Abstract
DNA methylation is involved in key cellular processes, including X-chromosome inactivation, imprinting and transcriptional silencing of specific genes and repetitive elements. DNA methylation patterns are frequently perturbed in human diseases such as imprinting disorders and cancer. The recent discovery that the three members of the TET protein family can convert 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC) has provided a potential mechanism leading to DNA demethylation. Moreover, the demonstration that TET2 is frequently mutated in haematopoietic tumours suggests that the TET proteins are important regulators of cellular identity. Here, we review the current knowledge regarding the function of the TET proteins, and discuss various mechanisms by which they contribute to transcriptional control. We propose that the TET proteins have an important role in regulating DNA methylation fidelity, and that their inactivation contributes to the DNA hypermethylation phenotype often observed in cancer.
Keywords: DNA methylation, 5-hydroxymethylcytosine, TET proteins, stem cells, cancer
See Glossary for abbreviations used in this article.
Glossary.
AID/APOBEC activation-induced deaminase/apolipoprotein B RNA-editing catalytic component
ChIP-seq chromatin immunoprecipitation followed by sequencing
DNMT DNA methyltransferase
PRC2 Polycomb repressive complex 2
shRNA short-hairpin RNA
UHRF1 Ubiquitin-like, with PHD and RING finger domains 1
Introduction
Embryonic stem cells (ESCs) are derived from the inner cell mass of blastocysts. They are pluripotent and have the potential to differentiate and generate cells of all three germ layers. During this process, epigenetic modifications contribute to defining the transcriptional programme that specifies cellular identity. An example of an epigenetic modification with essential roles during embryonic development is DNA methylation. In mammals, DNA methylation occurs predominantly at CpG dinucleotides where DNMTs mediate the transfer of a methyl group to cytosines, generating 5-methylcytosine (5mC; Goll & Bestor, 2005). Interestingly, almost 25% of 5mC in ESCs is associated with CHG or CHH (where H is A, C or T), that is, in a non-CG context (Lister et al, 2009). Whether this form of 5mC has different functions to 5mC in a CG context is not known and is not discussed further in this review. There are three enzymatically active mammalian DNMTs—DNMT3A, DNMT3B and DNMT1. DNMT3A and DNMT3B are de novo methyltransferases that establish DNA methylation patterns by targeting unmethylated CpG sites. DNMT1 acts primarily as a maintenance methyltransferase. It localizes to replication foci during S-phase, where it preferentially methylates hemi-methylated CpGs through its interaction with UHRF1 (Bostick et al, 2007; Sharif et al, 2007). In this way, DNA methylation patterns are efficiently preserved through cell division. The DNMTs are not required for ESC integrity or self-renewal (Tsumura et al, 2006). However, knockout studies in mice have shown that both establishment and maintenance of DNA methylation are essential for embryonic development (Li et al, 1992; Okano et al, 1999). In agreement with this, DNA methylation patterns undergo major changes during early embryonic development with a global loss of DNA methylation occurring in the zygote shortly after fertilization. The paternal genome is rapidly demethylated, while the maternal genome exhibits a gradual loss of methylation until the eight-cell stage. After this, the methylation patterns are re-established by de novo methylation in the developing embryo (reviewed in Reik et al, 2001).
