Abstract
The development of the central nervous system (CNS) relies on precisely orchestrated gene expression regulation. Dysregualtion of both genetic and environmental factors can affect proper CNS development and results in neurological diseases. Recent studies have shown that similar to protein coding genes, noncoding RNA molecules have a significant impact on normal CNS development and on causes and progression of human neurological disorders. In this review, we have highlighted discoveries of functions of noncoding RNAs, in particular microRNAs and long noncoding RNAs, in neural development and neurological diseases. Emerging evidence has shown that microRNAs play an essential role in many aspects of neural development, such as proliferation of neural stem cells and progenitors, neuronal differentiation, maturation and synaptogenesis. Misregulation of microRNAs is associated with some mental disorders and neurodegeneration diseases. In addition, long noncoding RNAs are found to play a role in neural development by regulating expression of protein coding genes. Therefore, examining noncoding RNA-mediated gene regulations has revealed novel mechanisms of neural development and provided new insights into the etiology of human neurological diseases.
Keywords: neural development, mental disorders, neurodegeneration diseases, noncoding RNAs, microRNAs (miRNAs), long noncoding RNAs (lncRNAs), neural stem cells
Introduction
Noncoding RNAs (ncRNAs) are functional RNA molecules that do not show protein translation capability. ncRNAs consist of ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), piwi-interacting RNAs (piRNAs), microRNAs (miRNAs), and long noncoding RNAs (lncRNAs) and so on. These ncRNAs have shown to play distinct but also conserved roles in normal development and pathological processes in invertebrates and vertebrates. Studies have reported that many miRNAs as well as lncRNAs are specifically expressed in the central nervous system (CNS) [1–5]. In this review, we will focus on revealing the functions of miRNAs and lncRNAs in the CNS development and neurological disorders.
miRNAs are ~22 nucleotides (nt) highly conserved small noncoding RNAs found in almost all eukaryotic cells [6]. Like coding genes, miRNAs are mainly transcribed by the RNA Polymerase II (Pol II) into long primary miRNA transcripts (pri-miRNAs), while a subset of miRNAs has been shown to be transcribed by RNA Pol III [7]. Pri-miRNAs are cleaved by Drosha, a class 2 RNase III enzyme, to produce short hairpin stem-loop structures, known as precursor miRNAs (pre-miRNAs) [8]. Pre-miRNAs are then processed into about 20–25 nucleotides double-stranded mature miRNAs by RNase III enzyme Dicer [9]. The duplex undergoes unwinding and the strand with the weakest base pairing at its 5′ terminus, together with Dicer and many associated proteins including Argonaute proteins, form an active RNA-induced silencing complex (RISC) [10, 11]. A miRNA recognizes and binds to the non-coding portion of target messenger RNAs (mRNAs), typically in the 3'-untranslated region (3'-UTR), by imperfect complementary sequence recognition, particularly the seed sequence of the mature miRNA [11, 12]. Once bound, the RISC can negatively regulate miRNA targets by degrading the mRNA or repressing its translation [6, 13]. miRNAs are able to target numerous mRNAs and so their regulatory potential is large. Moreover, several evidences indicate regulatory feedback loop between miRNA and their targets [14].
miRNAs are classified into intergenic and intragenic miRNAs, depending on their genomic location [15–17]. Intergenic miRNAs are located at intergenic regions (between genes). These miRNAs are usually transcribed using their own promoters. Intragenic miRNAs normally lie in intronic regions of coding genes with the same orientation. They are often transcribed together with the host genes. Additionally, some miRNAs can be co-transcribed as clusters [18].
Emerging evidence has demonstrated potential functions of lncRNAs in many biological events. The majority of the human genome has previously been considered as “junk” DNA, since only about 1.5% of the human genome, which occupies over 3 billion DNA base pairs, consists of protein coding genes [19]. A recent study of a large-scale complementary DNA (cDNA) sequencing project has shown that only one fifth of transcripts of the human genome are protein-coding genes, while the rest four fifths are RNA transcripts that don’t encode proteins [20]. These RNA transcripts are normally longer than 200 nucleotides, thus they are called lncRNAs. Except no open reading frame (ORF) found within lncRNAs, they share many features with coding mRNAs such as 5’ capping, and usually contain exons and introns [21]. However, some studies point out that lncRNAs may or may not be polyadenylated, and are often transcribed from either strand within a protein-coding gene [22]. Because of the huge number and different features of genomic location and orientation of lncRNAs, the potential functions of lncRNAs haven’t yet been well characterized. However, it is unlikely that lncRNAs are “junk” transcripts. lncRNAs have been shown to modulate gene transcription via different mechanisms. For example, some cis-antisense lncRNAs complementary to protein coding transcripts regulate expression of the coding genes [23–27]; some lncRNAs modulate the function of transcription factors by acting as co-regulators [26, 28–32]. Moreover, some studies have shown that lncRNAs are involved in epigenetic regulation such as chromatin and histone modification and X-chromosome inactivation [33–39].
Therefore, noncoding RNAs such as miRNAs and lncRNAs participate in regulating gene expression in many ways. The underlying mechanisms of noncoding RNA functions in normal development and under disease conditions are beginning to be uncovered.
miRNA functions in the CNS development, a Dicer ablation approach
Since Dicer is the key enzyme in miRNA processing, several studies have reported the global effects of miRNAs in the CNS development by ablating Dicer and in turn blocking biogenesis of all miRNAs in specific regions and at different developing stages in the CNS using tissue specific Cre lines.
Conditional deletion of Dicer from the mouse cerebral cortex using the Emx1-Cre line results in a significant reduction in cortical size, which is caused by reduced neural stem cell and neural progenitor pool, increased apoptosis and impaired neuronal differentiation [40–43]. Dicer ablation from the mouse CNS using the Nestin-Cre line has confirmed the cortical defects and revealed abnormal expansion and differentiation of oligodendrocytes in the spinal cord [43]. Dicer deletion from postmitotic neurons in the cortex and the hippocampus using calmoduln kinase II (CaMKII) promoter-driven Cre transgenic mice results in smaller cortex, enhanced cortical cell death, impaired axonal path finding and dendritic branching in the hippocampal CA1 region [44]. Ablation of Dicer in the hippocampus at different time points using Emx1-Cre, Nestin-Cre and Nex-Cre lines has revealed miRNA functions in hippocampal progenitor proliferation and differentiation [45]. Conditional deletion of Dicer in the dopaminoceptive neurons in the basal ganglia using a dopamine receptor-1 (DR-1) Cre line causes a reduced brain size and mass, and smaller striatal medium spiny neurons [46].
Since Wnt1-Cre is active in neural crest cells, apoptosis of neural crest -derived cells, and subsequently a loss of the enteric, sensory and sympathetic neurons are reported in Wnt1-Cre Dicer conditional knockout mice [47]. Another investigation of Wnt1-Cre mediated Dicer ablation has found malformation of the midbrain and cerebellum, reduced dopminergic neuron differentiation and malformation of the dorsal root ganglia [48]. Ablation of Dicer using the Olig1-Cre line does not affect motor neuron development, but reduces the number of astrocytes and oligodendrocyte progenitor cells (OPCs) in the spinal cord, and results in a great reduction of mature oligodendrocytes in the brain and an impaired myelination [49, 50]. Similarly, Dicer ablation using the 2’,3’-cyclic nucleotide 3’ phosphodiesterase (Cnp)-Cre line, which is active in the oligodendrocyte lineage, causes a great reduction of OPCs and mature oligodendrocytes [51].
