significant contribution of Nanohaloarchaea to hypersaline community composition is likely due to limitations of the tools routinely used to assess environmental microbial diversity, including laboratory culture, microscopy, amplification of 16S rRNA gene fragments, and sequence database similarity searches for unassembled metagenomic reads. The isolation of cultured strains from environmental habitats is known to exclude many organisms that are highly successful in their native habitats. It is therefore not surprising the 96 hypersaline archaeal isolates described to date do not include any Nanohaloarchaea. Repeated efforts to culture these microorganisms in our own laboratory have also been unsuccessful. Furthermore, cultivation-independent microbial diversity studies based on 16S rRNA gene amplification are known to suffer from primer bias (Sipos et al., 2007). Mismatches between Nanohaloarchaea and many commonly used universal primers may have impeded detection in earlier studies. Primers likely to have been particularly problematic are highlighted in Table 1 (Amann et al., 1990, 1995; Lane, 1991; DeLong, 1992; DasSarma and Fleischman, 1995; Ihara et al., 1997; Brunk and Eis, 1998; Daims et al., 1999; Grant et al., 1999; Baker et al., 2003; Raes and Bork, 2008). The exceptionally small size of Nanohaloarchaea compared with other halophilic microorganisms makes them difficult to visualize by microscopy in the absence of selective enrichment techniques or group-specific probes, and can prevent recovery during sample concentration procedures targeting larger microorganisms or smaller viruses (Rodriguez-Brito et al., 2010). Similar issues have been noted for other nano-sized archaea, identified solely by 16S rRNA gene sequencing (Casanueva et al., 2008; Gareeb and Setani, 2009).
The presence of ultrasmall, uncultivated novel archaeal lineages in natural environments may be a common occurrence. Nanohaloarchaea represent the third nano-sized archaeal lineage to be described. However, unlike the thermophilic Nanoarchaeum equitans (Huber et al., 2002) or the acidophilic ARMAN lineages (Baker et al., 2006, 2010), members of the Nanohaloarchaea appear to be free-living based on microscopic observations. The larger genomes of Nanohaloarchaea (approximately 1.2 Mbp) relative to other symbiotic/parasitic nano-sized archaea (ARMAN, <1 Mbp; Nanoarchaeum equitans, <0.5 Mbp) are consistent with a possible non-host associated lifestyle for this group. It is interesting to contemplate the environmental pressures selecting for the evolution of ultrasmall microorganisms with small genomes, and to consider the extent of an ultrasmall microbial biosphere. The realization that ultrasmall populations can comprise a significant fraction of the total microbial community, yet have eluded previous detection, suggests that historical estimates of microbial biomass and numerical abundance in natural environments may be substantially underestimated. This is particularly relevant in non-extreme habitats where the existence of ultrasmall microbial populations have not yet been described or investigated.
Routine metagenomic analysis methods currently rely on the expectation that undiscovered microorganisms will have a certain degree of similarity to those already known, creating a potential bias against novel discoveries. Although this study exposes limitations of commonly used microbial diversity assessment tools in the context of detecting novel archaea in hypersaline lakes, these limitations apply even more emphatically to other more complex microbial communities, which often contain elaborate mixed consortia of Bacteria, Archaea, Eukarya and viruses. This study reinforces the utility of community genomics and de novo sequence assembly as important methods for the detection and analysis of biological diversity.