CpG dinucleotides are found unevenly distributed throughout the genome. In general, CpGs are under-represented in mammals, presumably owing to the mutagenic properties of 5mC. However, certain regions of DNA contain a high density of CpG and are referred to as CpG islands. They often cover sites of transcriptional initiation and 60–70% of annotated gene promoters are associated with a CpG island, including most housekeeping genes, as well as tissue-specific and developmental genes (reviewed in Blackledge & Klose, 2011; Deaton & Bird, 2011). Studies on the genome-wide distribution of DNA methylation estimate that the majority of CpG sites are methylated, when not present in CpG islands (Li et al, 2010; Lister et al, 2009; Meissner et al, 2008; Mohn et al, 2008; Weber et al, 2007), including those present in intragenic regions, repetitive sequences and mobile elements. Methylation of intragenic regions has been proposed to inhibit cryptic transcription and correlate with gene expression, whereas methylation of repetitive and mobile sequences is central for genomic stability and host defence mechanisms (Weber & Schubeler, 2007). Although it occurs relatively rarely, methylation of CpG island promoters correlates with transcriptional silencing, which is thought to be mediated by a direct interference of transcription-factor binding or through the recruitment of repressive methyl-binding proteins such as MeCP2 (Bogdanovic & Veenstra, 2009). Comparative studies of DNA methylation in ESCs and differentiated tissues have shown that a fraction of CpG islands, which includes stem-cell-associated and germline-associated genes, as well as certain tissue-specific genes, becomes de novo methylated during development. This observation suggests that DNA methylation has a role both in the loss of pluripotency and in the following cellular specification (Meissner et al, 2008; Mohn et al, 2008). In addition to its vital roles during development, DNA methylation has also been associated with tumorigenesis as most cancer cells display aberrant DNA methylation patterns (reviewed in Jones & Baylin, 2007). This includes both global hypomethylation of the genome and promoter-specific hypermethylation. Global hypomethylation is believed to create genomic instability, whereas hypermethylation of CpG islands at gene promoters can lead to undesirable silencing of genes (Jones & Baylin, 2007). Direct silencing of tumour suppressor genes by DNA methylation is a well-characterized feature of cancer cells (Jones & Baylin, 2007). In addition, methylation events can affect lineage-specific differentiation, which can also promote tumorigenesis (Feinberg et al, 2006). To ensure proper embryogenesis and prevent the development of disease, it is therefore important to regulate tightly DNA methylation patterns. Still, little is known about the mechanisms adjusting methylation levels, and it is largely unknown what prevents the accumulation of DNA methylation at CpG islands. Importantly, despite an intense search, no direct DNA demethylase has been identified. However, the recent finding by Rao and colleagues that the ten-eleven translocation 1 (TET1) protein can catalyse the conversion of 5mC into 5-hydroxymethylcytosine (5hmC; Tahiliani et al, 2009) suggests a potential mechanism for active demethylation. In this review, we summarize several recent studies on the genome-wide distribution of TET1 and 5hmC in ESCs and discuss the potential roles of TET proteins and hydroxymethylation in the regulation of DNA methylation patterns and transcription.
The TET proteins and the putative functions of 5hmC
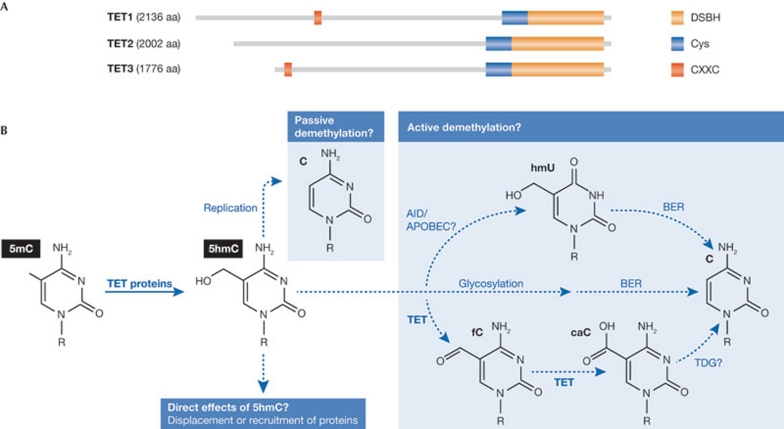
The TET protein family members—TET1, TET2 and TET3—are 2-oxoglutarate and Fe(II)-dependent dioxygenases that all have the capacity to convert 5mC into 5hmC in vitro and in vivo (Fig 1A; Ito et al, 2010; Ko et al, 2010; Tahiliani et al, 2009). In addition to the conserved iron-binding catalytic subunit, TET1 and TET3 also have a CXXC domain. This DNA-binding domain has previously been described as a CpG-binding motif, which could be involved in the recruitment of TET1 and TET3 to DNA. The TET proteins show tissue-specific differential expression with TET1 being mainly expressed in ESCs, whereas TET2 and TET3 are more ubiquitously expressed (Szwagierczak et al, 2010; Tahiliani et al, 2009).
Figure 1.
Possible biological roles of TET proteins and 5hmC. (A) The domain structure of human TET proteins. TET1–3 contain a cysteine (Cys)-rich region followed by the double-stranded β-helix (DSBH) fold characteristic of the 2OG-Fe(II) oxygenases and required for catalytic activity. TET1 and TET3 also contain a CXXC domain. (B) Several biological consequences of the TET-mediated conversion of 5mC to 5hmC can be envisioned. 5hmC might facilitate a passive demethylation that is replication-dependent or could be converted to cytosine through an active demethylation pathway. Finally, 5hmC might also have direct effects by displacing or recruiting effector proteins. 5hmC, 5-hydroxymethylcytosine; 5mC, 5-methylcytosine.