Conditional ablation of Dicer in specific regions in the CNS and in distinct cell lineages has demonstrated the importance of the miRNA-mediated pathway in the CNS development and in differentiation of different cell types. However, because Dicer is also involved in maintaining the heterochromatin assembly, likely by the short interfering RNA (siRNA) pathway, the CNS defects in Dicer knockout mice need to be carefully interpreted [52, 53]. Examining functions of individual miRNAs will help reveal precise roles of miRNAs in the CNS development.
miRNA functions in the development of neural stem cells and progenitors
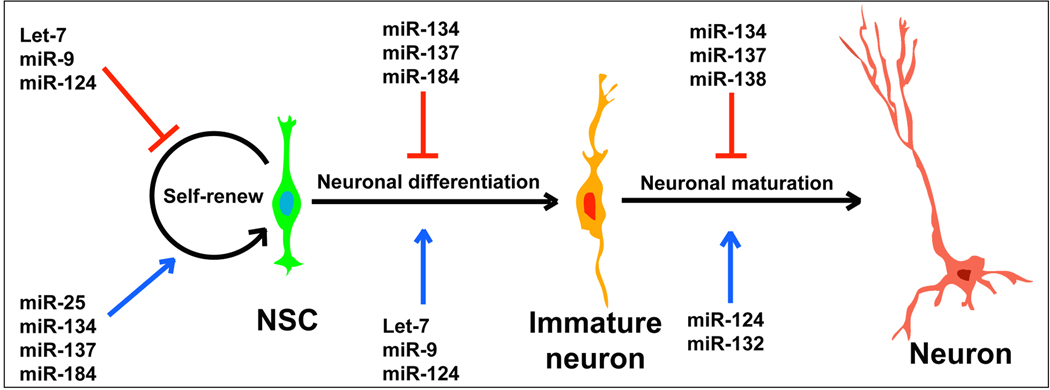
Neural stem cells (NSCs) and neural progenitors (NPs) are identified in both embryonic and adult CNS in mammals [54]. NSCs can self-renew and differentiate into multiple lineages such as neurons, astrocytes and oligodendrocytes [54, 55]. Both intrinsic factors, for instance transcription factors, epigenetic and post-transcriptional regulators, and extrinsic signals from the stem cell microenvironment play important roles in regulating self-renewal and differentiation of NSCs [55–57]. Several reports have shown that miRNAs are also essential for the development of NSCs and NPs in the CNS in vivo and in cultures (Fig. 1).
Figure 1.
A list of representative miRNAs that function on neural stem cell (NSC) self-renewal, neuronal differentiation and neuronal maturation.
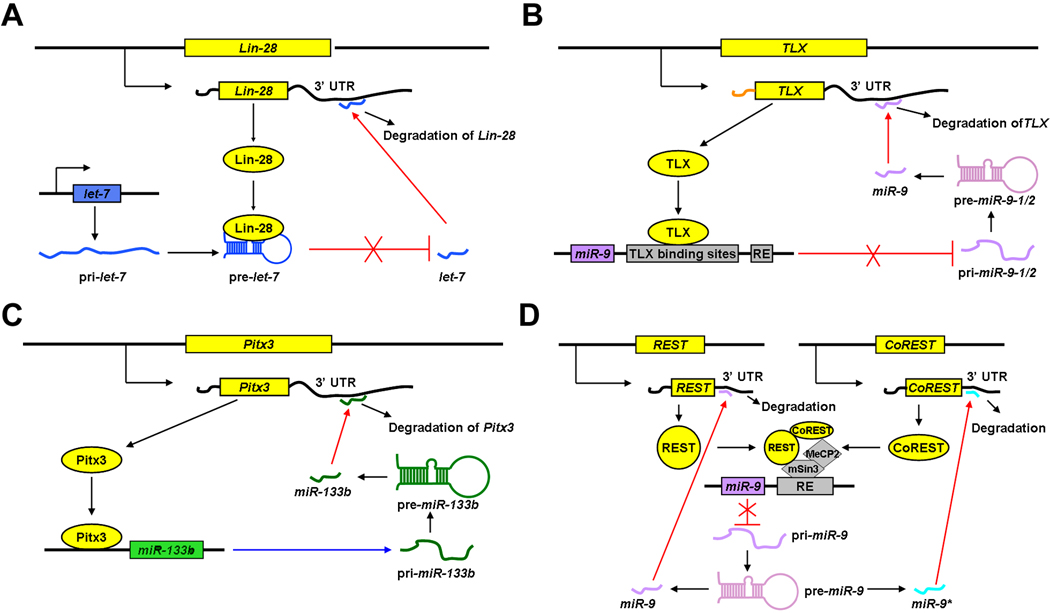
let-7, the first identified miRNA [58], has been shown to regulate NSC proliferation and differentiation by targeting the nuclear receptor TLX and the cell cycle regulator cyclin D1 [59]. Over-expression of let-7b inhibits NSC proliferation and enhances differentiation, while knockdown of let-7b promotes NSC proliferation [59]. It appears that the expression levels of let-7 in NSCs are controlled by a feedback regulation of Lin-28, a pluripotency factor that controls miRNA processing in NSCs [60]. Lin-28 binds to the let-7 precursor and inhibits its processing by Dicer. On the other hand, the expression of Lin-28 is repressed by let-7 and miR-125, allowing the maturation of let-7. This feedback loop reveals an autoregulation between miRNA let-7 and miR-125, and the transcription factor Lin-28 during NSC development [60] (Fig. 2A).
Figure 2.
A feedback regulation of miRNAs and their target genes. Protein coding genes inhibit (A, B, D) or promote (C) biogenesis of miRNAs, while miRNAs post-transcriptionally silence output of these proteins.
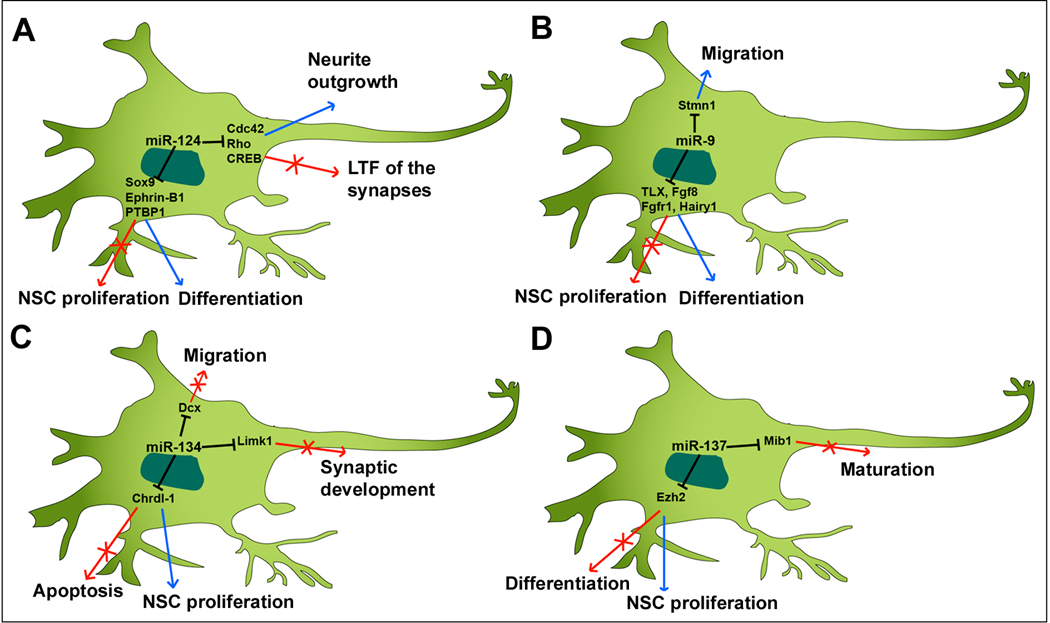
miR-124 is identified as a brain-enriched miRNA and its expression is upregulated during neuronal differentiation [61]. Over-expressing miR-124 in cultured NSCs and in embryonic cortical NPs using lenti-virus and in utero electroporation, respectively, promotes neurogenesis and stimulates cortical progenitor migration [62]. In the adult brain, NSCs are identified in the subventricular zone (SVZ). In cultured adult NSCs derived from the SVZ and in the SVZ in vivo, knocking down miR-124 causes an increase of dividing cells and a decrease of postmitotic neurons, while over-expressing miR-124 enhances neuronal differentiation [63]. Moreover, miR-124 regulates neuronal differentiation by suppressing Sox9 expression in adult NSCs [63]. A recent study has shown that miR-124 regulates neuronal differentiation through a mutual inhibition mechanism of Ephrin-B1 [64]. In addition, miR-124 promotes differentiation of NPs by modulating a network of nervous system-specific alternative splicing through suppressing expression of PTBP1, which encodes a global repressor of alternative pre-mRNA splicing [65]. Together, miR-124 plays a general role in promoting differentiation of embryonic and adult NSCs and NPs. It appears that miR-124 executes its function through repressing various targets (Fig. 3A).