Acknowledgments
We thank Sue Welch and Dawn Cardace for sample collection assistance at Lake Tyrrell; Mike Dyall-Smith for generous access to reagents and laboratory equipment; Cheetham Salt Works (Lake Tyrrell, Australia) and South Bay Salt Works, Chula Vista (San Diego, CA) for permission to collect samples; Brian Collins (USFWS) for help with sample collection at South Bay Salt Works; Matt Lewis and the J Craig Venter Institute for library construction and sequencing; Nerida Wilson for assistance with phylogenetic trees; and the US Department of Energy Joint Genomes Institute for genome annotation support via the Integrated Microbial Genome Expert Review (IMG-ER) resource. We also thank Farooq Azam (SIO/UCSD) for kindly permitting use of the Nikon confocal microscope purchased with support from the Gordon and Betty Moore Foundation. Funding for this work was provided by NSF award number 0626526 (JFB, KBH, EEA) and NIH award R21HG005107-02 (EEA). JAU was supported by a Fulbright-Conicyt fellowship. CBA is supported by an Action Thématique et Incitative sur Programme of the French Centre National de la Recherche Scientifique (CNRS). Work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract No DE-AC02-05CH11231.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allen EE, Banfield JF. Community genomics in microbial ecology and evolution. Nat Rev Microbiol. 2005;3:489–498. doi: 10.1038/nrmicro1157. [DOI] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD, et al. Enigmatic, ultrasmall, uncultivated archaea. Proc Natl Acad Sci USA. 2010;107:8806–8811. doi: 10.1073/pnas.0914470107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Tyson GW, Webb RI, Flanagan J, Hugenholtz P, Allen EE, et al. Lineages of acidophilic archaea revealed by community genomic analysis. Science. 2006;314:1933–1935. doi: 10.1126/science.1132690. [DOI] [PubMed] [Google Scholar]
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Benlloch S, Acinas SG, Anton J, Lopez-Lopez A, Luz SP, Rodriguez-Valera F. Archaeal biodiversity in crystallizer ponds from a solar saltern: culture versus PCR. Microb Ecol. 2001;41:12–19. doi: 10.1007/s002480000069. [DOI] [PubMed] [Google Scholar]
- Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis A, Kwan D, Thomas JR.2008Halophilic adaptations of proteinsIn: Siddiqui KS, Thomas T (eds). Protein Adaptation in Extremophiles Nova Biomedical Books: New York; 71–104. [Google Scholar]
- Bolhuis H, Palm P, Wende A, Falb M, Rampp M, Rodriguez-Valera F, et al. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics. 2006;7:169. doi: 10.1186/1471-2164-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branciamore S, Gallori E, Di Giulio M. The basal phylogenetic position of Nanoarchaeum equitans (Nanoarchaeota) Front Biosci. 2008;13:6886–6892. doi: 10.2741/3196. [DOI] [PubMed] [Google Scholar]
- Brochier C, Bapteste E, Moreira D, Philippe H. Eubacterial phylogeny based on translational apparatus proteins. Trends Genet. 2002;18:1–5. doi: 10.1016/s0168-9525(01)02522-7. [DOI] [PubMed] [Google Scholar]
- Brunk CF, Eis N. Quantitative measure of small-subunit rRNA gene sequences of the kingdom korarchaeota. Appl Environ Microbiol. 1998;64:5064–5066. doi: 10.1128/aem.64.12.5064-5066.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML. Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl Environ Microbiol. 2004;70:5258–5265. doi: 10.1128/AEM.70.9.5258-5265.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanueva A, Galada N, Baker GC, Grant WD, Heaphy S, Jones B, et al. Nanoarchaeal 16S rRNA gene sequences are widely dispersed in hyperthermophilic and mesophilic halophilic environments. Extremophiles. 2008;12:651–656. doi: 10.1007/s00792-008-0170-x. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Clarke K, Gorley R. Primer v6: User Manual/Tutorial. PRIMER-E: Plymouth, UK; 2006. [Google Scholar]
- Cuadros-Orellana S, Martin-Cuadrado AB, Legault B, D'Auria G, Zhaxybayeva O, Papke RT, et al. Genomic plasticity in prokaryotes: the case of the square haloarchaeon. ISME J. 2007;1:235–245. doi: 10.1038/ismej.2007.35. [DOI] [PubMed] [Google Scholar]
- Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- DasSarma S, Fleischman EM.1995Archaea: A Laboratory Manual - HalophilesVol 1.Cold Spring Harbor Laboratory Press: Plainview, NY [Google Scholar]
- DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demergasso C, Casamayor EO, Chong G, Galleguillos P, Escudero L, Pedros-Alio C. Distribution of prokaryotic genetic diversity in athalassohaline lakes of the Atacama Desert, Northern Chile. FEMS Microbiol Ecol. 2004;48:57–69. doi: 10.1016/j.femsec.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Fuchs BM, Glockner FO, Wulf J, Amann R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 2000;66:3603–3607. doi: 10.1128/aem.66.8.3603-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi S, Yoshimune K, Wakayama M, Moriguchi M, Nishikawa K. Unique amino acid composition of proteins in halophilic bacteria. J Mol Biol. 2003;327:347–357. doi: 10.1016/s0022-2836(03)00150-5. [DOI] [PubMed] [Google Scholar]