Interestingly, whereas levels of 5mC are relatively constant, 5hmC intensities vary significantly between tissues, with the highest levels reported for specific cell types of the brain (Globisch et al, 2010; Kriaucionis & Heintz, 2009). ESCs also have relatively high levels of 5hmC, which decrease during differentiation (Globisch et al, 2010; Koh et al, 2011; Szwagierczak et al, 2010; Tahiliani et al, 2009).
Several biological functions of TET-mediated conversion of 5mC into 5hmC can be envisioned (Fig 1B). The fact that de novo methyltransferase activity is low in differentiated tissues, taken together with the observation that 5hmC is found in relatively high levels in certain tissues, suggests that 5hmC in some tissues has a low turnover. In these situations, 5hmC might function by altering the local chromatin environment through the recruitment or displacement of proteins. Favouring this model, it has been reported that most 5mC-binding proteins do not recognize 5hmC (Jin et al, 2010; Valinluck et al, 2004) and thereby presumably dissociate from DNA when 5mC is converted into 5hmC. However, it is also apparent that the generation of 5hmC could be involved in the demethylation of DNA. First, this could be through a passive mechanism in which the mark, in contrast to 5mC, would not be maintained through DNA replication. Passive DNA demethylation would be particularly relevant in rapidly dividing cells such as ESCs. The fact that brain cells, which are not rapidly dividing, have high levels of 5hmC, supports this model. Moreover, it has been shown that DNMT1 methylates 5hmC-containing DNA with a lower efficiency to 5mC hemi-methylated DNA (Valinluck & Sowers, 2007).
Second, production of 5hmC could be an intermediate in an active demethylation pathway that ultimately replaces 5mC with cytosine in non-dividing cells. This has been proposed to involve specific DNA repair mechanisms such as deamination mediated by the AID/APOBEC family of cytidine deaminases, converting hmC into hmU followed by base excision repair (BER) or 5hmC glycosylation followed by BER (reviewed in Guo et al, 2011). In addition, the hypothesis that generation of 5hmC is an intermediate in an enzymatic pathway of active demethylation was recently supported by studies demonstrating the existence of formylcytosine and carboxylcytosine in mammalian DNA (Fig 1B; He et al, 2011; Ito et al, 2011; Pfaffeneder et al, 2011). These new cytosine modifications can be generated by two successive oxidation reactions of 5hmC catalysed by the TET proteins (He et al, 2011; Ito et al, 2011), raising the possibility that the TET proteins might be involved in several steps in converting 5mC to cytosine. As the TET proteins cannot convert carboxylcytosine to cytosine, a decarboxylase or a glycosylase might be involved in this step. In agreement with this, depletion of thymidine-DNA glycosylase (TDG) leads to accumulation of carboxylcytosine in mouse ESCs (He et al, 2011), and other studies have shown that TDG is required for DNA demethylation (Cortellino et al, 2011).
Functional studies on Tet1 in mouse ESCs
Since the discovery of TET proteins, several studies have focused on elucidating the role of hydroxymethylation and TET proteins in ESCs, with the main focus on Tet1. A well-characterized event of stem cell differentiation is the transcriptional silencing of pluripotency factors such as Oct4 and Nanog, which is partly mediated through DNA methylation of their promoters. The high levels of Tet1 and 5hmC in ESCs compared with that in differentiated cells suggests that Tet1 could act as a regulator of self-renewal by preventing DNA methylation of pluripotency genes by hydroxymethylation. This model was reinforced by a study demonstrating impaired self-renewal in Tet1-depleted mouse ESCs (Ito et al, 2010). The authors suggested that reduced Nanog expression caused the phenotype, presumably mediated by increased DNA methylation of the Nanog promoter. The importance of Tet1 in ESC maintenance, however, remains to be clarified. Other studies did not find changes in cell morphology or expression of pluripotency factors after knockdown of Tet1 (Koh et al, 2011; Williams et al, 2011; Xu et al, 2011). Moreover, a recent study showed that Tet1-knockout ESCs expressed normal levels of pluripotency factors and displayed a normal morphology (Dawlaty et al, 2011). In agreement with this, the Tet1-knockout mice were viable and fertile, although some had a smaller body size at birth (Dawlaty et al, 2011).