Figure 3.
miRNAs, such as miR-124 (A), miR-9 (B), miR-134 (C) and miR-137 (D), control proliferation and apoptosis of neural stem cells and progenitors, neuronal differentiation, migration, and maturation and synaptogenesis by targeting various genes.
miR-9 is another CNS-enriched miRNA. miR-9 is shown to inhibit NSC proliferation but promote differentiation through a feedback regulation of a nuclear receptor TLX [66] (Fig. 2B). In human embryonic stem cell (ESC) derived NPs, miR-9 is shown to have a positive effect on proliferation and to promote migration by directly targeting Stmn1, which increases microtubule instability [67]. The opposite effect of miR-9 on proliferation is perhaps caused by differential physical contacts of miR-9 with target genes and the different culture systems.
In the CNS of Xenopus, miR-9 function is required for the development of the forebrain and hindbrain through different mechanisms: progenitors lacking miR-9 in the forebrain undergo apoptosis, but miR-9 deficient progenitors in the hindbrain fail to exit the cell cycle [68]. In zebrafish, miR-9 promotes differentiation of NPs that give rise to neurons at the midbrain-hindbrain domain and controls the organization of the midbrain-hindbrain boundary by targeting several genes in the Fgf signaling, such as fgf8-1 and fgfr1 [69]. In the chick spinal cord, miR-9 defines a subtype of motor neurons that project axons to the axial muscles by specifically targeting FoxP1 [70]. In the mouse brain, miR-9 function is demonstrated by the generation of miR-9-2 and miR-9-3 double knockout mice. miR-9 double mutants show reduced cortical layers, disordered migration of interneurons, and misrouted thalamocortical axons and corticofugal axon projections, suggesting an important role of miR-9 in NP proliferation, differentiation and migration during brain development [71]. Moreover, it appears that miR-9 regulates multiple target genes, including Foxg1, Pax6 and Gsh2, which have shown to be essential in cortical development [71]. Therefore, miR-9 plays an important role in many aspects of the CNS development, such as neuronal differentiation and migration, and cell type specification (Fig. 3B).
The above studies have demonstrated that the major role of let7, miR-124 and miR-9 is to induce differentiation of NSCs and NPs into specific cell types. miRNAs that promote proliferation of NSCs and NPs have also been identified (Fig. 1). miR-134 plays a role in enhancing proliferation of cortical NPs by targeting doublecortin (Dcx) and/or Chordin-like 1 (Chrdl-1) [72] (Fig. 3C). miR-25 is shown to be a major player in the miR-106-25 cluster in neural development. Overexpression of miR-25 but not miR-106b and miR-93 promotes adult NP proliferation [73]. Interestingly, the expression of the miR-106-25 cluster is regulated by FoxO3, a transcription factor maintaining the NSC population [74].
miRNAs regulate target gene expression at a post-transcriptional level. Recent work has shown that miRNAs themselves are also controlled by epigenetic regulators in the NSC development. miR-137, whose expression is co-regulated by DNA methyl-CpG-binding protein (MeCP2) and transcriptional factor Sox2, is shown to modulate adult NSC proliferation and cell fate determination by targeting Ezh2, a histone methyltransferase and Polycomb group protein [75] (Fig. 3D). Ectopic expression of miR-137 in adult NSCs enhances the proliferation, while knockdown of miR-137 promotes the differentiation of adult NSCs [75]. miR-184 expression is suppressed by methyl-CpG binding protein 1 (MBD1), and miR-184 promotes adult NSC proliferation by repressing the expression of Numb-like (Numbl) [76].
Taken together, self-renewal, proliferation and differentiation of NSCs and NPs are controlled by complex gene regulation networks that consist of both protein coding genes and noncoding miRNAs. During proliferation and differentiation of NSCs and NPs, one miRNA can have multiple target genes (Figs. 1 and 3). The availability of physical contacts and the binding affinity of a miRNA and its targets perhaps determine interactions of the miRNA with the specific targets. The interactions of miRNAs and their target genes eventually produce proper protein output of key factors that control precise specification of distinct cell types in different regions of the CNS, and ensure accurate CNS functions.
miRNA regulation of neuronal maturation and synapse formation
Neuronal maturation, neurite outgrowth and synaptogenesis allow the assembly of a functional CNS to control complex behaviors. miRNAs have been shown to play a critical role in these progresses.
miR-124 functions not only in neuronal differentiation but also in neurite outgrowth and synaptogenesis (Fig. 3A). In primary cortical neurons, ectopic miR-124 expression enhances neurite outgrowth [77]. Cdc42 and Rac1 belong to the Rho family and have been shown to regulate actin filaments and microtubules [78]. miR-124 regulates neurite outgrowth by inhibiting Cdc42 expression and altering the localization of Rac1 in mouse P19 embryonic carcinoma cells [77]. In the nervous system of Aplysia californica, miR-124 is highly expressed in the sensory-motor synapse [79]. Over-expression of miR-124 results in a remarkable reduction of long-term facilitation (LTF) of the synapses, while inhibition of miR-124 shows the opposite phenotype. The regulation of miR-124 on long-term plasticity is through targeting cAMP response element-binding (CREB), a cellular transcriptional factor [79]. Furthermore, miR-124a is shown to be required for hippocampal axogenesis through suppression of LIM homeobox 2 (Lhx2) [80].
miR-137 has been reported to promote NSC proliferation and inhibit neuronal differentiation [75]. A recent study has shown that miR-137 negatively regulates neuronal maturation (Fig. 3D). Ectopic expression of miR-137 inhibits neuronal maturation, dendritic morphogenesis and spine development both in cultured hippocampal neurons and in mouse brains. Conversely, lack of miR-137 promotes neuronal maturation [81]. miR-137 regulates neuronal maturation by targeting Mind bomb 1 (Mib1), an ubiquitin ligase, since ectopic expression of Mib1 enhances dendritic elongation and rescues the defects in neuronal maturation caused by miR-137 [81].
miR-133b is specifically expressed in dopaminergic neurons (DNs) in midbrains and is deficient in the midbrain of Parkinson’s disease patients [82]. miR-133b is shown to inhibit the maturation and function of DNs by targeting paired-like homeodomain transcription factor Pitx3. In Pitx3 deficient mouse model, miR-133b expression is significantly reduced; suggesting that miR-133b expression is under the regulation of Pitx3 [82, 83]. These reports indicate a negative feedback regulation between Pitx3 and miR133b in balancing the development of DNs [82] (Fig. 2C).
Several studies have shown the importance of miRNAs in synaptogenesis. The miR-132/miR-212 cluster is reported to mediate the dendritic growth and spine formation [84]. Conditional deletion of the miR-132/miR-212 cluster results in a great reduction in the dendrite length and spine density. Additionally, miR-132 over-expression transgenic mice show decreased dendritic spine density and impaired novel object recognition memory [85]. As a predicted target of miR-132, MeCP2 is down-regulated in miR-132 transgenic mice [85]. Furthermore, miR-132 is identified as a fragile X mental retardation protein (FMRP)-associated miRNA, and affects neuronal morphology and modifies synaptic strength in a FMRP-dependent manner [86].
miR-134 is a brain-specific miRNA and is localized to the synapto-dendritic compartment of rat hippocampal neurons. miR-134 is found to negatively regulate the size of dendritic spines, which are the postsynaptic sites of excitatory synaptic transmission, by targeting and repressing Limk1 [87] (Fig. 3C). Based on a large-scale screening in the synapto-dendritic compartment in primary hippocampal neurons, miR-138 is found to be highly expressed in synaptosomes, a biochemical fraction that is important for synaptic protein synthesis and storage [88]. In a single cell fluorescent sensor assay, inhibition of miR-138 leads to a significant increase of spinal volume without changing the synaptic puncta number [89]. These functional regulations of miR-138 on synaptic development and synaptic transmission are related to its target acyl-protein thioesterase 1 (APT1), whose activity is required for spine enlargement [89].