- Gareeb A, Setani M. Assessment of alkaliphilic haloarchaeal diversity in Sua pan evaporator ponds in Botswana. Afr J Biotechnol. 2009;8:259–267. [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Goldberg SM, Johnson J, Busam D, Feldblyum T, Ferriera S, Friedman R, et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc Natl Acad Sci USA. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S, Grant WD, Jones BE, Kato C, Li L. Novel archaeal phylotypes from an East African alkaline saltern. Extremophiles. 1999;3:139–145. doi: 10.1007/s007920050109. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hallsworth JE, Yakimov MM, Golyshin PN, Gillion JL, D'Auria G, de Lima Alves F, et al. Limits of life in MgCl2-containing environments: chaotropicity defines the window. Environ Microbiol. 2007;9:801–813. doi: 10.1111/j.1462-2920.2006.01212.x. [DOI] [PubMed] [Google Scholar]
- Hrdy I, Hirt RP, Dolezal P, Bardonova L, Foster PG, Tachezy J, et al. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. A new phylum of archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Tyson GW. Microbiology: metagenomics. Nature. 2008;455:481–483. doi: 10.1038/455481a. [DOI] [PubMed] [Google Scholar]
- Ihara K, Umemura T, Katagiri I, Kitajima-Ihara T, Sugiyama Y, Kimura Y, et al. Evolution of the archaeal rhodopsins: evolution rate changes by gene duplication and functional differentiation. J Mol Biol. 1999;285:163–174. doi: 10.1006/jmbi.1998.2286. [DOI] [PubMed] [Google Scholar]
- Ihara K, Watanabe S, Tamura T. Haloarcula argentinensis sp. nov. and Haloarcula mukohataei sp. nov., two new extremely halophilic archaea collected in Argentina. Int J Syst Bacteriol. 1997;47:73–77. doi: 10.1099/00207713-47-1-73. [DOI] [PubMed] [Google Scholar]
- Jiang H, Dong H, Zhang G, Yu B, Chapman LR, Fields MW. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl Environ Microbiol. 2006;72:3832–3845. doi: 10.1128/AEM.02869-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König H, Nusser E, Stetter KO. Glycogen in methanolobus and methanococcus. FEMS Microbiol Lett. 1985;28:265–269. [Google Scholar]
- König H, Skorko R, Zillig W, Reiter W-D. Glycogen in thermoacidophilic archaebacteria of the genera Sulfolobus, Thermoproteus, Desulfurococcus, and Thermococcus. Arch Microbiol. 1982;132:297–303. [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M (eds).Nucleic Acid Techniques in Bacterial Systematics Wiley: Chichester; New York; 115–175. [Google Scholar]
- Lasken RS. Single-cell genomic sequencing using multiple displacement amplification. Curr Opin Microbiol. 2007;10:510–516. doi: 10.1016/j.mib.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang J, Ni F, Dong X, Han B, Han D, et al. Genome wide exploration of the origin and evolution of amino acids. BMC Evol Biol. 2010;10:77. doi: 10.1186/1471-2148-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo I, Denef VJ, Verberkmoes NC, Shah MB, Goltsman D, DiBartolo G, et al. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature. 2007;446:537–541. doi: 10.1038/nature05624. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Moreira D, Lopez-Lopez A, Rodriguez-Valera F. A novel haloarchaeal-related lineage is widely distributed in deep oceanic regions. Environ Microbiol. 2001;3:72–78. doi: 10.1046/j.1462-2920.2001.00162.x. [DOI] [PubMed] [Google Scholar]
- Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M. probeCheck—a central resource for evaluating oligonucleotide probe coverage and specificity. Environ Microbiol. 2008;10:2894–2898. doi: 10.1111/j.1462-2920.2008.01706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2009a;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009b;25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- Matte-Tailliez O, Brochier C, Forterre P, Philippe H. Archaeal phylogeny based on ribosomal proteins. Mol Biol Evol. 2002;19:631–639. doi: 10.1093/oxfordjournals.molbev.a004122. [DOI] [PubMed] [Google Scholar]
- Maturrano L, Santos F, Rossello-Mora R, Anton J. Microbial diversity in Maras salterns, a hypersaline environment in the Peruvian Andes. Appl Environ Microbiol. 2006;72:3887–3895. doi: 10.1128/AEM.02214-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- Mongodin EF, Nelson KE, Daugherty S, Deboy RT, Wister J, Khouri H, et al. The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci USA. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu MB, Martinez-Garcia M, Santos F, Pena A, Guven K, Anton J. Prokaryotic diversity in Tuz Lake, a hypersaline environment in Inland Turkey. FEMS Microbiol Ecol. 2008;65:474–483. doi: 10.1111/j.1574-6941.2008.00510.x. [DOI] [PubMed] [Google Scholar]
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- Ochsenreiter T, Pfeifer F, Schleper C. Diversity of archaea in hypersaline environments characterized by molecular-phylogenetic and cultivation studies. Extremophiles. 2002;6:267–274. doi: 10.1007/s00792-001-0253-4. [DOI] [PubMed] [Google Scholar]