Sidebar A | In need of answers.
Do the TET proteins have major roles in regulating transcription?
Are the TET proteins regulating DNA methylation fidelity?
Do the TET proteins and 5hmC contribute to DNA demethylation?
Does 5hmC have a signalling function?
How does loss of function of TET2 lead to cancer development?
How are TET proteins recruited to specific DNA-binding sites?
Do the TET proteins have functionally redundant functions?
To gain more insight into the functional role of Tet1 in mouse ESCs, several laboratories mapped the genome-wide occupancy of Tet1 (Williams et al, 2011; Wu et al, 2011a; Xu et al, 2011). A recent comparison of the reported data sets (Wu & Zhang, 2011) shows that 90% of the reported Tet1 target genes (around 10,000) identified in the various studies overlap. Tet1 mainly binds to gene-rich regions, with the highest preference for transcription start sites (TSSs) and less intense binding throughout gene bodies. Moreover, when promoters are grouped on the basis of their CpG content, Tet1 mainly associates with promoters of high CpG content (Williams et al, 2011; Wu et al, 2011a; Xu et al, 2011). The correlation between Tet1 binding and CpG density also appeared outside promoter regions and indicates that the association of Tet1 with DNA could be facilitated by CpG-rich motifs. This finding is supported by the fact that Tet1 has a CXXC domain, which was recently suggested to be responsible for Tet1 DNA binding ability (Xu et al, 2011; Zhang et al, 2010).
In ESCs, promoters have previously been characterized according to CpG content, DNA methylation and histone modifications (Fouse et al, 2008; Meissner et al, 2008; Mikkelsen et al, 2007; Mohn et al, 2008). Promoters with high CpG content are largely DNA hypomethylated and virtually all are associated with histone 3 lysine 4 trimethylation (H3K4me3). These findings correlate with studies showing that de novo DNA methylation is selectively inhibited by methylation of H3K4 (Ooi et al, 2007; Otani et al, 2009). Consistent with the high association of Tet1 with CpG-rich regions, most Tet1-bound promoters are H3K4me3-positive. Interestingly, a high fraction of the Tet1 target genes were also associated with H3K27me3, and therefore also positive for binding of the PRC2 (Williams et al, 2011; Wu et al, 2011a). Gene ontology analysis of Tet1 target genes revealed enrichment for genes involved in basic housekeeping processes and those regulating differentiation and development, which is in agreement with Tet1 being associated with either H3K4me3 or bivalent H3K4me3/H3K27me3-positive genes, respectively.
Genome-wide studies of 5hmC
The genome-wide distribution of 5hmC in ESCs was investigated by several laboratories using different approaches. 5hmC-specific antibodies were used to determine the genome-wide distribution of 5hmC in ESCs by DNA immunoprecipitation followed by sequencing (DIP-seq; Ficz et al, 2011; Williams et al, 2011; Wu et al, 2011b; Xu et al, 2011), and Pastor et al developed two novel methods for detection of 5hmC localization that are based on enzymatic or chemical conversion of 5hmC (Pastor et al, 2011). Reassuringly, the different methods and independent 5hmC antibodies gave similar results (Wu & Zhang, 2011). Coinciding with Tet1 binding, 5hmC is enriched in gene-rich regions. Overlay of 5hmC-positive regions and Tet1 binding demonstrated a significant, although not complete, overlap both genome-wide and at promoters (Williams et al, 2011; Wu et al, 2011b; Xu et al, 2011). Similarly to 5hmC, 5mC is also enriched throughout gene bodies, indicating that the two marks often coexist. However, differences in the distribution of 5mC and 5hmC were also observed. The 5hmC modification is rarely found in heterochromatic regions such as repetitive sequences, which are known to have high 5mC levels (Ficz et al, 2011; Pastor et al, 2011; Williams et al, 2011). Conversely, 5hmC is enriched at promoter regions and TSSs (Ficz et al, 2011; Pastor et al, 2011; Williams et al, 2011; Wu et al, 2011b; Xu et al, 2011), whereas 5mC is generally depleted from these regulatory areas. Interestingly, at genome-wide levels, 5hmC localizes to regions with higher CpG density than 5mC (Williams et al, 2011). This is also the case for promoter regions, where 5hmC seems to associate preferentially with promoters of intermediate or high CpG content (Pastor et al, 2011; Williams et al, 2011; Wu et al, 2011b), indicating that if 5mC was present at these sites, it would be rapidly converted to 5hmC. This way, Tet1 might contribute to maintaining the hypomethylated signature of CpG islands in ESCs.
