Abstract
Indole signaling is one of the putative universal signaling networks in bacteria. We have investigated the use of desformylflustrabromine (dFBr) derivatives for the inhibition of biofilm formation through modulation of the indole-signaling network in E. coli and S. aureus. We have found dFBr derivatives that are 10-1000 times more active than indole itself, demonstrating that the flustramine family of indolic natural products represent a privileged scaffold for the design of molecules to control pathogenic bacterial behavior.
The emergence of resistance to multiple antimicrobial agents in pathogenic bacteria has become a significant global public health threat. In 2005, almost 95,000 people acquired methicillin resistant Staphylococcus aureus (MRSA) infections in the United States, of which nearly 19,000 people died-more than die annually from medical complications of HIV/AIDS.1 Without developing innovative approaches to combat these multidrug resistant pathogens, many fields of medicine such as surgery and care of the critically ill will be severely affected. This situation is so dire that Infectious Disease Society of America has recently issued a call to action for the medical community.2
One promising approach to controlling bacterial infections is to develop small molecules that attenuate bacterial behaviors that are detrimental to the human host.3, 4 Examples of these behaviors include the production of virulence factors and biofilm formation. Such an approach, which operates through a non-microbicidal mechanism, would be highly desirable, as this would not exert evolutionary pressure on the microorganisms to adapt and become resistant. Seminal approaches in this area include the use of acyl homoserine lactone derivatives,5, 6 brominated furanones,7-9 and modulators of autoinducer-2 (AI-2)10-14 (Figure 1).
Figure 1.
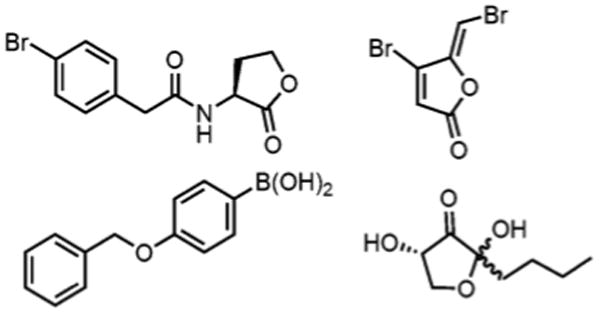
Synthetic small molecules that control bacterial behavior.
Indole signaling is involved in the regulation of a number of bacterial behaviors.15-17 Our group has recently become interested in harnessing this signaling pathway to control these behaviors through the design of small molecule indole derivatives. We have chosen indole signaling because indole is a putative universal signal (along with AI-2) amongst diverse bacteria.16 Eighty-five species of bacteria have been documented to produce indole, while both indole positive and indole negative strains of bacteria will change behavior based upon the extracellular presence of indole. For example, indole will modulate biofilm formation,17 virulence,18 antibiotic resistance,19 and acid tolerance,20 all key behaviors of pathogenic bacteria. Therefore, highly active indole derivates have the potential to modulate pathogenic behavior in wide swaths of bacteria (both Gram-positive and Gram-negative) and represent a potentially powerful approach to controlling pathogenic bacterial behavior in vivo. To the best of our knowledge, however, highly active modulators of indole signaling have not been developed, nor have molecular design principles been elucidated to augment the activity of indole in the context of bacterial indole signaling.
Our group has had success employing marine natural products as structural templates for the design of small molecules that control biofilm formation and antibiotic resistance.21, 22 Applying this approach to indole signaling, we are interested in investigating the potential of flustramine derivatives to control biofilm formation. The flustramines are a group of indole-derived natural products isolated from the North Sea bryozoans Flustra foliacea.23-25 This family consists of eleven secondary metabolites, six pyrroloindoline and five indolic alkaloids (representative members depicted in Figure 2). It was noted that these specific bryozoans contained no microbial settling on the distal part of the zooid, implying that they possess a chemical defense system geared towards controlling bacterial behavior.
Figure 2.
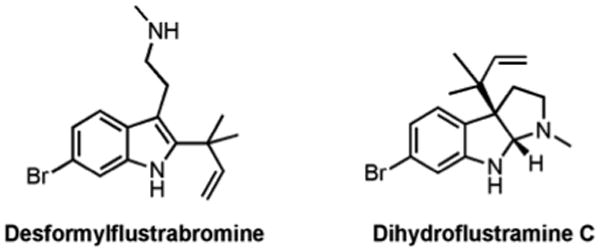
Representative members of the flustramine family.
