EMBO Reports (2011) 12: 11, 1127–1134. doi:; DOI: 10.1038/embor.2011.190
Animal cells use a group of innate immune sensors to detect viral invasion. The RIG-I-like receptor (RLR) family of RNA helicases is largely responsible for detecting replicating viral RNAs in the cytosol of most, if not all, cells infected with RNA viruses (Rehwinkel & Reis e Sousa, 2010). Two RLRs, RIG-I and MDA5, recognize different types of viral RNA, and thus launch antiviral immune responses against different families of RNA virus. RLRs belong to the helicase superfamily 2 (SF2), and have conserved motifs in their two SF2 domains (Hel1 and Hel2), as well as RNA-dependent ATPase activity (Fairman-Williams et al, 2010). The binding of viral RNA to the carboxy-terminal domain (CTD) of RLRs has been proposed to trigger a conformational change that exposes the amino-terminal caspase activation and recruitment domains (CARDs), which then activate the mitochondrial adaptor protein MAVS to induce type-I interferons and other antiviral molecules. Four recent studies, including one published in a recent issue of EMBO reports by the Hopfner group, provide a detailed view of how RIG-I binds to RNA and the conformational changes that lead to its activation (Fig 1; Civril et al, 2011; Jiang et al, 2011; Kowalinski et al, 2011; Luo et al, 2011).
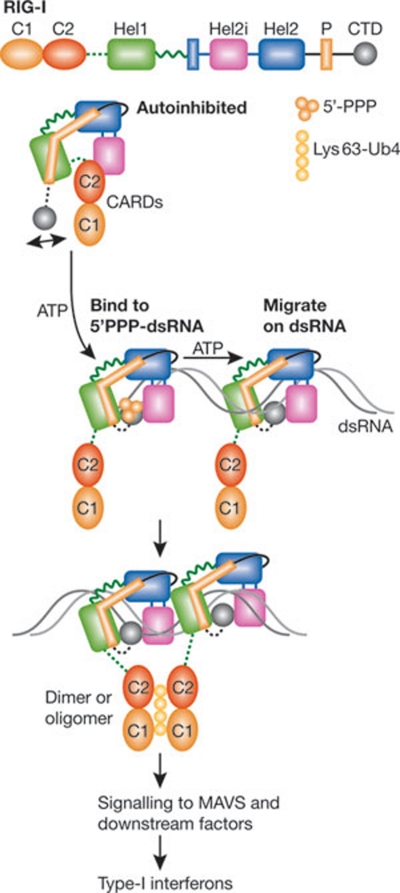
Figure 1.
Structural insights on dsRNA-mediated activation of RIG-I. RIG-I is an intracellular nanomachine that recognizes viral RNAs and elicits an innate immune response. In the absence of viral infection, RIG-I is autoinhibited, and the access of the CARDs, which are important for signal transduction, is limited sterically by the neighbouring helicase insertion domain (Hel2i) and the C2–Hel1 linker. The CTD recognizes 5′-triphosphate in dsRNA and initiates the binding of the helicase domains to the first turn of the double helix, where an ATP-binding site is assembled. ATP hydrolysis is required for the migration of RIG-I along the duplex RNA. The liberated CARDs bind Lys 63 polyubiquitin chains and stimulate the massive aggregation of the mitochondrial protein MAVS, which ultimately leads to the induction of type I interferons and other antiviral molecules. C1/C2, CARD1/CARD2, caspase activation and recruitment domain 1/2; CTD, carboxy-terminal domain; dsRNA, double-stranded RNA; Hel1/Hel2, helicase 1/helicase 2; P, pincer domain.
