Abstract
Histone lysine methylation has been implicated in epigenetic regulation of transcription. Using stable-isotope labelling and quantitative mass spectrometry, we analysed the dynamics of histone lysine methylation. Here we report that histone methylation levels are transiently reduced during S phase and are gradually re-established during subsequent cell cycle stages. However, despite the recovery of overall methylation levels before the next S phase, the methylation levels of parental and newly incorporated histones differ significantly. In addition, histone methylation levels are maintained at steady states by both restriction of methyltransferase activity and the active turnover of methyl groups in cells undergoing an extended G1/S phase arrest. Finally, we propose a ‘buffer model’ that unifies the imprecise inheritance of histone methylation and the faithful maintenance of underlying gene silencing.
Keywords: histone, methylation, mitotic inheritance
Introduction
Histone lysine methylation is involved in various chromatin-templated biological processes (Zhang & Reinberg, 2001; Shilatifard, 2006; Bannister & Kouzarides, 2011; Yun et al, 2011). Certain histone methylation states such as histone H3 trimethylated at Lys 9 (H3K9me3) and H3K27me3 are critical for classic epigenetic phenomena, including position-effect variegation (Rea et al, 2000) and polycomb silencing (Cao et al, 2002; Czermin et al, 2002; Kuzmichev et al, 2002; Muller et al, 2002). However, when and how newly synthesized histones acquire these ‘epigenetic’ lysine methylation, and whether this step is mediated by a faithful inheritance mechanism, remain largely unknown (Wu & Zhu, 2011; Zhu & Reinberg, 2011).
Stable-isotope labelling by amino acids in cell-culture-based quantitative mass spectrometry (MS; Ong et al, 2002) has provided a powerful tool to study the dynamics of histone methylation in vivo (Bonenfant et al, 2007; Pesavento et al, 2008; Scharf et al, 2009; Sweet et al, 2010; Zee et al, 2010). These reports have provided important insight into the dynamics of histone lysine methylation. However, the kinetics of the establishment of the two most ‘epigenetic’ histone lysine methylation states, H3K9me3 and H3K27me3, remains unknown, because the matrix-assisted laser desorption/ionization–time of flight-based quantification used in a previous attempt (Scharf et al, 2009) could not distinguish the large quantity of isobaric ions derived from the histone polypeptides that contained different modifications with identical molecular masses (for example, H3K9me3 or H3K9ac and H3K27-R40 peptides that contained K27me3/K36me0, H3K27me0/K36me3, K27me1/K36me2 or K27me2/K36me1).
Here we report that for most methylation states, including H3K9me3 and H3K27me3, newly deposited H3.1 histones did not reach levels similar to the parental histones even after one entire cell cycle, which suggests that ‘epigenetic’ histone lysine methylation is not maintained in a precise manner during mitotic division.
Results And Discussion
Establishing lysine methylation on new histones
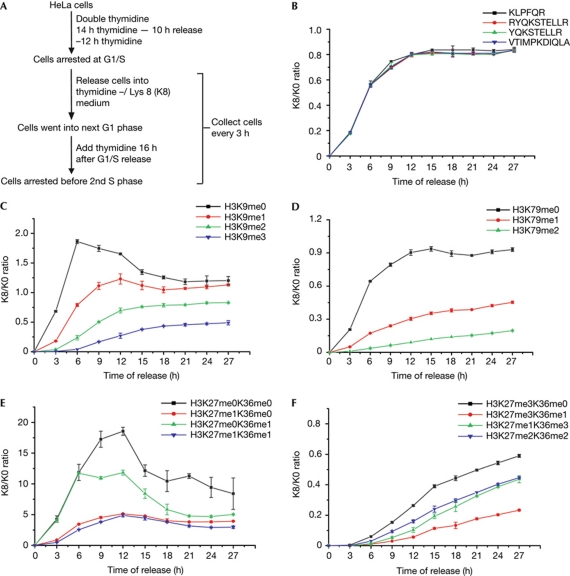
To track the formation of lysine methylation on newly synthesized H3.1 canonical histones, we conducted experiments according to the scheme illustrated in Fig 1A. In brief, HeLa cells were arrested at the G1/S phase boundary by double thymidine blocking and were released in labelling media that contained [13C6, 15N2] L-lysine (abbreviated as K8). H3.1 histones were purified (supplementary Fig S1 online) at various time points and subjected to MS analysis. To prevent cells from entering into the next S phase, thymidine was added 18 h after G1/S release (Fig 1A). We used liquid chromatography/MS/MS, which efficiently separated most of the isobaric peptides with reverse-phase chromatography (Yuan et al, 2011), and an Orbitrap mass spectrometer allowed high mass accuracy, enabling discrimination between acetyl and trimethyl groups.