Functions of miRNAs in neurite outgrowth and synaptogenesis suggest that miRNAs may directly regulate local translation of target proteins that are essential for these biological events [90]. The transportation of miRNAs to a subcellular location in neurons, and the turnover of miRNAs on the binding sites of target genes during neuronal maturation and synaptogenesis still remain interesting research topics.
miRNAs in neurological disorders
Misregulation of miRNAs is associated with the pathogenesis of different human neurological diseases such as neurodevelopmental disorders, neurodegeneration diseases and affective mental disorders (Table 1).
Table 1.
miRNAs involved in neurological disorders.
| Neurological disorders |
miRNAs | Proven targets |
Potential mechanisms | References |
|---|---|---|---|---|
| Tourette's syndrome | miR-189 | SLITRK1 | Mutation at the binding site for miR-189 in the target 3’UTR | [91] |
| Fragile X syndrome | miR-19b, miR-302b*, miR-323-3p | FMRP | Repression of FMR1 expression level | [93] |
| Schizophrenia | miR-181b, miR-219 |
VSNL1, GRIA2#CaMKIIγ | Upregulation of miR-181b in Schizophrenia NMDA signaling and miR-219 forming a feedback loop in Schizophrenia |
[98] [99] |
| Alzheimer's disease (AD) | miR-107 miR-29a/b miR-298, miR-328 miR-101 miR-146a |
BACE1 BACE1 BACE1 APP CFH |
Downregulation of miR-107 and increase of its target BACE1 in AD Downregulation of miR-29a/b and increase of its target BACE1 in AD Repression of BACE1 expression and accumulation of Aβ Repression of APP expression and accumulation of Aβ Downregulation of miR-146a in AD |
[105] [106] [108] [109] [110] |
| Huntington's disease (HD) | miR-9 miR-9* miR-34b |
REST CoREST ? |
Upregulation of miR-9/9* in HD A potential biomarker for HD |
[114] [115] |
| Parkinson’s disease (PD) | miR-7 miR-153 miR-433 |
α-synuclein FGF20 |
Repression of α-synuclein accumulation A SNP in the miR-433 binding site of FGF20 3’UTR in AD |
[116] [118] |
| Prion disease | miR-191 miR-342-3p, miR-320, let-7b, miR-328, miR-128, miR-139-5p, miR-146a, miR-337-3p, miR-338-3p |
EGR1 ? |
De-regulation after infection with mouse-adapted scrapie | [119] |
| Down’s syndrome (DS) | miR-155, miR-802 miR-125b |
MeCP2 CDKN2A |
Upregulation of miR-155 and miR-802 in DS and suppression of MeCP2 Upregulation of miR-125b in DS and suppression of CDKN2A |
[124] [125] |
| Stroke | miR-21 miR-124a |
FASLG JAG1 |
Upregulation of miR-125b in Stroke and suppression of FASLG Downregulation of miR-124a after Stroke |
[127] [128] |
| Epilepsy | miR-146a | ? | Upregulation after TLE | [129] |
| Depression and bipolar disorder (BD) | miR-182 miR-30e miR-134 miR-16 miR-34a |
ADCY6, CLOCK, DSIP ? ? SERT GRM7 |
Polymorphism in the rs76481776 of the pre-miR-182 Polymorphism in the ss178077483 of the pre-miR-30e A potential biomarker for bipolar disorders Upregulation of miR-16 after SSRI treatment Downregulation of miR-34a after treatment using mood stabilizers |
[130] [131] [133] [134] [135] |
miRNAs have been proven to play a critical role in the etiology of human neuropsychiatric disorders. Variations of the SLITRK1 gene in the 3’-UTR binding site for human miR-189 is detected in DNA samples of patients with Tourette's syndrome (TS), a developmental neuropsychiatric disorder [91]. The Fragile X syndrome, a common human mental retardation, is caused by a loss of RNA-binding protein FMRP. FMRP is found to be associated with the miRNA processing pathway [92]. miR-19b, miR-302b* and miR-323-3p have been reported to repress the expression of FMRP, suggesting a role of miRNAs in the pathogenesis of Fragile X syndrome [93]. A double-stranded RNA-binding protein DGCR8 is involved in miRNA processing, and functions as an essential Drosha co-factor [94]. DGCR8 is commonly deleted in patients with DiGeorge syndrome, characterized by a variety of defects, including schizophrenia and microcephaly [95]. Abnormal miRNA biogenesis may contribute to the etiology of DiGeorge syndrome [96].
Schizophrenia is a syndrome of reality distortion along with alterations in thought, emotional processing, volition and cognition. Several miRNAs are found more highly expressed in the prefrontal cortex of schizophrenic patients than normal subjects [97]. A report has shown that miR-181b is upregulated, while its target genes calcium sensor gene visinin-like 1 (VSNL1) and ionotropic AMPA glutamate receptor subunit (GRIA2), which are associated with schizophrenia, are downregulated in the superior temporal gyrus of schizophrenic patient brain samples [98]. The N-methyl-D-aspartate (NMDA) glutamate receptor signaling regulates neurotransmission and synaptic plasticity, and executes many brain functions. Dysfunction of the NMDA receptor signaling pathway results in cognitive disorders, which show symptoms of schizophrenia and autism. miR-219 is shown to specifically target calcium/calmodulin-dependent protein kinase II γ subunit (CaMKIIγ), a component of the NMDA receptor signaling [99]. Knockdown of miR-219 in the mouse brain alters the behavioral responses associated with disrupted NMDA receptor signaling [99]. Furthermore, autism spectrum disorder (ASD) is a neural developmental disorder featured in an impaired communication ability and social interaction, as well as repetitive or restricted behavior. miRNA expression profiles of ASD patients have been performed to explore a potential role of miRNAs in the pathogenesis of Autism [100–102].
Alzheimer's disease (AD) is a neurodegenerative disease that is the most common form of dementia. It includes changes in memory and functions of the hippocampus. Comparisons in the hippocampus of fetal, adult and AD brains have identified miRNAs that are highly expressed in the fetal hippocampus but differentially regulated in AD patients, for instance an upregulation of miR-9 and miR-128 in AD patients [103]. Another expression profiling analysis of miRNAs in short post-mortem interval (PMI) human brains has revealed an upregulation of miR-9, miR-125b and miR-146a in the temporal lobe in the AD patient brains [104].
Several miRNAs are found downregulated in AD brain tissues, such as miR-15a and the miR-107 and miR-29a/b-1 cluster [105–107]. The expression of miR-107 is greatly decreased even in early stages of the AD patients [105]. β-site amyloid precursor protein-cleaving enzyme (BACE1), which contributes to the accumulation of amyloid-β peptides (Aβ), the key molecule in the pathogenesis of AD, has been identified as the target of miR-107 [105]. During the AD progression, the expression of BACE1 is upregulated but miR-107 expression is decreased [105]. Moreover, the miR-29a/b-1 cluster is significantly decreased in AD patients accompanied by an abnormally high expression of BACE1. In a cell culture system, the miR-29a/b-1 cluster is shown to negatively regulate the expression of BACE1 and the accumulation of Aβ peptide [106]. In addition, miR-298 and miR-328 are reported to repress the expression of BACE1 [108]. Furthermore, the expression of amyloid precursor protein (APP) and accumulation of Aβ is negatively regulated by miR-101 in cultured hippocampal neurons, suggesting a potential role of miR-101 in AD progression [109]. miR-146a, which is regulated by the NF-κB signaling, is upregulated in AD brains, and miR-146a targets complement factor H (CFH), a repressor of the inflammatory response of the brain [110].