…full-length RIG-I structure in the absence of RNA [and] the helicase structures in RNA-bound states reveal clear snapshots of the RNA-induced conformational rearrangement
Although using different fragments of the RNA sensor, all four groups determined the crystal structures of RIG-I helicase domains. In addition, Kowalinski and colleagues obtained the structure of full-length RIG-I from duck in the ligand-free state. Comparison of the full-length RIG-I structure in the absence of RNA with the helicase structures in RNA-bound states reveals clear snapshots of the RNA-induced conformational rearrangement. However, before we discuss these beautiful structures, it is important to note that the RNA ligands able to activate RIG-I to induce type I interferons usually contain 5′-triphosphate (5′-ppp), a signature present in replicating viral RNAs but not in host cellular RNAs, which normally contain 5′-modifications—such as a 5′-cap in mRNA (Rehwinkel & Reis e Sousa, 2010). The RNAs used in the structural studies are short double-stranded RNAs (dsRNAs) lacking 5′-ppp, and probably would not induce type I interferons. In addition, we still lack a crystal structure of RNA-bound full-length RIG-I. Therefore, the recent structures of RNA-bound RIG-I helicase might not represent its active form. Furthermore, even after bound to 5′-ppp dsRNA, RIG-I remains inactive until its N-terminal CARDs bind to Lys 63-linked polyubiquitin chains that are not anchored to any cellular protein (Zeng et al, 2010). Thus, to distinguish the dsRNA-bound RIG-I from fully active RIG-I, we refer to the conformation of dsRNA-bound RIG-I as the ‘competent state’.
In the absence of RNA, full-length RIG-I adopts an autoinhibited conformation (Kowalinski et al, 2011). Two CARDs, joined head-to-tail, form one rigid unit that is attached to a unique insertion domain in Hel2 (Hel2i) through an extensive interaction interface between Hel2i and the second CARD (C2). The C2–Hel2i interaction on the two apposing surfaces is mediated by salt bridges and hydrophobic interaction patches and is therefore energetically stable. Neither the structure of the CARDs nor of the Hel2i seems to change upon interaction. This interface has significant overlap with the dsRNA–Hel2i interaction interface, suggesting that during the RNA-triggered transition, extra energy is required to disrupt the C2–Hel2i interface and enable a dsRNA to enter its binding site. On the other hand, the ubiquitin E3 ligase TRIM25, which synthesizes Lys 63 polyubiquitin chains and is required for RIG-I activation, has been shown to interact with the first CARD (C1) from the same side where Hel2i contacts the C2 domain, suggesting that Hel2i could exert steric hindrance to polyubiquitination. Furthermore, a key residue for both polyubiquitination and polyubiquitin binding, Lys 172, is located at the aqueous surface that faces the Hel1 domain, with the initial segment of the C2–Hel1 linker positioned above it so that Asp 186 can form a salt bridge with Lys 172. This arrangement led the Cusack group to propose that both Hel2i and the C2–Hel1 present strong steric hindrance at Lys 172 (Kowalinski et al, 2011). Because the binding of Lys 63 polyubiquitin is critical for RIG-I to reach a fully active state, the spatial constraints to the CARDs seem to be crucial for keeping the ligand-free RIG-I inhibited. In support of this model, a point mutation at Phe 540, located at the C2–Hel2i interface, renders RIG-I constitutively active. Notably, Phe 540 is not conserved in MDA5, which might thus be regulated through a different mechanism.
The autoinhibited RIG-I maintains a high degree of flexibility that probably facilitates its surveillance for viral RNAs. The ligand-free helicase domain has significant flexibility, with the Hel1 domain wobbling around the Hel2/Hel2i. Attachment of the CARDs to the Hel2i does not seem to impose much restriction to Hel1 due to the long C2–Hel1 linker. Similarly, the CTD at the end of the second pincer helix was disordered in the crystal structure of the full-length RIG-I, and should have a high degree of flexibility in the ligand-free state. As the receptacle of the RIG-I sensor, the CTD is therefore free to sample the surrounding space for viral RNAs, and binding of a viral RNA to the CTD will lead to a transitory RNA-bound autoinhibited state, which quickly switches to the more stable competent state. As major conformational changes in Hel1 and Hel2—as well as the disruption of the C2–Hel2i interaction—are required for exiting the autoinhibited state, the significant energetic gain from the protein–dsRNA interactions must be consumed to overcome the transition energy barrier and stabilize the competent state. Additionally, because the conformational switch to the competent state is accompanied by the assembly of individual motifs into an active ATPase/helicase, ATP binding might further shift the balance toward the competent state.