Figure 1.
Establishment of lysine methylation on new histones. (A) Experimental scheme. (B) Incorporation of newly synthesized H3.1 histones after release from G1/S-phase arrest, which is indicated by the K8/K0 curve of H3 backbone peptides. (C–F) Establishment of lysine methylation on newly synthesized H3.1 histones versus the parental H3.1 histones, which is indicated by the K8/K0 curve of the methylated H3 peptides. (C) The K8/K0 curve of the H3:K9–R17 methylation moieties. (D) The K8/K0 curve of the H3:E73–R83 methylation moieties. (E) The K8/K0 curve of the H3:K27–R40 peptides with lower methylation states. (F) The K8/K0 curve of the H3:K27–R40 peptides with higher methylation states. All experiments in this figure and subsequent figures were repeated three times, and the error bars represent the standard deviation.
At 9 h after release, the K8/K0 (where K0 is non-labelled L-lysine) ratio of H3.1 backbone peptides reached a plateau of about 0.8 (the ratio did not reach 1 owing to the residual K0 from the intracellular amino-acid pool; Fig 1B), which is in accordance with the cell cycle progression determined by fluorescence-activated cell sorting analysis (supplementary Fig S2 online). Surprisingly, even for cells collected 27 h after release, which were blocked at the next G1/S-phase boundary, a striking difference for the methylation levels between the parental histones and newly synthesized histones remained (Fig 1C–F; supplementary Fig S3 online). The K8/K0 ratio for lower methylation states was generally higher than the backbone peptides, and the K8/K0 ratio for the higher methylation states, including H3K9me3 and H3K27me3, was much lower than that for the backbone peptides (Fig 1C–F). Therefore, we concluded that ‘epigenetic’ lysine methylation of newly synthesized H3 histones does not reach the same levels of old H3 histones even after one entire cell cycle. The only exception was H3K9me2, which reached levels comparable between new and old histones with a fast kinetics that follows H3 histone incorporation (Fig 1C; supplementary Fig S4 online). This is an interesting observation, especially in the context that G9a, the main histone methyltransferase that catalyses H3K9me2 (Tachibana et al, 2002), is localized to the replication fork via its interaction with DNMT1 (Esteve et al, 2006).
Cell cycle maintenance of the histone methylation
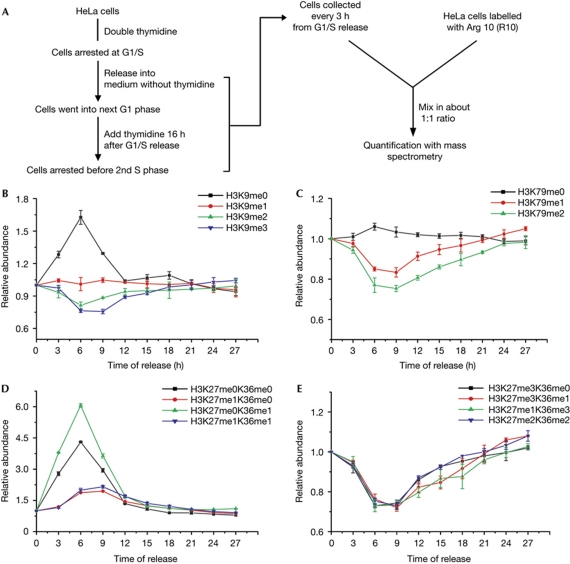
The ‘maturation’ of lysine methylation on newly synthesized histones is apparently a slow event (Fig 1). To compensate for the dilution of histone methylation levels, the old histones should be further methylated and the total methylation level should recover. We quantified the histone lysine methylation levels throughout the cell cycle as depicted in Fig 2A. In brief, H3.1 histones were prepared from unlabelled cells at different cell cycle stages and were mixed with an external reference, which was [13C6, 15N4] L-arginine (R10)-labelled H3.1 histones from unsynchronized cells. The relative abundance of the distinct histone methylation states at each time point was calculated according to the R0/R10 ratio, which was normalized to the R0/R10 ratio of the backbone peptides.