The dependency of 5hmC signal intensity on the levels of 5mC and Tet1 expression was investigated in several studies. DIP-seq of 5hmC in Dnmt triple knockout (TKO) ESCs revealed that most of the 5hmC signal was lost in cells devoid of DNA methylation, confirming that the existence of 5hmC is dependent on pre-existing 5mC (Ficz et al, 2011; Williams et al, 2011). Also, depletion of Tet1 led to a significant decrease in 5hmC levels consistent with the known enzymatic activity of Tet1 (Williams et al, 2011; Wu et al, 2011b; Xu et al, 2011). Estimations of 5hmC levels at selected 5hmC and Tet1-positive genes show 20–40% reduction in 5hmC signal after depletion of Tet1 (Williams et al, 2011; Xu et al, 2011), which is in agreement with studies on changes of global 5hmC levels in Tet1 knockout and in Tet1 shRNA-treated ESCs (Koh et al, 2011; Xu et al, 2011; Dawlaty et al, 2011). There were modest effects of Tet1 depletion on DNA methylation in ESCs. Globally, only slight increases in 5mC levels were detected after Tet1 knockdown or knockout (Dawlaty et al, 2011; Ficz et al, 2011; Williams et al, 2011; Wu et al, 2011b; Xu et al, 2011). In this regard, the relative abundance of 5mC compared with 5hmC should be noted. With an estimated 45 5hmC per 1,000 5mC in ESCs (Ito et al, 2011), changes in 5hmC levels would not lead to detectable changes in global 5mC levels. Numerous studies did, however, detect gene-specific increases in the levels of 5mC after Tet1 depletion, which was associated with a concomitant loss of 5hmC. These results show that Tet1 does indeed regulate DNA methylation levels at certain genes (Dawlaty et al, 2011; Koh et al, 2011; Williams et al, 2011; Wu et al, 2011a; Xu et al, 2011).
The fact that there is not a complete overlap in 5hmC-positive regions and Tet1 binding and the rather limited effect of Tet1 depletion on 5hmC and 5mC levels both suggest that other pathways or proteins have important roles in regulating 5hmC levels in ESCs. Considering that Tet2 is also expressed in ESCs, there could be a functional redundancy between Tet1 and Tet2, by which Tet2 might partly compensate for the loss of Tet1.
The function of Tet1 and 5hmC in transcriptional regulation
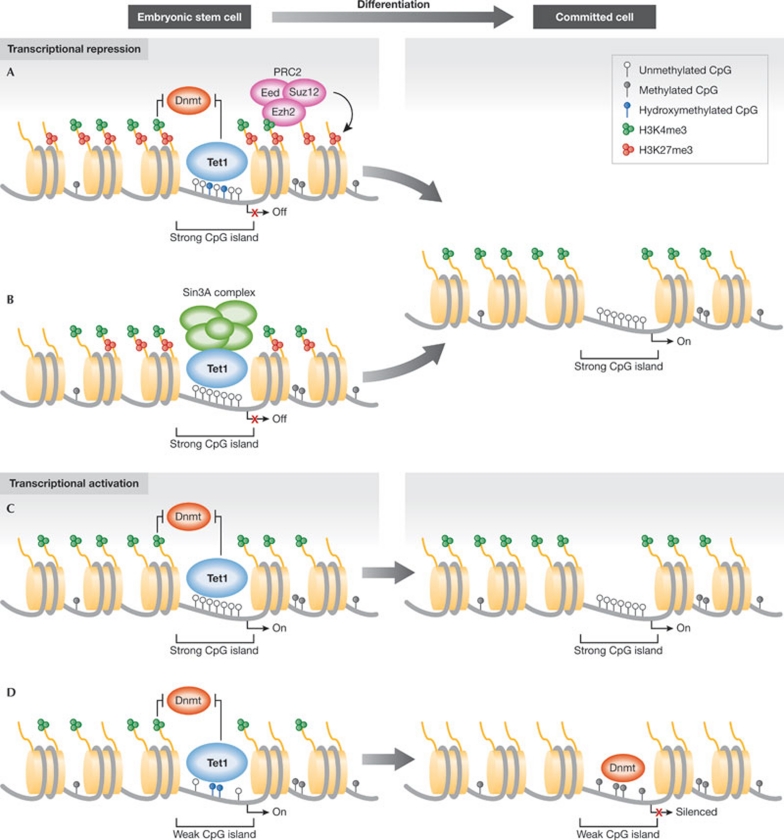
With the characterization of the enzymatic activity of TET proteins, it was anticipated that they would regulate transcription by adjusting levels of DNA methylation at promoters. This was further supported by the observation that both Tet1 and 5hmC localize to TSSs. However, gene-expression profiling of Tet1-depleted ESCs showed that less than 10% of Tet1 target genes change expression after Tet1 depletion (Williams et al, 2011; Wu et al, 2011a). Unexpectedly, it was also found that the number of genes downregulated was similar to, or even lower than, the number of genes upregulated after Tet1 depletion, indicating that Tet1 could also have repressive effects on gene transcription. In agreement with this, a large fraction of Tet1-bound promoters are also occupied by PRC2 (Williams et al, 2011; Wu et al, 2011a), and two studies found a significant