We first investigated desformylflustrabromine (dFBr) as a potential modulator of bacterial behavior by assaying for its ability to inhibit biofilm formation. dFBr was synthesized as previously reported26 and tested for the ability to modulate Escherichia coli and S. aureus biofilm formation (representative Gram-negative and Gram-positive strains) using the crystal violet reporter assay. dFBr inhibited E. coli and S. aureus biofilm formation with IC50 values of 174 μM and 70 μM respectively. Follow up growth curve analysis indicated that dFbr was inhibiting E. coli biofilm formation through a toxic mechanism, while early growth delay (4 hours) was observed with S. aureus (bacterial density was equivalent to untreated control at 6, 8, and 24 hours). Given the goal of identifying non-toxic modulators of bacterial behavior, we posited that we could modulate the structure of dFBr to develop indole derivatives that inhibited biofilm development in both bacterial strains through a non-toxic mechanism analogous to indole itself. To achieve this aim, we set out to systematically probe each of the four areas of highlighted diversity for their impact on activity (Figure 3). The importance of the bromine (Region A) was probed by synthesizing and assaying debromodesformylflustrabromine (Figure 4). This compound was found to be essentially devoid of activity. The reverse prenyl group (Region B) was modulated by employing a Grandberg Fischer indolization to introduce C-2 substituents, while substituents on the aliphatic nitrogen (Region C) were introduced by alkylating nosyl-protected indole 1 followed by installation of the reverse prenyl group, bromination, and deprotection. Finally, substituents on the indole nitrogen (Region D) were introduced through protected tryptamine derivative 2 to probe the importance of the N-H bond and steric/electronic constraints (an inclusive list of all analogues screened is provided in the SI). Each derivative was then assayed for the ability to modulate E. coli and S. aureus biofilm formation.
Figure 3.
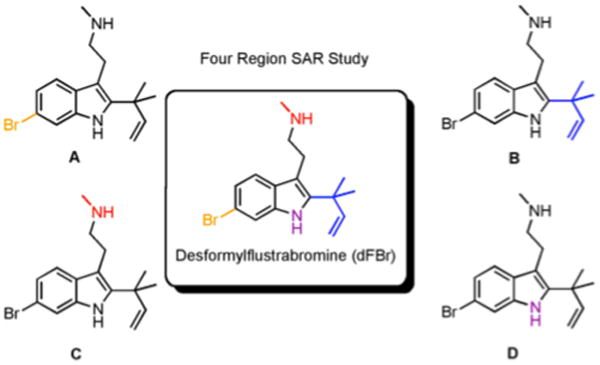
Four regions within the dFBr scaffold that can be rapidly modulated to deliver compounds that control bacterial.
Figure 4.
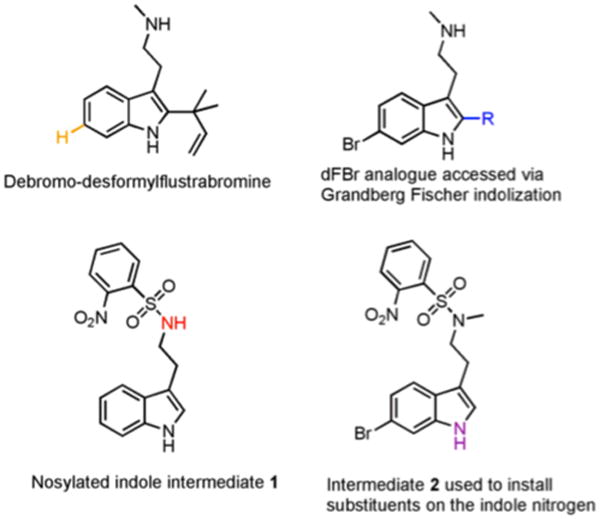
Derivatives and intermediates used to access dFBr analogues.