Figure 2.
Fluctuation of global histone lysine methylation during the cell cycle. (A) Experimental scheme. (B–E) Fluctuation in global lysine methylation levels on H3.1 histones during the cell cycle, which is indicated by the relative abundance curve of methylated H3 peptides. For each methylation state, the abundance at time zero was designated as one. (B) The relative abundance curve of the H3:K9–R17 methylation moieties. (C) The relative abundance curve of the H3:E73–R83 methylation moieties. (D) The relative abundance curve of the H3:K27–R40 peptides with lower methylation states. (E) The relative abundance curve of the H3:K27–R40 peptides with higher methylation states.
The levels of most methylation states were significantly altered during the S phase. All unmethylated states increased in abundance immediately after the onset of DNA replication and reached peak levels about 6 h after release (Fig 2B–D; supplementary Fig S5 online), which is consistent with previous observations that most histones were devoid of lysine methylation before deposition (Loyola et al, 2006). The levels of the unmethylated states began to decrease at the late S phase (9 h after release), and the methylation levels reached a plateau stage 15 h after release, which was before entering into the next G1 phase (Fig 2B–D; supplementary Fig S5 online). For the higher methylation states (H3K9me2/3, H3K79me2, H3K27me2/3 and H3K36me2/3), the overall levels first dropped to 70–80% during the S phase before recovering to the initial levels. All of the high methylation states that were measured showed similar kinetics, which suggests that it is a general event that replication-dependent histone deposition transiently dilutes the histone methylation levels. Our observations are consistent with previous reports that studied the dynamics of H3K79 methylation (Feng et al, 2002) and H3K27me3 (Aoto et al, 2008) during the cell cycle in mammalian cells with western blot analysis.
Histone methylation maintenance in arrested cells
Newly deposited histones do not obtain the same level of histone lysine methylation as the parental histones after one round of the cell cycle (Fig 1). However, the total levels of most lysine methylation states seem to be maintained (Fig 2). Therefore, the next question is whether such maintenance is a coincidence of the cell cycle duration and the time necessary for recovering the methylation levels, or whether it is a consequence of an active mechanism that restricts the methylation levels. To examine this question, we investigated the histone methylation levels in non-cycling cells that were undergoing an extended G1/S-phase arrest.
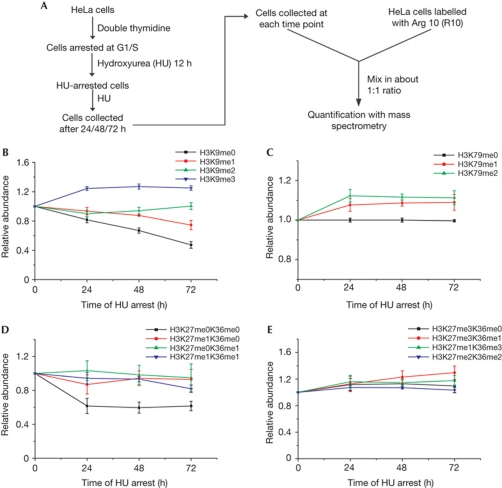
After double thymidine blocking, HeLa cells were subjected to an extra 12 h of hydroxyurea (HU) blocking to ensure tight S-phase arrest. These cells were further arrested with HU for 0, 24, 48 and 72 h to study the methylation dynamics in non-cycling cells. Cells further treated with HU at the 0- and 24-h time points showed comparable amounts of sub-G1 cells with untreated cells; the sub-G1 cells increased to about 5% at the 48- and 72-h time points, reflecting a small percentage of apoptotic cells (supplementary Fig S6 online). H3.1 histones were prepared from cells at these time points and R10-labelled H3.1 histones were used as an external reference to monitor histone methylation levels (Fig 3A).