enrichment of 5hmC at bivalent promoters (Pastor et al, 2011; Wu et al, 2011a). Moreover, Wu et al (2011a) showed a substantial overlap between genes that were upregulated by Tet1 depletion and genes upregulated in cells not expressing the PRC2 complex protein Eed, suggesting a potential role of Tet1 in PRC2-mediated repression (Wu et al, 2011a). They further demonstrated that the chromatin-binding ability of PRC2 members was decreased in Tet1-depleted cells, supporting a role of Tet1 in PRC2 recruitment. As a stable interaction between Tet1 and PRC2 could not be demonstrated (Williams et al, 2011; Wu et al, 2011a), Tet1 might indirectly facilitate PRC2 chromatin binding by decreasing DNA methylation levels at PRC2 target genes (Fig 2A). Moreover, by preventing stable methylation of Polycomb target genes, which are enriched for genes involved in cell-fate decisions, Tet1 might be important for keeping the plasticity of these genes during development.
Figure 2.
Potential roles of Tet1 in transcriptional regulation. (A,B) Tet1 could be involved in transcriptional repression in ESCs through the recruitment of repressive complexes. Tet1 has been proposed to facilitate PRC2 binding indirectly by reducing DNA methylation at PRC2 target genes (A). Moreover, Tet1 could mediate transcriptional repression by directly recruiting the Sin3A co-repressor complex to a subset of its target genes (B). During differentiation, Tet1 is downregulated, which would allow for the repressed genes to be activated. (C,D) Tet1 might also contribute to transcriptional activation by preventing DNA methylation. At strong CpG islands that rarely undergo DNA methylation, Tet1 binding might act as a failsafe mechanism to remove aberrant DNA methylation (C). However, Tet1 also binds to weak CpG islands that have been reported to become de novo DNA-methylated during differentiation. They often also display high levels of 5hmC, indicating that Tet1, by converting 5mC into 5hmC, ensures the timely methylation and silencing of these target genes during differentiation (D). 5hmC, 5-hydroxymethylcytosine; ESC, embryonic stem cell; PRC, Polycomb repressive complex; Tet1, ten-eleven translocation 1.
Another mechanism by which Tet1 could contribute to transcriptional repression is through its association with the Sin3A co-repressor complex (Williams et al, 2011): Tet1 has been shown to interact with the Sin3A complex; Tet1 has displayed a significant overlap with Sin3A on target genes; and the recruitment of Sin3A to a subset of these genes was dependent on Tet1 expression. Moreover, a significant overlap between genes that are de-repressed by Tet1 or Sin3A knockdown was observed, indicating that transcriptional repression by Tet1 is mediated through the recruitment of Sin3A (Fig 2B). Interestingly, increased expression of the Tet1 and Sin3A target genes were also observed when Tet1 was depleted in Dnmt TKO ESCs, suggesting that the repressive function of Tet1 is independent of its catalytic activity and indicating the possibility that TET proteins could also have catalytic independent functions.