Huntington's disease (HD) is a genetic disorder with programmed degeneration of brain neurons. Patients have behavioral and motor symptoms and cognitive decline leading to dementia. The expression profile of miRNAs in two transgenic mouse models of HD has shown that miRNAs such as miR-22, miR-29c and miR-128 are downregulated [111]. In human samples, the expression of miR-9/9*, miR-29b and miR-124a are downregulated, while miR-132 is upregulated with increasing HD grade compared to controls [112]. Previous studies indicate that aberrant cellular distribution of RE1-silencing transcription factor (REST) is one of the molecular mechanisms underlying HD etiology. The REST repressor complex, including mSin3, REST co-repressor 1 (CoREST) and MeCP2, regulate expression levels of miR-9/9*, miR-124a and miR-132 [113]. Interestingly, miR-9 and miR-9* have targeting effects on REST and CoREST, respectively, suggesting REST silencing complex and miR-9/9* form a negative feedback loop during HD development [114] (Fig. 2D). Furthermore, miR-34b is elevated in plasma from HD patients even before the symptom onsets, suggesting miR-34b a potential biomarker for HD diagnosis [115].
Parkinson’s disease (PD) results from a loss of dopaminergic neurons in the substantia nigra. It involves abnormalities in movement variably accompanied by sensory, mood and cognitive changes. Alpha-synuclein accumulation is related to the pathogenesis of PD. A study has shown that miR-7 and miR-153 negatively regulate α-synuclein in primary neurons [116]. In the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced neurotoxin PD mouse model, miR-7 is downregulated, suggesting that miR-7 may protect cells from oxidative stress [117]. Moreover, a recent study has demonstrated that a single nucleotide polymorphism (SNP) in the 3'-UTR of fibroblast growth factor 20 (FGF20) destroys a target site for miR-433. Thus, miR-433 is linked to an increased risk of AD by indirectly causing overexpression of α-synuclein [118].
miRNAs are also associated with prion-induced neurodegeneration [119, 120]. In the mouse brains infected with mouse-adapted scrapie, miRNAs such as miR-342-3p, miR-320 and let-7b are greatly upregulated, while miR-337-3p and miR-338-3p are downregulated during the disease process [119]. Among potential targets of these miRNAs predicted by the computational algorithms, 119 genes are also de-regulated during the prion-induced neurodegeneration [119].
Down’s syndrome (DS), also called trisomy 21, is a genetic disease caused by an extra copy of entire or part of the human chromosome 21. DS patients usually have physical growth abnormality, heart defects, leukemia, low-cognitive ability and Alzheimer’s disease [121]. Since DS is caused by an extra copy of the chromosome 21, which leads to over-expression of many genes located on it, it is possible that miRNAs located on the chromosome 21 play a role during the DS pathological process by negatively regulating expression of target genes and subsequently contribute to the symptoms of DS. Indeed miRNAs that are located on the chromosome 21, including let-7c, miR-99a, miR-125b-2, miR-155 and miR-802, are found upregulated in the fetal hippocampus and heart samples from DS patients, while their target genes are found decreased [122, 123]. Gain and loss of function of miR-155 and miR-802 have shown that they target MeCP2, which is downregulated in the DS brain [124]. In addition, an upregulation of miR-125b and downregulation of its target CDKN2A are found in advanced AD and DS brains [125].
Cerebral infarction (Stroke) is related to a disturbance in the blood supply to the brain. miRNA expression profiling in young stroke patients has shown 138 upregulated miRNAs and 19 downregulated miRNAs [126]. miR-21 is found to be upregulated in neurons in the ischemic boundary zone after stroke in rat brains. miR-21 protects ischemic neurons, after oxygen and glucose deprivation, from cell death by targeting tumor necrosis factor α family member FAS ligand gene (FASLG), a cell death inducer [127]. Recent study has shown that miR-124a is downregulated in neural progenitor cells of the SVZ after stroke [128]. By targeting Notch signaling ligand Jagged-1 (JAG1), miR-124a impairs precursor proliferation and promotes neuronal differentiation after stroke [128].
In the temporal lobe epilepsy (TLE) rat model as well as in human TLE, the expression of miR-146a is found upregulated and is expressed in reactive astrocytes in the hippocampus, mainly in the regions where neuronal cell loss and reactive gliosis occur [129]. This observation implies the possible involvement of miR-146a in the modulation of the astroglial inflammatory response during the TLE processes and provides a potential target for clinical treatment of TLE.
Depression is an affective mental disorder with a low mood, a loss of interests in activities, and sometimes, a cognitive impairment. A study has shown an association of the T allele of the rs76481776 polymorphism of pre-miR-182 with the late insomnia in major depression (MD) patients [130]. Overexpression of miR-182 causes reduction of miR-182 targeting genes Adenylate cyclase type 6 (ADCY6), Circadian Locomotor Output Cycles Kaput (CLOCK) and Delta sleep-inducing peptide (DSIP), which are involved in molecular clock regulation, suggesting that miR-182 might contribute to the dysregulation of circadian rhythms in the MD patients with insomnia [130]. The polymorphism in the ss178077483 of pre-miR-30e is also found to be associated with a higher risk of the MD disorder [131].
A miRNA expression profiling analysis in the prefrontal cortex of patients with bipolar disorder (BD), also known as manic-depressive disorder, has identified differentially expressed miRNAs [132]. miR-134 is suggested as a biomarker of mania episodes in BD, based on an analysis of expression levels of miRNAs in the circulating blood before and after the treatment of bipolar mania patients [133]. In addition, chronic treatment with selective serotonin reuptake inhibitor (SSRI) fluoxetine in mice results in an upregulation of miR-16 and downregulation of SERT, which is a target of miR-16, suggesting that miRNAs participate in the therapeutic action of SSRI [134]. Several miRNAs are found to be dysregulated after the chronic treatment with mood stabilizers Lithium and Valproate [135]. Among these miRNAs, miR-34a expression is reduced after the treatment, while its target metabotropic glutamate receptor 7 (GRM7) is upregulated [135]. These studies suggest a role of miRNAs in the clinical therapy of antidepression medications.
In summary, these studies indicate that similar to coding genes, misregulation of miRNAs also contributes to the pathogenesis of neurodegeneration diseases and mental disorders (Table 1). miRNA expression profiles using patient samples have shown that expression levels of many miRNAs are altered. Thus, it is a daunting task to figure out major miRNAs that are directly associated with the causes and progression of neurological diseases. Moreover, it is likely that neurological disorders are the result of altered networks of gene expression regulations of coding genes and noncoding RNAs.
lncRNAs in the CNS development
Based on in situ hybridization data from the Allen Brain Atlas (http://www.brain-map.org/), 849 lncRNAs from diverse locations in the genome have shown distinct expression patterns in different regions, cell types and subcellular localization in adult mouse brains [4]. Similar to miRNAs, lncRNAs are also classified into intergenic and intragenic populations. Even though functions of lncRNAs in the CNS development are largely unclear, studies of intragenic lncRNAs that are associated with protein-coding genes in the genome are emerging. Bioinformatic investigation of genomic locations and comparisons of expression patterns of lncRNAs and their adjacent protein coding transcripts have implied a potential cis-regulation mechanism of lncRNAs in modulating the transcription of coding genes in the developing and adult mouse brains [136].