The most potent analogue synthesized was derivative 3 (Figure 5), which recorded and IC50 of ca. 5.9 μM against S. aureus and an IC50 of 53 μM against our E. coli strain. Compound 4 was also found to be moderately effective, exhibiting IC50 values of 80 μM and 65 μM against E. coli and S. aureus respectively. Follow up growth curve and colony count analysis indicated that both compounds were modulating biofilm development via a non-microbicidal mechanism.
Figure 5.
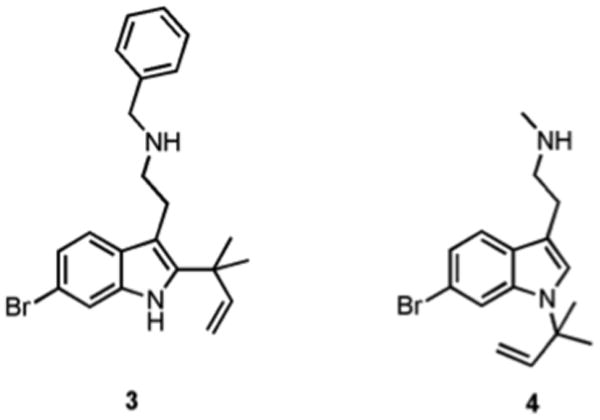
Lead dFBr analogues 3 and 4.
To establish that the biofilm inhibition activity of the dFBr analogue 3 is occurring via the indole signaling pathway, we compared the activity of 3 to that of indole against the E. coli strain BW25113, an isogenic sdiA knockout mutant, and an isogenic tnaA knockout mutant as a function of temperature (25 °C and 37 °C). Indole signaling pathways have been most widely studied in E. coli and it has been shown that the transcriptional regulator SdiA is involved in the control of biofilm formation by indole in E. coli at 37 °C, though the exact mechanism by which this occurs has not yet been elucidated.17, 27 Indole signaling in E. coli has been primarily observed at temperatures lower than 37 °C and it has been demonstrated that the addition of exogenous indole reduces biofilm formation to a greater extent at 25 °C than at 37 °C.
Analogous to previous reports, we found that biofilm formation by the wild type strain was reduced in the presence of indole in a dose dependant manner at 25 °C, while a lesser effect was observed at 37 °C (65% inhibition in the presence of 1 mM indole at 25 °C compared to 38% at 37 °C) (Figure 6). Similarly biofilm formation by the tnaA mutant, which lacks the ability to produce indole and should therefore exhibit a greater response to the addition of exogenous indole, exhibited a dose-dependant response that was amplified at 25 °C compared to 37 °C (74% inhibition in the presence of 1 mM indole at 25 °C compared to 25% at 37 °C). Against the sdiA mutant, indole addition resulted in a considerable response at 25 °C, reducing biofilm formation by 87% at 1 mM, while the response observed at 37 °C was reduced from that exhibited by the wild type (33% inhibition for the wild type compared to 9% for the sdiA mutant at 250 μM).
Figure 6.
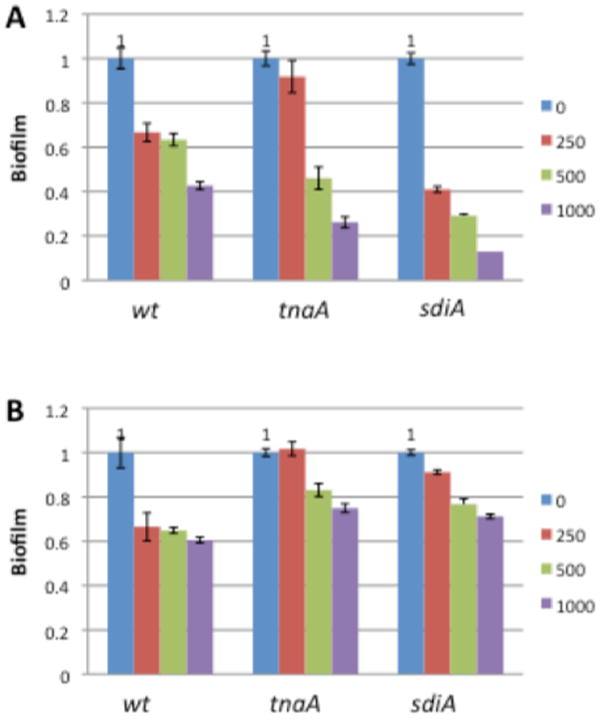
Biofilm inhibition activity of indole against wild-type and knockout E. coli strains: (A) 25 _C; (B) 37 _C. Concentrations in μM.