The transcriptional activator effects of Tet1 were less profound than the observed repressive functions. No direct correlation between genes that were downregulated after Tet1 depletion and genes with profound changes in 5hmC and 5mC levels were observed (Williams et al, 2011). Furthermore, we found that most of the transcriptional activating effects of Tet1 were also detected in the Dnmt TKO cells (Williams et al, 2011), suggesting that many of these events are indirect effects of Tet1 depletion. These results are in agreement with the idea that DNA methylation at promoters acts as a mechanism that maintains inactive genes in a silenced state; however, the absence of DNA methylation does not lead to gene activation per se, but rather renders the gene permissive for activation. This is also apparent from studies comparing gene expression in wild-type and Dnmt TKO ESCs, in which only a minor fraction of the methylated genes becomes upregulated by the loss of DNA methylation (Fouse et al, 2008). However, as described above, it seems that 5hmC is enriched at several promoters with intermediate CpG content. These weak CpG islands are prone to undergo methylation during differentiation (Mohn et al, 2008; Weber et al, 2007), suggesting that Tet1 ensures that these promoters remain unmethylated in the undifferentiated ESCs. This observation indicates that, besides acting as a failsafe mechanism to prevent stochastic methylation of strong CpG islands at housekeeping genes (Fig 2C), Tet1 also protects numerous weak CpG island genes that are predisposed to undergo methylation. During differentiation, downregulation of Tet1 would allow for DNA methylation to occur at these promoters leading to transcriptional silencing (Fig 2D).
Concluding remarks and perspectives
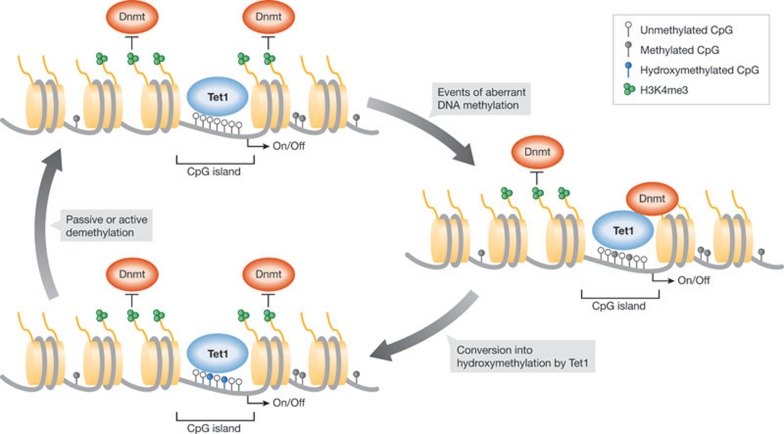
In summary, extensive studies on Tet1 and 5hmC in mouse ESCs have shed light on their functional role in regulating DNA methylation and transcription. Genome-wide identification of Tet1-binding sites illustrated that Tet1 binds to a large number of genes with the highest signal intensity around TSSs. Most Tet1-bound promoters contain a CpG island and are mainly unmethylated in ESCs, suggesting that Tet1 is a major player in maintaining CpG islands free of methylation. This is further supported by the finding that 5hmC also localizes to TSSs and is enriched in regions of higher CpG density than 5mC, indicating that 5mC is converted into 5hmC by Tet1 specifically at CpG islands. Therefore, we propose that Tet1 could be an important regulator of DNA methylation fidelity. Normally, CpG islands are thought to be protected from DNA methylation by high levels of H3K4me3 that inhibit the recruitment of de novo Dnmts; however, deregulation of H3K4 methylation or increased DNMT activity might occasionally lead to sporadic DNA methylation of CpG islands (Fig 3). We propose that the TET proteins would convert such methylation events into 5hmC, which could be followed by passive or active demethylation restoring the unmethylated state of CpG islands. These events could be difficult to detect by 5mC- and 5hmC-DIP for two reasons: first, the available antibodies have limited sensitivity and therefore the experiments are restricted to detecting regions with dense methylation or hydroxymethylation; and second, sporadic DNA methylation is predominantly a stochastic event that only occurs at the same loci in few cells in the total cell population.
Figure 3.
The function of Tet1 in DNA methylation fidelity. Tet1 associates with the CpG islands of a large number of genes. If aberrant DNA methylation occurs at these promoters, conversion into hydroxymethylation by Tet1 could facilitate passive or active DNA demethylation. In this way, Tet1 regulates DNA methylation fidelity by preventing the accumulation of aberrant DNA methylation at CpG islands.