The RNA-bound RIG-I helicase structure (competent state) shows an extensive reorganization from the autoinhibited conformation. The two helicase-CTD–dsRNA structures show that CTD and helicase form a ring around the duplex RNA (Jiang et al, 2011; Luo et al, 2011). If the structures of the helicase–RNA complex and the helicase alone are aligned using Hel2i as the static reference, the Hel1 domain swings about 50° and its centre of weight moves almost 35 Å so that, in its new position, it interacts with the backbone of dsRNA (Fig 1). Because the second helix of the pincer domain lies on the surface of Hel1, the RNA-induced relocation of Hel1 makes the intersecting angle between the two pincer helices substantially smaller than that in the ligand-free state. All these changes create an active ATPase and lead to close protein–RNA contacts.
The CTD of RIG-I has been shown to bind 5′-ppp of dsRNA, which triggers the translocation of RIG-I along dsRNA in an ATP-dependent manner (Myong et al, 2009). The structures show that the presence of the CTD in the RNA-interacting ring is necessary for the helicase domain to be able to translocate along the 3′-strand. In the two helicase-CTD–dsRNA structures, the CTD binds at the 5′-terminus and therefore needs to switch away from its bound position for RIG-I to initiate translocation. Although RIG-I has a helicase domain, the Marcotrigiano group compared their helicase-CTD–dsRNA structure with that of the hepatitis C virus NS3 helicase–single-stranded DNA complex and saw that RIG-I lacks the unwinding phenylalanine-loop motif, suggesting that it might not have unwinding activity (Jiang et al, 2011). On the basis of these observations, it is likely that, in the translocating mode, the helicase domain maintains the contact with the 3′-strand and the CTD keeps its interaction with the 5′-strand, so that the helicase-CTD ring will slide along the 3′-strand even when there is no unwinding of the duplex.
The CARDs in the competent state of RIG-I are accessible for binding Lys 63 polyubiquitin, which then induces massive aggregation of MAVS, an event that activates and propagates the antiviral signalling cascade (Hou et al, 2011). Although the structures of RNA-bound RIG-I helicase domains show no evidence of RIG-I dimerization or oligomerization, it remains possible that RIG-I could dimerize or oligomerize in the presence of a 5′-ppp RNA that is able to induce interferons. Alternatively, many RIG-Is might bind to long dsRNA as beads on a string. Because the RIG-I footprint is ∼9 nucleotides, close to one helical repeat of a duplex, even without protein–protein interaction, adjacent translocating RIG-I molecules might be approximately parallel to each other and their CARDs could interact with Lys 63 polyubiquitin chains (Fig 1), further strengthening the RIG-I oligomers. These RIG-I oligomers might be more effective in promoting MAVS aggregation through CARD interaction. Considering the RIG-I footprint on dsRNA, it is noteworthy that a minimum of 19 bp of dsRNA containing 5′-ppp is required to induce type I interferons (Schlee et al, 2009).
RIG-I and its SF2 members have multiple conserved sequence motifs (Fairman-Williams et al, 2010). It is thus likely that the structural description for a ligand-dependent conformational switch in the RIG-I helicase domain will have general implications to the SF2 superfamily. In fact, there are more than 30 SF2 helicases, which include Ago2, Dicer, the Swi2/Snf2 family and DEAD-box family. The structural insights gained from the RIG-I helicase domain will provide a framework for understanding the SF2 helicase functions in RNA interference, chromatin remodelling, DNA repair and other important cellular activities.
References
- Civril F et al. (2011) EMBO Rep 12: 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther U-P, Jankowsky E (2010) Curr Opin Struct Biol 20: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F et al. (2011) Cell 146: 448–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F et al. (2011) Nature 479: 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E et al. (2011) Cell 147: 423–435 [DOI] [PubMed] [Google Scholar]
- Luo D et al. (2011) Cell 147: 409–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S et al. (2009) Science 323: 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Reis e Sousa C (2010) Science 327: 284–286 [DOI] [PubMed] [Google Scholar]
- Schlee M et al. (2009) Immunity 31: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W et al. (2010) Cell 141: 315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]