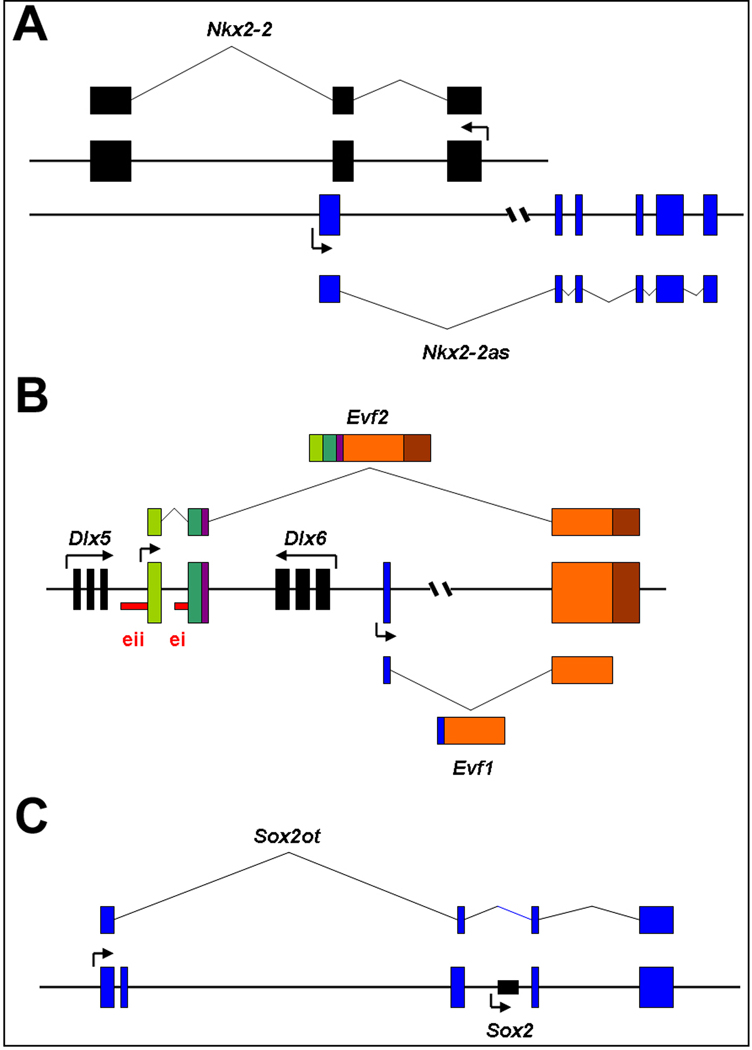
Nkx2.2 antisense (Nkx2.2as) is an antisense lncRNA to Nkx2.2 gene, which is expressed in the developing mammalian forebrain and is required for oligodendrocyte development [137] (Fig. 4A). Ectopic expression of Nkx2.2as in NSCs in cultures induces oligodendrocyte differentiation through an upregulation of the Nkx2.2 mRNA level, suggesting that Nkx2.2as regulates NSC differentiation, at least partially, by promoting Nkx2.2 expression [25].
Figure 4.
Regulatory functions of representative long noncoding RNAs (lncRNAs) on expression of protein coding genes. A. Nkx2.2 antisense (Nkx2.2as) is an antisense lncRNA to Nkx2.2 gene and promotes Nkx2-2 expression. B. Evf2 is transcribed from the intergenic region between the Dlx-5 and Dlx-6 loci, and is overlapped with Dlx-5/6 enhancer i (ei) and enhancer ii (eii) sequences. Evf2 acts as a transcriptional co-activator of Dlx-2 and activates the Dlx5/6 enhancer. C. Sox2 overlapping transcript Sox2OT is a lncRNA containing Sox2 gene and shares the same transcriptional orientation with Sox2. Sox2OT is expressed in the neurogenic region of the adult mouse brain
Embryonic ventral forebrain-1 (Evf1) is a 2.7 kb lncRNA transcribed upstream of the mouse Dlx-6 gene [138]. As an alternatively spliced form of Evf1, Evf2 is transcribed from the intergenic region between the Dlx-5 and Dlx-6 loci, and is overlapped with the conserved Dlx-5/6 intergenic enhancer [26, 139] (Fig. 4B). Dlx-6 is a homeobox containing transcription factor and plays an important role in forebrain neurogenesis [140]. Induced by the Sonic hedgehog (Shh) signaling pathway, Evf2 has been proven to function as a transcriptional co-activator of Dlx-2 and activates the Dlx5/6 enhancer during forebrain development [26]. Deletion of Evf2 results in a reduction of GABAergic interneurons and impaired synaptic inhibition in the developing hippocampus [141].
Sox2 is a transcription factor and plays a key role in the maintenance of the undifferentiating state of embryonic and adult NSCs [142]. Sox2 overlapping transcript Sox2OT is a lncRNA containing Sox2 gene and shares the same transcriptional orientation with Sox2 (Fig. 4C). Similar to Sox2, Sox2OT is stably expressed in mouse embryonic stem cells and down-regulated during differentiation. Sox2OT is expressed in the neurogenic region of the adult mouse brain, and is dynamically regulated during vertebrate CNS development, implying its role in regulating NSC self-renewal and neurogenesis [4, 143].
Retinal noncoding RNA 2 (RNCR2), an intergenic lncRNA also known as Gomafu and Miat, is an abundant polyadenylated RNA in the developing retina [144]. RNCR2 is highly expressed in both mitotic and postmitotic retinal progenitors. Knockdown of RNCR2 leads to an increase of amacrine cells and Müller glial cells in postnatal retina. Mis-localization of RNCR2 from nuclear to cytoplasm photocopies the effects caused by RNCR2 knockdown, suggesting that RNCR2 is required for retinal precursor cell specification [145].
lncRNAs in neurological disorders
Scattered studies have shown that lncRNAs are also involved in neurobiological disorders. Sox2OT has been identified as one of a potential biomarkers of AD for both early and late stages [146]. BC200 RNA is a 200 nt noncoding RNA that is expressed in the nervous system [147]. In human nervous system, BC200 RNA is expressed by a subpopulation of neurons analogous to BC1 RNA, a 152 nt noncoding RNA localized in the dendrite-rich neuropil areas [147]. BC200 RNA and BC1 are likely regulators of local protein expression in the postsynaptic dendritic microdomains [148]. A study has shown that BC200 RNA is upregulated in the frontal cortex of the AD brains compared to the age-matched controls. The expression level of BC200 RNA is increased in parallel with the AD progression [149]. However, in another independent work in a rigorously controlled study of short post-mortem AD brains, BC200 RNA is found to be significantly decreased [150].
Human accelerated region 1 (HAR1) belongs to HAR family, which contains a set of 49 segments of the human genome that are conserved through vertebrate evolution but strikingly different in humans [151]. HAR1 is located on the long arm of the chromosome 20 and overlaps with lncRNA HAR1F, which is expressed in Cajal-Retzius cells in the human fetal cortex [151]. The expression of HAR1 is inhibited by transcription repressor REST in brain samples of HD patients, suggesting a potential role of HAR1 in neurodegeneration disorders [152].
Perspectives
It has been well accepted that noncoding RNAs, in particular miRNAs, play a critical role in neural development and functions. Identification of region and cell type specific miRNAs in the CNS will help determine actions of individual miRNAs in the CNS development. Moreover, investigation of how specific expression of miRNAs is regulated during development by identifying miRNA promoters and binding transcription factors will reveal the transcriptional logic of miRNA expression and functions. Because one miRNA has multiple targets, it remains a challenge to dissect the networks of miRNA-targets in the CNS development.
The association of miRNAs and human neurological diseases still requires comprehensive investigation. Because of the fast development of miRNA synthesis and delivery techniques, it is promising to use miRNAs as diagnostic biomarkers and as new therapeutic tools for neurological disorders. Moreover, to better understand the etiology of miRNA-mediated human neurological diseases, one needs to develop proper genetic tools to generate animal models with altered miRNA expressions.
There is no doubt that examining lncRNAs functions in neural development and under disease conditions will remain an exciting research focus. Genome wide RNA sequencing and bioinformatic analysis will help identify region and cell type specific lncRNAs in the CNS and their association with protein coding genes in the genome. Examining how lncRNAs manipulate the gene expression will continue to reveal novel gene regulation mechanisms in neural development.
Acknowledgments
We thank Drs. Dolores Malaspina, Rajiv Ratan and Quanhong Ma for critical reading of the manuscript and providing thoughtful comments. Owing to space limitations, we apologize for being unable to cite many excellent papers in this field. This work was supported by the Ellison Medical Foundation (T. S.), an award from the Hirschl/Weill-Caulier Trust (T. S.) and an R01-MH083680 grant from the NIH/NIMH (T. S.).