- Oh D, Porter K, Russ B, Burns D, Dyall-Smith M. Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds. Extremophiles. 2010;14:161–169. doi: 10.1007/s00792-009-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev. 1999;63:334–348. doi: 10.1128/mmbr.63.2.334-348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J Ind Microbiol Biotechnol. 2002a;28:56–63. doi: 10.1038/sj/jim/7000176. [DOI] [PubMed] [Google Scholar]
- Oren A.2002bHalophilic Microorganisms and Their Environments Kluwer Academic: Dordrecht; Boston; xxi, 575pp. [Google Scholar]
- Oren A. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 2008;4:2. doi: 10.1186/1746-1448-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A, Arahal DR, Ventosa A. Emended descriptions of genera of the family Halobacteriaceae. Int J Syst Evol Microbiol. 2009;59:637–642. doi: 10.1099/ijs.0.008904-0. [DOI] [PubMed] [Google Scholar]
- Pagaling E, Wang H, Venables M, Wallace A, Grant WD, Cowan DA, et al. Microbial biogeography of six salt lakes in Inner Mongolia, China, and a salt lake in Argentina. Appl Environ Microbiol. 2009;75:5750–5760. doi: 10.1128/AEM.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Bag SK, Das S, Harvill ET, Dutta C. Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 2008;9:R70. doi: 10.1186/gb-2008-9-4-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler J, Glockner FO, Schonuber W.2001Fluorescence in situ Hybridization with rRNA-targeted oligonucleotide probesIn: Paul JH (ed),Methods in MicrobiologyVol 30.Academic Press: San Diego; 207–226. [Google Scholar]
- Podell S, Gaasterland T. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol. 2007;8:R16. doi: 10.1186/gb-2007-8-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Gaasterland T, Allen EE. A database of phylogenetically atypical genes in archaeal and bacterial genomes, identified using the DarkHorse algorithm. BMC Bioinformat. 2008;9:419. doi: 10.1186/1471-2105-9-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P, Wolf YI, Koonin EV. Search for a ‘Tree of Life' in the thicket of the phylogenetic forest. J Biol. 2009;8:59. doi: 10.1186/jbiol159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol. 2008;6:693–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- Ram RJ, Verberkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC, II, et al. Community proteomics of a natural microbial biofilm. Science. 2005;308:1915–1920. [PubMed] [Google Scholar]
- Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol. 2007;62:142–160. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang Z. Phylogenetic inference using whole genomes. Annu Rev Genomics Hum Genet. 2008;9:217–231. doi: 10.1146/annurev.genom.9.081307.164407. [DOI] [PubMed] [Google Scholar]
- Rhodes ME, Fitz-Gibbon ST, Oren A, House CH. Amino acid signatures of salinity on an environmental scale with a focus on the Dead Sea. Environ Microbiol. 2010;12:2613–2623. doi: 10.1111/j.1462-2920.2010.02232.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010;4:739–751. doi: 10.1038/ismej.2010.1. [DOI] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Sabet S, Diallo L, Hays L, Jung W, Dillon JG. Characterization of halophiles isolated from solar salterns in Baja California, Mexico. Extremophiles. 2009;13:643–656. doi: 10.1007/s00792-009-0247-1. [DOI] [PubMed] [Google Scholar]
- Sime-Ngando T, Lucas S, Robin A, Tucker KP, Colombet J, Bettarel Y, et al. 2010Diversity of virus-host systems in hypersaline Lake Retba, Senegal Environ Microbioldoi: 10.1111/j.1462-2920.2010.02323.x [DOI] [PubMed]
- Sipos R, Szekely AJ, Palatinszky M, Revesz S, Marialigeti K, Nikolausz M. Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targetting bacterial community analysis. FEMS Microbiol Ecol. 2007;60:341–350. doi: 10.1111/j.1574-6941.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- Susko E, Roger AJ. On reduced amino acid alphabets for phylogenetic inference. Mol Biol Evol. 2007;24:2139–2150. doi: 10.1093/molbev/msm144. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Verhees CH, Kengen SW, Tuininga JE, Schut GJ, Adams MW, De Vos WM, et al. The unique features of glycolytic pathways in Archaea. Biochem J. 2003;375:231–246. doi: 10.1042/BJ20021472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall DP, Deluca T. Ortholog detection using the reciprocal smallest distance algorithm. Methods Mol Biol. 2007;396:95–110. doi: 10.1007/978-1-59745-515-2_7. [DOI] [PubMed] [Google Scholar]
- Walsby AE. Gas vesicles. Microbiol Rev. 1994;58:94–144. doi: 10.1128/mr.58.1.94-144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes P, Simmons SL, Denef VJ, Banfield JF. The dynamic genetic repertoire of microbial communities. FEMS Microbiol Rev. 2009;33:109–132. doi: 10.1111/j.1574-6976.2008.00144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, Xie G, Copeland A, Gonzalez JM, Han C, Kiss H, et al. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9:R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.