Figure 3.
Maintenance of overall histone lysine methylation levels in cells under extended G1/S-phase arrest. (A) Experimental scheme. (B–E) Maintenance of global lysine methylation levels on H3.1 histones in long-term-arrested cells, which is indicated by the relative abundance curve of the methylated H3 peptides. For each methylation state, the abundance at 12 h of hydroxyurea (HU) arrest was designated as one. (B) The relative abundance curve of the H3:K9–R17 methylation moieties. (C) The relative abundance curve of the H3:E73–R83 methylation moieties. (D) The relative abundance curve of the H3:K27–R40 peptides with lower methylation states. (F) The relative abundance curve of the H3:K27–R40 peptides with higher methylation states.
The levels of most methylation states on H3.1 histones continued to change during the first 24 h of additional HU arrest. Generally, the lower methylation states tended to decrease, whereas the higher methylation states tended to increase, indicating a further conversion from the lower methylation states to the higher methylation states (Fig 3B–E; supplementary Fig S7 online). However, during the 24- to 72-h extended G1/S-phase arrest, histone methylation levels tended to reach a steady state. Among the 15 distinct H3 lysine methylation states, only 4 methylation states (H3K9me0, H3K9me1, H3K27me0/K36me1 and H3K27me3/K36me1) showed slightly more than a 10% change in abundance, and the alteration per unit time was significantly lower when compared with dividing cells (Fig 3B–E). During 24–72 h, we observed about 30% decrease in H3K9me0, a 10% decrease in H3K9me1, a 10% increase in H3K9me2 and almost no change in H3K9me3 (Fig 3B). Therefore, we determined the relative abundance of H3:K9–R17 peptides bearing distinct modifications and found that H3K9me0/1/2/3 accounted for about 13%, 7.5%, 32.6% and 47.1%, respectively (supplementary Fig S8 online). These results explain why a big decrease in lower methylation states only led to a small increase in the higher methylation states.
Overall, in cells experiencing an extended G1/S-phase arrest, upon ‘maturation’ of the methylation states, the levels of higher methylation states tend to be maintained at relatively stable levels, which suggests the existence of active mechanisms that restrict the activity of histone methyltransferases and/or that promote active histone demethylation.
Turnover of histone methylation
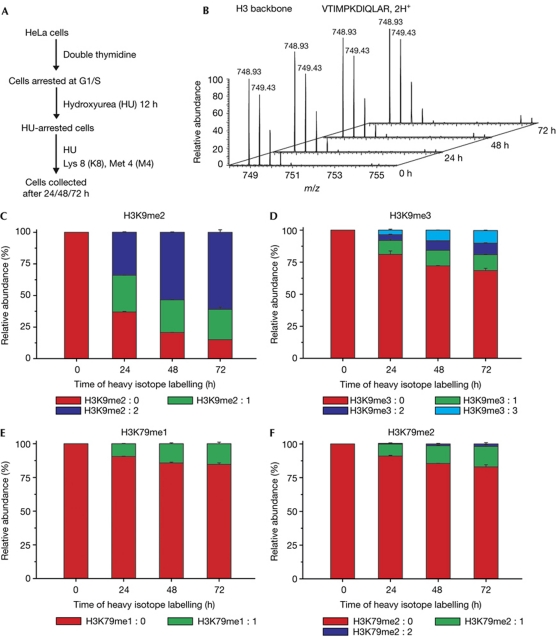
To investigate how histone methylation levels were maintained in cells undergoing an extended G1/S-phase arrest, the turnover of histone methylation in arrested cells was examined. As illustrated in Fig 4A, cells were sequentially treated with double thymidine and HU as previously described. After 12 h of HU arrest, cells were shifted into labelling media that contained HU, K8 and [13C, D3] L-methionine (abbreviated as M4). We examined the K8/K0 ratio of a backbone peptide from the replication-dependent histone variant H3.1, which was 0% K8-labelled even after 72 h of labelling (Fig 4B). This result indicated that there was a complete inhibition of replication-dependent chromatin assembly during HU treatment, which also indicated that repair-coupled histone deposition was too low to impact our analysis. Methionine is the precursor of S-adenosyl methionine, which is the sole donor of methyl groups in mammalian cells. By quantifying the levels of newly installed methyl groups, which were 13CD3-labelled and 4 Da heavier than regular methyl groups, we were able to observe the turnover of methyl groups without being influenced by the deposition of newly synthesized histones. The meX:Y naming system (Zee et al, 2010) was used to describe the methylation species, where X is the number of total methyl groups and Y is the number of heavy-isotope-labelled ones. For example, H3K9me2:0 represents unlabelled dimethylated H3K9, and H3K9me2:1 represents dimethylated H3K9 with one heavy methyl group.