Compound 3 was much more active as a biofilm inhibitor compared to indole against all three strains, exhibiting comparable activity at 10-100 μM to that of indole at 250-1000 μM (Figure 7). As predicted, 3 displayed similar activity trends as a function of temperature to indole, affecting biofilm formation to a greater degree at 25 °C than at 37 °C for the mutant strains. Furthermore, the addition of 3 resulted a much greater effect on biofilm formation by the wild type than by the sdiA mutant at 37 °C (82% inhibition for the wild type compared to 49% for the sdiA mutant at 100 μM), indicating that, as for indole, the biofilm inhibitory activity of 3 is partially dependent upon SdiA at elevated temperatures.
Figure 7.
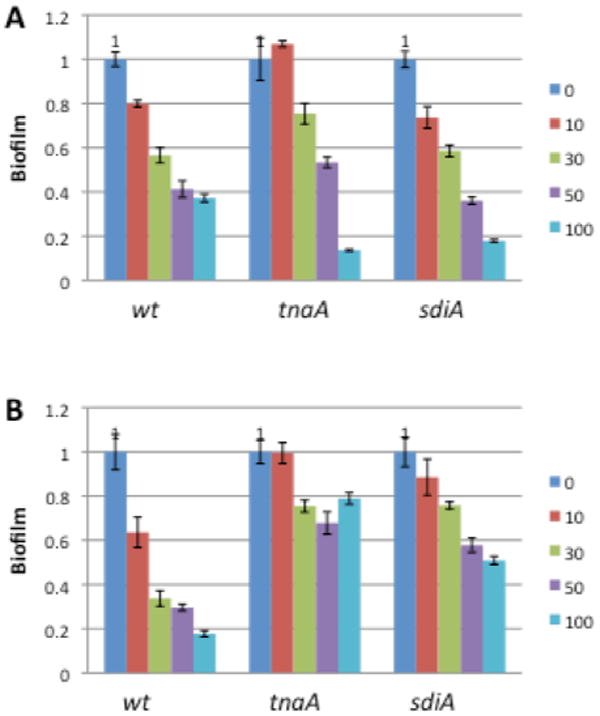
Biofilminhibition activity of compound 3 againstwild-type and knockout E. coli strains: (A) 25 _C; (B) 37 _C. Concentrations in μM.
In conclusion, we have employed desformylflustrabromine (dFBr) as a structural template to design indolic derivatives that are non-microbicidal inhibitors of biofilm formation. Lead compound 3 is 10-1000 times more active than indole. Mechanistic studies in wt and knockout E. coli strains have demonstrated that, identical to indole itself, the activity of lead compound 3 is dependent on temperature, SdiA, and TnaA, thus suggesting that the anti-biofilm activity of 3 may be occurring via modulation of indole-based signaling pathways. The fact that 3, as for indole, retains some activity against the sdiA mutant suggests that factors other than SdiA are involved in biofilm regulation by indole in E. coli. Indeed indole has been shown to affect expression of 59 genes in biofilm bacteria at 37 °C, including yceK, which was shown to affect biofilm formation.27 Given that indole is a putative universal signal amongst diverse bacteria, 3 can be employed as a probe to further investigate the effects of manipulating indole signaling pathways in vitro and in vivo, as a mechanistic probe to deconvolute indole signaling in both indole positive and indole negative bacteria, and ultimately the therapeutic potential of controlling pathogenic bacterial behavior in vivo.
Supplementary Material
Acknowledgments
Funding Sources: The authors would like to thank the National Institutes of Health (GM055769 to JC and CM) for support of this work.
Footnotes
Supporting Information. Synthetic methods and compound characterization are provided for all new compounds. Representative 1H NMR and 13C NMR spectra are also provided. Protocols for biofilm inhibition assays and representative dose response curves and growth curves are provided for lead compounds against all bacterial strains studied. (http://pubs.acs.org/page/jacsat/submission/authors.html).
Author Contributions: The manuscript was written by CAB, RJW, and CM. All authors have given approval to the final version of the manuscript.