References
- 1.Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8(8):R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. Rna. 2008;14(3):432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, Garcia-Moreno F, Molnar Z, Margulies EH, Ponting CP. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71(4):605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. Transcriptional control of gene expression by microRNAs. Cell. 2010;140(1):111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 7.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 8.Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol. 2006;342:33–47. doi: 10.1385/1-59745-123-1:33. [DOI] [PubMed] [Google Scholar]
- 9.Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 10.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8(1):23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 11.Neilson JR, Sharp PA. Small RNA regulators of gene expression. Cell. 2008;134(6):899–902. doi: 10.1016/j.cell.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5(9):R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284(27):17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao FB. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 2008;31(1):20–26. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 16.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 17.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 18.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 20.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 21.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 22.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67(8):3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 24.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2010;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tochitani S, Hayashizaki Y. Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem Biophys Res Commun. 2008;372(4):691–696. doi: 10.1016/j.bbrc.2008.05.127. [DOI] [PubMed] [Google Scholar]
- 26.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20(11):1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451(7175):202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454(7200):126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440(7083):556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 30.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 31.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414(6861):317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414(6861):322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311(5764):1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- 34.Mazo A, Hodgson JW, Petruk S, Sedkov Y, Brock HW. Transcriptional interference: an unexpected layer of complexity in gene regulation. J Cell Sci. 2007;120(Pt 16):2755–2761. doi: 10.1242/jcs.007633. [DOI] [PubMed] [Google Scholar]
- 35.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denisenko O, Shnyreva M, Suzuki H, Bomsztyk K. Point mutations in the WD40 domain of Eed block its interaction with Ezh2. Mol Cell Biol. 1998;18(10):5634–5642. doi: 10.1128/mcb.18.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30(2):167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 38.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20(10):1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135(23):3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawase-Koga Y, Low R, Otaegi G, Pollock A, Deng H, Eisenhaber F, Maurer-Stroh S, Sun T. RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J Cell Sci. 2010;123(Pt 4):586–594. doi: 10.1242/jcs.059659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersson T, Rahman S, Sansom SN, Alsio JM, Kaneda M, Smith J, O'Carroll D, Tarakhovsky A, Livesey FJ. Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS One. 2010;5(10):e13453. doi: 10.1371/journal.pone.0013453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238(11):2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28(17):4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q, Bian S, Hong J, Kawase-Koga Y, Zhu E, Zheng Y, Yang L, Sun T. Timing specific requirement of microRNA function is essential for embryonic and postnatal hippocampal development. PLoS One. 2011 doi: 10.1371/journal.pone.0026000. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci U S A. 2008;105(14):5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zehir A, Hua LL, Maska EL, Morikawa Y, Cserjesi P. Dicer is required for survival of differentiating neural crest cells. Dev Biol. 2010;340(2):459–467. doi: 10.1016/j.ydbio.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol. 2010;2(3):152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- 49.Zheng K, Li H, Zhu Y, Zhu Q, Qiu M. MicroRNAs are essential for the developmental switch from neurogenesis to gliogenesis in the developing spinal cord. J Neurosci. 2010;30(24):8245–8250. doi: 10.1523/JNEUROSCI.1169-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65(5):612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Budde H, Schmitt S, Fitzner D, Opitz L, Salinas-Riester G, Simons M. Control of oligodendroglial cell number by the miR-17-92 cluster. Development. 2010;137(13):2127–2132. doi: 10.1242/dev.050633. [DOI] [PubMed] [Google Scholar]
- 52.Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6(8):784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 53.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Temple S. The development of neural stem cells. Nature. 2001;414(6859):112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 55.Gage FH. Mammalian neural stem cells. Science. 2000;287(5457):1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 56.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135(9):1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 57.Arsenijevic Y. Mammalian neural stem-cell renewal: nature versus nurture. Mol Neurobiol. 2003;27(1):73–98. doi: 10.1385/MN:27:1:73. [DOI] [PubMed] [Google Scholar]
- 58.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 59.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A. 2010;107(5):1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 61.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 62.Maiorano NA, Mallamaci A. Promotion of embryonic cortico-cerebral neuronogenesis by miR-124. Neural Dev. 2009;4:40. doi: 10.1186/1749-8104-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12(4):399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arvanitis DN, Jungas T, Behar A, Davy A. Ephrin-B1 reverse signaling controls a posttranscriptional feedback mechanism via miR-124. Mol Cell Biol. 2010;30(10):2508–2517. doi: 10.1128/MCB.01620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27(3):435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16(4):365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6(4):323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev Cell. 2011;20(1):19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11(6):641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- 70.Otaegi G, Pollock A, Hong J, Sun T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J Neurosci. 2011;31(3):809–818. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31(9):3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-Specific Modulation of Cortical Neuronal Development by Mmu-miR-134. Cereb Cortex. 2011 doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- 73.Brett JO, Renault VM, Rafalski VA, Webb AE, Brunet A. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY) 2011;3(2):108–124. doi: 10.18632/aging.100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5(5):527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189(1):127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, Zhao X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6(5):433–444. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314(14):2618–2633. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279(5350):509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 79.Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63(6):803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 14(9):1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 81.Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, Teng ZQ, Luo Y, Peng J, Bordey A, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28(6):1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317(5842):1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nunes I, Tovmasian LT, Silva RM, Burke RE, Goff SP. Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc Natl Acad Sci U S A. 2003;100(7):4245–4250. doi: 10.1073/pnas.0230529100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, Mandel G, Goodman RH. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci U S A. 2010;107(47):20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hansen KF, Sakamoto K, Wayman GA, Impey S, Obrietan K. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS One. 2010;5(11):e15497. doi: 10.1371/journal.pone.0015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65(3):373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 88.Bai F, Witzmann FA. Synaptosome proteomics. Subcell Biochem. 2007;43:77–98. doi: 10.1007/978-1-4020-5943-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11(6):705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64(6):871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 91.Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310(5746):317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 92.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7(2):113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 93.Yi YH, Sun XS, Qin JM, Zhao QH, Liao WP, Long YS. Experimental identification of microRNA targets on the 3' untranslated region of human FMR1 gene. J Neurosci Methods. 2010;190(1):34–38. doi: 10.1016/j.jneumeth.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 94.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 95.Digilio MC, Marino B, Cappa M, Cambiaso P, Giannotti A, Dallapiccola B. Auxological evaluation in patients with DiGeorge/velocardiofacial syndrome (deletion 22q11.2 syndrome) Genet Med. 2001;3(1):30–33. doi: 10.1097/00125817-200101000-00007. [DOI] [PubMed] [Google Scholar]