Figure 4.
Turnover of methyl groups on histone lysine residues in cells undergoing an extended G1/S-phase arrest. (A) Experimental scheme. (B) Mass spectra of an H3 backbone peptide at time points of 0, 24, 48 and 72 h, which indicated no incorporation of newly synthesized H3.1 histones. (C) The percentage of H3K9me2 peptides that contained a different composition of light and heavy methyl groups. (D) The percentage of H3K9me3 peptides that contained a different composition of light and heavy methyl groups. (E) The percentage of H3K79me1 peptides that contained a different composition of light and heavy methyl groups. (F) The percentage of H3K79me2 peptides that contained a different composition of light and heavy methyl groups.
We focused our analysis on the H3K9 and H3K79 methylation states, as a combination of light and heavy methyl groups on peptides containing both K27 and K36 resulted in insurmountable heterogeneity. Interestingly, the turnover rate of distinct methylation types was significantly different. For H3K9me2, a remarkable shift from light to heavy methyl groups was observed (Fig 4C; supplementary Fig S9 online). At 24 h, there were about 34% H3K9me2:2, 29% H3K9me2:1 and 37% H3K9me2:0. Considering that there was about 10% increase in total H3K9me2 during 24–72 h (Fig 3B), at 72 h we would expect to detect about 40% H3K9me2:2, 26% H3K9me2:1 and 34% H3K9me2:0, if there were no active turnover of H3K9me2. However, the actual data at 72 h were about 61% H3K9me2:2, 24% H3K9me2:1 and 15% H3K9me2:0, indicating a robust active turnover of H3K9me2. In contrast, the turnover rate of H3K9me3 was much slower. During the plateau stage (24–72 h, as shown in Fig 3B), the proportion of H3K9me3:0/3:1/3:2/3:3 levels only changed from 81%:11%:4%:4% to 69%:12%:9%:10% (Fig 4D).
The turnover of H3K79me1/2 was even more limited. As observed in the plateau stage of overall abundance (24–72 h in Fig 3C), the proportion of H3K79me1:0/1:1 changed from 91%:9% to 85%:15%, whereas that of H3K79me2:0/2:1/2:2 changed from 91%:9%:0% to 83%:15%:1% (Fig 4E,F).
Our data indicate that the levels of H3K9me2 were maintained in a dynamic equilibrium by methylation and demethylation, whereas the maintenance of H3K9me3 and H3K79me1/2 largely relied on inhibiting the activity of histone methyltransferases.
An interesting question is whether the quiescent cells might share similar mechanisms to control the levels of histone modifications. We attempted to conduct similar experiments using HeLa cells arrested with serum starvation or T24 bladder carcinoma cells arrested with contact inhibition (Jin et al, 1997). Unfortunately, we did not achieve 100% cell cycle arrest in these attempts, because we observed the deposition of certain levels of newly synthesized H3.1 histones (supplementary Fig S10 online). We were unable to go further using these two systems, because the incorporation of newly synthesized histones in a portion of unarrested cells would jeopardize our interpretation. Similar experiments in other systems that can fully arrest the cells would be interesting for future study.