References
- 1.Klein E, Smith DL, Laxminarayan R. Emerg Infect Diseases. 2007;13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J., Jr Clin Infect Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 3.Bjarnsholt T, Givskov M. Philos Trans R Soc London B Biol Sci. 2007;362:1213–222. doi: 10.1098/rstb.2007.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen TB, Givskov M. Int J Med Microbiol. 2006;296:149–161. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Geske GD, Wezeman RJ, Siegel AP, Blackwell HE. J Am Chem Soc. 2005;127:12762–12763. doi: 10.1021/ja0530321. [DOI] [PubMed] [Google Scholar]
- 6.Smith KM, Bu YG, Suga H. Chem Biol. 2003;10:81–89. doi: 10.1016/s1074-5521(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 7.Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, Rice SA, Eberl L, Molin S, Hoiby N, Kjelleberg S, Givskov M. Microbiology. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
- 8.Wu H, Song Z, Hentzer M, Andersen JB, Molin S, Givskov M, Hoiby N. J Antimicrob Chemother. 2004;53:1054–1061. doi: 10.1093/jac/dkh223. [DOI] [PubMed] [Google Scholar]
- 9.Hjelmgaard T, Persson T, Rasmussen TB, Givskov M, Nielsen J. Bioorg Med Chem. 2003;11:3261–3271. doi: 10.1016/s0968-0896(03)00295-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang BH, Ni NT, Choudhary G, Peng HJ, Li MY, Chou HT, Lu CD, Gilbert ES. Chem Biol Drug Des. 2009;74:51–56. doi: 10.1111/j.1747-0285.2009.00834.x. [DOI] [PubMed] [Google Scholar]
- 11.Janda KD, Lowery CA, Abe T, Park J, Eubanks LM, Sawada D, Kaufmann GF. J Am Chem Soc. 2009;131:15584–15585. doi: 10.1021/ja9066783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janda KD, Lowery CA, Park J, Kaufmann GF. J Am Chem Soc. 2008;130:9200–9201. doi: 10.1021/ja802353j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meijler MM, McKenzie KM, Lowery CA, Qi LW, Janda KD. Abstr Papers Am Chem Soc. 2005;229:U218–U218. [Google Scholar]
- 14.Sintim HO, Smith JAI, Wang JX, Nguyen-Mau SM, Lee V. Chem Comm. 2009;45:7033–7035. doi: 10.1039/b909666c. [DOI] [PubMed] [Google Scholar]
- 15.Wood TK, Hong SH, Ma Q. Trends BiotechoL. 2011;29:87–94. doi: 10.1016/j.tibtech.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Lee JH. FEMS Microbiol Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 17.Lee JT, Wood TK, Jayaraman A. BMC Microbiol. 2007;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartlett DH, Mueller RS, Beyhan S, Saini SG, Yildiz FH. J Bacterial. 2009;191:3504–3516. doi: 10.1128/JB.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishino K, Nikaido E, Yamaguchi A. J Bacteriol. 2007;189:9066–9075. doi: 10.1128/JB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peti W, Lee J, Page R, Garcia-Contreras R, Palermino JM, Zhang XS, Doshi O, Wood TK. J Mol Biol. 2007;373:11–26. doi: 10.1016/j.jmb.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards JJ, Melander C. Antiinfect Agents Med Chem. 2009;8:295–314. [Google Scholar]
- 22.Rogers SA, Huigens RW, Cavanagh J, Melander C. Antimicrob Agents Chemother. 2010;54:2112–2118. doi: 10.1128/AAC.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carle JS, Christophersen C. J Org Chem. 1981;46:3440–3443. [Google Scholar]
- 24.Carle JS, Christophersen C. J Org Chem. 1980;45:1586–1589. [Google Scholar]
- 25.Carle JS, Christophersen C. J Am Chem Soc. 1979;101:4012–4013. [Google Scholar]
- 26.Lindel T, Brauchle L, Golz G, Bohrer P. Org Lett. 2007;9:283–286. doi: 10.1021/ol0627348. [DOI] [PubMed] [Google Scholar]
- 27.Wood TK, Lee J, Zhang XS, Hegde M, Bentley WE, Jayaraman A. JSME J. 2008;2:1007–1023. doi: 10.1038/ismej.2008.54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