- 96.Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008 doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 97.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8(2):R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17(8):1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- 99.Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, Sales N, Willoughby D, Elmen J, Hansen HF, et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci U S A. 2009;106(9):3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, Lao K, Kosik KS. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9(3):153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- 101.Sarachana T, Zhou R, Chen G, Manji HK, Hu VW. Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med. 2010;2(4):23. doi: 10.1186/gm144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seno MM, Hu P, Gwadry FG, Pinto D, Marshall CR, Casallo G, Scherer SW. Gene and miRNA expression profiles in autism spectrum disorders. Brain Res. 2011;1380:85–97. doi: 10.1016/j.brainres.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 103.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007;18(3):297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 104.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer's disease temporal lobe neocortex. Neurosci Lett. 2009;459(2):100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 105.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28(5):1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105(17):6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hebert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, Sergeant N, Buee L, De Strooper B. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet. 2010;19(20):3959–3969. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 108.Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284(4):1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vilardo E, Barbato C, Ciotti M, Cogoni C, Ruberti F. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J Biol Chem. 2010;285(24):18344–18351. doi: 10.1074/jbc.M110.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008;283(46):31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, Park KH, Jung KH, Lee SK, Kim M, et al. Altered microRNA regulation in Huntington's disease models. Exp Neurol. 2011;227(1):172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 112.Johnson R, Buckley NJ. Gene dysregulation in Huntington's disease: REST, microRNAs and beyond. Neuromolecular Med. 2009;11(3):183–199. doi: 10.1007/s12017-009-8063-4. [DOI] [PubMed] [Google Scholar]
- 113.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103(7):2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28(53):14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gaughwin PM, Ciesla M, Lahiri N, Tabrizi SJ, Brundin P, Bjorkqvist M. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington's disease. Hum Mol Genet. 2011;20(11):2225–2237. doi: 10.1093/hmg/ddr111. [DOI] [PubMed] [Google Scholar]
- 116.Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285(17):12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106(31):13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82(2):283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3(11)::e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pogue AI, Dua P, Eicken C, Hill JM, Lukiw WJ. Up-regulation of microRNA-146a (miRNA-146a), a marker for inflammatory neurodegeneration, in sporadic Creutzfeldt-Jakob disease (sCJD) and Gerstmann-Straussler Scheinker (GSS) syndrome. Journal of Toxicology and Environmental Health. 2011 doi: 10.1080/15287394.2011.618973. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roizen NJ, Patterson D. Down's syndrome. Lancet. 2003;361(9365):1281–1289. doi: 10.1016/S0140-6736(03)12987-X. [DOI] [PubMed] [Google Scholar]
- 122.Kuhn DE, Nuovo GJ, Martin MM, Malana GE, Pleister AP, Jiang J, Schmittgen TD, Terry AV, Jr, Gardiner K, Head E, et al. Human chromosome 21-derived miRNAs are overexpressed in down syndrome brains and hearts. Biochem Biophys Res Commun. 2008;370(3):473–477. doi: 10.1016/j.bbrc.2008.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 123.Elton TS, Sansom SE, Martin MM. Trisomy-21 gene dosage over-expression of miRNAs results in the haploinsufficiency of specific target proteins. RNA Biol. 2010;7(5):540–547. doi: 10.4161/rna.7.5.12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kuhn DE, Nuovo GJ, Terry AV, Jr, Martin MM, Malana GE, Sansom SE, Pleister AP, Beck WD, Head E, Feldman DS, et al. Chromosome 21-derived microRNAs provide an etiological basis for aberrant protein expression in human Down syndrome brains. J Biol Chem. 2010;285(2):1529–1543. doi: 10.1074/jbc.M109.033407. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 125.Pogue AI, Cui JG, Li YY, Zhao Y, Culicchia F, Lukiw WJ. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci Lett. 2010;476(1):18–22. doi: 10.1016/j.neulet.2010.03.054. [DOI] [PubMed] [Google Scholar]
- 126.Tan KS, Armugam A, Sepramaniam S, Lim KY, Setyowati KD, Wang CW, Jeyaseelan K. Expression profile of MicroRNAs in young stroke patients. PLoS One. 2009;4(11):e7689. doi: 10.1371/journal.pone.0007689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Buller B, Liu X, Wang X, Zhang RL, Zhang L, Hozeska-Solgot A, Chopp M, Zhang ZG. MicroRNA-21 protects neurons from ischemic death. Febs J. 2010;277(20):4299–4307. doi: 10.1111/j.1742-4658.2010.07818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu XS, Chopp M, Zhang RL, Tao T, Wang XL, Kassis H, Hozeska-Solgot A, Zhang L, Chen C, Zhang ZG. MicroRNA Profiling in Subventricular Zone after Stroke: MiR-124a Regulates Proliferation of Neural Progenitor Cells through Notch Signaling Pathway. PLoS One. 2011;6(8):e23461. doi: 10.1371/journal.pone.0023461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, van Vliet EA, Baayen JC, Gorter JA. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur J Neurosci. 2010;31(6):1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- 130.Saus E, Soria V, Escaramis G, Vivarelli F, Crespo JM, Kagerbauer B, Menchon JM, Urretavizcaya M, Gratacos M, Estivill X. Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum Mol Genet. 2010;19(20):4017–4025. doi: 10.1093/hmg/ddq316. [DOI] [PubMed] [Google Scholar]
- 131.Xu Y, Liu H, Li F, Sun N, Ren Y, Liu Z, Cao X, Wang Y, Liu P, Zhang K. A polymorphism in the microRNA-30e precursor associated with major depressive disorder risk and P300 waveform. J Affect Disord. 2010;127(1–3):332–336. doi: 10.1016/j.jad.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 132.Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, van den Oord EJ, Riley BP, Kendler KS, Vladimirov VI. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124(1–3):183–191. doi: 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rong H, Liu TB, Yang KJ, Yang HC, Wu DH, Liao CP, Hong F, Yang HZ, Wan F, Ye XY, et al. MicroRNA-134 plasma levels before and after treatment for bipolar mania. J Psychiatr Res. 2011;45(1):92–95. doi: 10.1016/j.jpsychires.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 134.Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science. 2010;329(5998):1537–1541. doi: 10.1126/science.1193692. [DOI] [PubMed] [Google Scholar]
- 135.Zhou R, Yuan P, Wang Y, Hunsberger JG, Elkahloun A, Wei Y, Damschroder-Williams P, Du J, Chen G, Manji HK. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology. 2009;34(6):1395–1405. doi: 10.1038/npp.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5(8) doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Price M, Lazzaro D, Pohl T, Mattei MG, Ruther U, Olivo JC, Duboule D, Di Lauro R. Regional expression of the homeobox gene Nkx-2.2 in the developing mammalian forebrain. Neuron. 1992;8(2):241–255. doi: 10.1016/0896-6273(92)90291-k. [DOI] [PubMed] [Google Scholar]
- 138.Kohtz JD, Fishell G. Developmental regulation of EVF-1, a novel non-coding RNA transcribed upstream of the mouse Dlx6 gene. Gene Expr Patterns. 2004;4(4):407–412. doi: 10.1016/j.modgep.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 139.Zerucha T, Stuhmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JL, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20(2):709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang Y, Dye CA, Sohal V, Long JE, Estrada RC, Roztocil T, Lufkin T, Deisseroth K, Baraban SC, Rubenstein JL. Dlx5 and Dlx6 regulate the development of parvalbumin-expressing cortical interneurons. J Neurosci. 2010;30(15):5334–5345. doi: 10.1523/JNEUROSCI.5963-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12(8):1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15(1):7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 143.Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, Mattick JS. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. Rna. 2009;15(11):2013–2027. doi: 10.1261/rna.1705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2(9):E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol. 2010;10:49. doi: 10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Arisi I, D'Onofrio M, Brandi R, Felsani A, Capsoni S, Drovandi G, Felici G, Weitschek E, Bertolazzi P, Cattaneo A. Gene expression biomarkers in the brain of a mouse model for Alzheimer's disease: mining of microarray data by logic classification and feature selection. J Alzheimers Dis. 2011;24(4):721–738. doi: 10.3233/JAD-2011-101881. [DOI] [PubMed] [Google Scholar]
- 147.Tiedge H, Chen W, Brosius J. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci. 1993;13(6):2382–2390. doi: 10.1523/JNEUROSCI.13-06-02382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sonenberg N, Lomakin IB, Tiedge H. Dendritic BC1 RNA: functional role in regulation of translation initiation. J Neurosci. 2002;22(23):10232–10241. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104(25):10679–10684. doi: 10.1073/pnas.0701532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lukiw WJ, Handley P, Wong L, Crapper McLachlan DR. BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD) Neurochem Res. 1992;17(6):591–597. doi: 10.1007/BF00968788. [DOI] [PubMed] [Google Scholar]
- 151.Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443(7108):167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 152.Johnson R, Richter N, Jauch R, Gaughwin PM, Zuccato C, Cattaneo E, Stanton LW. The Human Accelerated Region 1 noncoding RNA is repressed by REST in Huntington's disease. Physiol Genomics. 2010;41:269–274. doi: 10.1152/physiolgenomics.00019.2010. [DOI] [PubMed] [Google Scholar]