A ‘buffer model’ for epigenetic inheritance
Histone modifications that carry epigenetic information should be appropriately inherited between cell generations to maintain cell fate (Martin & Zhang, 2007; Probst et al, 2009; Margueron & Reinberg, 2010; Xu & Zhu, 2010). However, the levels of classic ‘epigenetic’ histone modifications, such as H3K9me3 and H3K27me3, were not precisely transmitted from ‘parental’ histones to newly deposited histones during mitotic division (Fig 1). This indicates that cells lack the machinery to discriminate newly deposited histones from parental histones in the processes that establish the lysine methylation. During S phase, a transient decrease in global levels of higher methylation states was observed (Fig 2). A similar transient decrease in H3K9me2 was also reported at the fission yeast centromeric heterochromatin determined by chromatin immunoprecipitation analysis (Chen et al, 2008). These collectively suggest that a transient reduction of repressive modification levels is a general event that many repressed genes would encounter during S phase. Together with our recent findings that histone lysine methylation is not required to exist symmetrically (Chen et al, 2011) and that the canonical H3.1 histone assembly pathway does not involve semiconservative segregation of the (H3-H4)2 tetramers (Xu et al, 2010), these results led to a critical question: how can cells maintain target gene repression without a mechanism that can precisely transmit the repressive histone modifications?
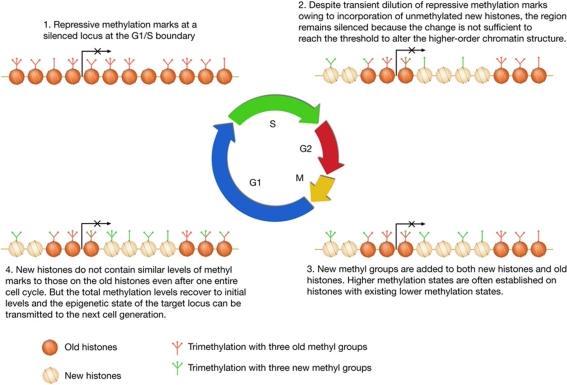
We propose that gene silencing is probably the consequence of an accumulation of repressive methylation in broad regions that might range from several kilobases to tens of kilobases (Boyer et al, 2006; Lee et al, 2006; Schwartz et al, 2006), rather than methylation at only a few ‘important’ nucleosomes, such as nucleosomes around transcriptional start sites, because transcriptional silencing is probably caused by reduced chromatin accessibility owing to higher-order chromatin structure formation (Li & Reinberg, 2011), which is a regional event rather than a local event. An accumulation of repressive methylation marks above a certain threshold, instead of uniform methylation within the domain, would be sufficient to maintain the gene silencing. Therefore, a fluctuation of the methylation levels within these domains can be tolerated without affecting the silencing of the target genes (Fig 5). Indeed, despite a transient drop in H3K9me2/3 and H3K27me2/3 levels during S phase (Fig 2), derepression of H3K9me2 targets, such as Mage-A2, and H3K27me2/3 targets, such as the HOX genes, was not observed during S phase (Tachibana et al, 2002; Bar-Joseph et al, 2008).
Figure 5.
A ‘buffer model’ for mitotic inheritance of repressive histone methylation marks.
With such a model, we abandon the idea that histone methylation should be faithfully maintained at near mononucleosome resolution to ensure its role in repressing epigenetically silenced genes. Instead, we propose that repressive histone lysine methylation asserts its functions in a way that is reminiscent of a ‘buffer’ system, which can tolerate transient fluctuations without disrupting its function.
Finally, we would like to point out that we think this ‘buffer’ model is dedicated to epigenetic silencing. For the ‘active’ modifications, it has been proposed that they might be maintained as the consequence of active transcription (Henikoff & Shilatifard, 2011).
Methods
Histone preparation for MS analysis. Chromatin histones were isolated from cells using an acid extraction (Shechter et al, 2007). H3.1 histones were purified by reverse-phase liquid chromatography using a SOURCE™ 15RPC ST 4.6/100 (GE) column for subsequent analysis. For experiments using R10-labelled histones as the external reference, R0- and R10-labelled H3.1 histones were mixed at a 1:1 ratio.
H3.1 histones were propionylated and then digested with trypsin according to previous reports (Garcia et al, 2007; Yuan et al, 2011).
Quantitative mass spectrometry analysis. Peptides were separated with a homemade analytical capillary column (50 μm × 12 cm) that was packed with C18 reverse-phase material and then electrosprayed into an LTQ-Orbitrap mass spectrometer (Thermo). Data were acquired in an information-dependent mode, and a full MS scan was collected from m/z 300 to 2,000 with a resolution of R=60,000. The top six abundant ions were fragmented by collision-induced dissociation, and the fragmented ions were scanned with an ion trap. MS/MS spectra for all distinct methylation states were verified according to our previous reports (Chen et al, 2011; Yuan et al, 2011).