Although transcriptional changes were observed after Tet1 knockdown, it is notable that most Tet1 target genes do not change expression status by Tet1 depletion. As mentioned previously, DNA methylation is not assumed to be an on–off switch rapidly regulating transcriptional changes, and transcriptional repression often precedes DNA methylation. Therefore, silencing of a promoter by sporadic DNA methylation would require the accumulation of DNA methylation over time. It is appealing to assume that a loss of TET proteins could contribute to the gene-specific hypermethylation that is often observed in cancer. This is strongly supported by several studies showing that human TET2 is frequently mutated in a variety of haematological malignancies (Delhommeau et al, 2009; Jankowska et al, 2009; Langemeijer et al, 2009), and that these mutations cause loss-of-function of the protein (Ko et al, 2010). Moreover, Figueroa et al demonstrated that TET2 mutations in patients with acute myeloid leukaemia were associated with a DNA hypermethylation phenotype (Figueroa et al, 2010). Recent analyses of Tet2-knockout mice provide further evidence for a role of Tet2 in the regulation of normal haematopoiesis (Ko et al, 2011; Li et al, 2011; Moran-Crusio et al, 2011; Quivoron et al, 2011). Importantly, Tet2-deficient mice also have an increased susceptibility to myeloid malignancies, suggesting a tumour suppressor function of Tet2. However, it should be mentioned that one study reported a correlation between TET2 loss-of-function and global DNA hypomethylation, which could promote cancer development (Ko et al, 2010). Therefore it cannot be ruled out that TET2 might also indirectly control DNA methylation, for instance, through regulation of DNMT activity.
In general, it is intriguing to hypothesize that loss of the TET proteins leads to the accumulation of stochastic aberrant DNA methylation at CpG islands that would occasionally provide cells with a growth advantage. This would lead to clonal expansion of the cells and could, in combination with other genomic transformations, promote cancer development. It will be important, in the near future, to directly test this hypothesis by showing a direct link between the loss of TET activity, accumulation of DNA methylation at specific promoters and subsequent cancer progression.
Jesper Christensen, Kristine Williams & Kristian Helin
Acknowledgments
We thank members of the Helin lab for discussions. The work in the Helin lab was supported by the Excellence Program of the University of Copenhagen and grants from the Danish Cancer Society, the Novo Nordisk Foundation, the Danish Medical Research Council, The Lundbeck Foundation and the Danish National Research Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Blackledge NP, Klose R (2011) CpG island chromatin: a platform for gene regulation. Epigenetics 6: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, Veenstra GJ (2009) DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma 118: 549–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 [DOI] [PubMed] [Google Scholar]
- Cortellino S et al. (2011) Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM et al. (2011) Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9: 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F et al. (2009) Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S (2006) The epigenetic progenitor origin of human cancer. Nat Rev Genet 7: 21–33 [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473: 398–402 [DOI] [PubMed] [Google Scholar]
- Figueroa ME et al. (2010) Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G (2008) Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2: 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T (2010) Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 5: e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H (2011) Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle 10: 2662–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF et al. (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, O'Keefe CL, Ganetzky R, McDevitt MA, Maciejewski JP (2009) Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood 113: 6403–6410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP (2010) Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 38: e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB (2007) The epigenomics of cancer. Cell 128: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M et al. (2010) Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A (2011) Ten-eleven-translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA 108: 14566–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP et al. (2011) Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8: 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM et al. (2009) Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 41: 838–842 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Li Y et al. (2010) The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol 8: e1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai C, Wang J, Zhang W, Petersen BE, Yang FC, Xu M (2011) Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118: 4509–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R et al. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A et al. (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D (2008) Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K et al. (2011) Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Ooi SK et al. (2007) DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M (2009) Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep 10: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA et al. (2011) Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473: 394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T (2011) The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl 50: 7008–7012 [DOI] [PubMed] [Google Scholar]
- Quivoron C et al. (2011) TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20: 25–38 [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J (2001) Epigenetic reprogramming in mammalian development. Science 293: 1089–1093 [DOI] [PubMed] [Google Scholar]
- Sharif J et al. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912 [DOI] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H (2010) Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res 38: e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A et al. (2006) Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11: 805–814 [DOI] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC (2007) Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res 67: 946–950 [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC (2004) Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res 32: 4100–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Schubeler D (2007) Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin Cell Biol 19: 273–280 [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D (2007) Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K (2011) TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y (2011) Tet1 and 5-hydroxymethylation: A genome-wide view in mouse embryonic stem cells. Cell Cycle 10: 2428–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y (2011a) Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y (2011b) Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25: 679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y et al. (2011) Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42: 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG (2010) TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res 20: 1390–1393 [DOI] [PubMed] [Google Scholar]