Mascot search results were analysed with MSquant, and the ratio of heavy/light peptide pairs was calculated based on the extracted ion chromatogram (Mortensen et al, 2010).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We appreciate Mengqiu Dong for providing the LTQ-Orbitrap mass spectrometer and Ning Yang for the colour illustration. The Chinese Ministry of Science and Technology grants 2011CB812700 and 2011CB965300 supported this work.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aoto T, Saitoh N, Sakamoto Y, Watanabe S, Nakao M (2008) Polycomb group protein-associated chromatin is reproduced in post-mitotic G1 phase and is required for S phase progression. J Biol Chem 283: 18905–18915 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph Z, Siegfried Z, Brandeis M, Brors B, Lu Y, Eils R, Dynlacht BD, Simon I (2008) Genome-wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells. Proc Natl Acad Sci USA 105: 955–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonenfant D, Towbin H, Coulot M, Schindler P, Mueller DR, van Oostrum J (2007) Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol Cell Proteomics 6: 1917–1932 [DOI] [PubMed] [Google Scholar]
- Boyer LA et al. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI (2008) Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Chen X, Xiong J, Xu M, Chen S, Zhu B (2011) Symmetrical modification within a nucleosome is not required globally for histone lysine methylation. EMBO Rep 12: 244–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S (2006) Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 20: 3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y (2002) Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 12: 1052–1058 [DOI] [PubMed] [Google Scholar]
- Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF (2007) Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc 2: 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A (2011) Histone modification: cause or cog? Trends Genet 27: 389–396 [DOI] [PubMed] [Google Scholar]
- Jin Y, Xu XL, Yang MC, Wei F, Ayi TC, Bowcock AM, Baer R (1997) Cell cycle-dependent colocalization of BARD1 and BRCA1 proteins in discrete nuclear domains. Proc Natl Acad Sci USA 94: 12075–12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI et al. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Reinberg D (2011) Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev 21: 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G (2006) PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell 24: 309–316 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D (2010) Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11: 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y (2007) Mechanisms of epigenetic inheritance. Curr Opin Cell Biol 19: 266–272 [DOI] [PubMed] [Google Scholar]
- Mortensen P et al. (2010) MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J Proteome Res 9: 393–403 [DOI] [PubMed] [Google Scholar]
- Muller J et al. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386 [DOI] [PubMed] [Google Scholar]
- Pesavento JJ, Yang H, Kelleher NL, Mizzen CA (2008) Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol 28: 468–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G (2009) Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10: 192–206 [DOI] [PubMed] [Google Scholar]
- Rea S et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Scharf AN, Barth TK, Imhof A (2009) Establishment of histone modifications after chromatin assembly. Nucleic Acids Res 37: 5032–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V (2006) Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB (2007) Extraction, purification and analysis of histones. Nat Protoc 2: 1445–1457 [DOI] [PubMed] [Google Scholar]
- Shilatifard A (2006) Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269 [DOI] [PubMed] [Google Scholar]
- Sweet SM, Li M, Thomas PM, Durbin KR, Kelleher NL (2010) Kinetics of re-establishing H3K79 methylation marks in global human chromatin. J Biol Chem 285: 32778–32786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M et al. (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhu B (2011) Split decision: why it matters? Front Biol 6: 88–92 [Google Scholar]
- Xu M, Zhu B (2010) Nucleosome assembly and epigenetic inheritance. Protein Cell 1: 820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B (2010) Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 328: 94–98 [DOI] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B (2011) H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286: 7983–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Wu J, Workman JL, Li B (2011) Readers of histone modifications. Cell Res 21: 564–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA (2010) In vivo residue-specific histone methylation dynamics. J Biol Chem 285: 3341–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15: 2343–2360 [DOI] [PubMed] [Google Scholar]
- Zhu B, Reinberg D (2011) Epigenetic inheritance: uncontested? Cell Res 21: 435–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.