This work reports on a genetic approach to unravel the developmental function of the group α Aurora kinases by analyzing a viable double mutant. The results show that α Aurora kinases are functionally divergent from the β Aurora kinase and suggest that α Aurora kinases regulate formative division plane orientation throughout development.
Abstract
To establish three-dimensional structures/organs, plant cells continuously have to adapt the orientation of their division plane in a highly regulated manner. However, mechanisms underlying switches in division plane orientation remain elusive. Here, we characterize a viable double knockdown mutant in Arabidopsis thaliana group α Aurora (AUR) kinases, AUR1 and AUR2, (aur1-2 aur2-2), with a primary defect in lateral root formation and outgrowth. Mutant analysis revealed that aur1-2 aur2-2 lateral root primordia are built from randomly oriented cell divisions instead of distinct cell layers. This phenotype could be traced back to cytokinesis defects and misoriented cell plates during the initial anticlinal pericycle cell divisions that give rise to lateral root primordia. Complementation assays showed that the Arabidopsis α group Aurora kinases are functionally divergent from the single β group member AUR3 and that AUR1 functions in division plane orientation prior to cytokinesis. In addition to defective lateral root patterning, aur1-2 aur2-2 plants also show defects in orienting formative divisions during embryogenesis, divisions surrounding the main root stem cell niche, and divisions surrounding stomata formation. Taken together, our results put forward a central role for α Aurora kinases in regulating formative division plane orientation throughout development.
INTRODUCTION
Plants require strict control over cell division orientation to initiate de novo organogenesis and to establish their overall shape. Recent work showed that proliferative division planes can be accurately predicted based on the interplay between the cytoskeleton and the cell shape, whereby the division plane is selected from a competition between several minimal area configurations (Besson and Dumais, 2011). Much progress has been made in understanding how established proliferative division planes in land plant cells are marked and maintained throughout mitosis (Rasmussen et al., 2011). However, although several proteins have been identified with a role in establishing specific divisions leading to daughter cells with different cell fates (formative divisions) throughout plant development (De Smet and Beeckman, 2011; Rasmussen et al., 2011), mechanisms driving formative division plane establishment in plant cells remain elusive. Animal cells achieve asymmetrical divisions by translating polarizing cues into an asymmetric distribution of polarity regulators like the partitioning defective complex (Gönczy, 2008). Aurora kinase A-dependent phosphorylation of polarity determinants plays an important role in these formative divisions in Drosophila melanogaster (Hutterer et al., 2006; Wirtz-Peitz et al., 2008; Johnston et al., 2009; Ogawa et al., 2009). Although Aurora kinase homologs have been found in plant genomes, homologs of the partitioning defective complex and other animal polarizing proteins have not, indicating that plant cells use different mechanisms to establish cellular asymmetry (Rasmussen et al., 2011). Nevertheless, the role of Aurora-dependent phosphorylation in establishing asymmetry might be conserved.
Aurora kinases function as key regulators of mitosis. Yeasts have a single Aurora kinase, whereas metazoans and land plants have at least two members (Demidov et al., 2005; Kawabe et al., 2005). Animal Auroras can be clustered into two functionally divergent groups consisting of Aurora A versus Aurora B and C. Aurora A functions in early mitotic events and bipolar spindle formation (Barr and Gergely, 2007; Macůrek et al., 2008; Sardon et al., 2008; Seki et al., 2008; Johnston et al., 2009), while Aurora B is part of the chromosomal passenger complex that relocalizes dynamically throughout mitosis (Carmena et al., 2009). Aurora B, the catalytic subunit of the chromosomal passenger complex, regulates various functions along this path, including kinetochore maturation, chromosome biorientation and spindle assembly checkpoint control, central spindle organization, and cytokinesis (Ruchaud et al., 2007; Fuller et al., 2008; Kelly and Funabiki, 2009; Song et al., 2009). Aurora C has a predominant function in human testis but can complement Aurora B when exogenously expressed (Slattery et al., 2009).
Arabidopsis thaliana Aurora kinases can also be subdivided in two groups. The α-group consists of AUR1 and AUR2, while the β-group consists of AUR3. AUR1 and AUR2 show a dynamic localization pattern, reminiscent of AURORA B, whereas AUR3 accumulates at pericentromeric chromosomal regions (Demidov et al., 2005; Kawabe et al., 2005). Arabidopsis Aurora kinases have been shown to phosphorylate Ser-10 of HISTONE H3 in concert with posttranslational modifications of neighboring residues (Demidov et al., 2005, 2009; Kawabe et al., 2005). Chemical inactivation of Aurora, interfering with this phosphorylation, delays metaphase chromosome alignment and causes lagging anaphase chromosomes without inhibiting mitotic progression (Kurihara et al., 2006, 2008; Demidov et al., 2009). However, nothing is known about their developmental role.
Here, we provide evidence that the two α Auroras have redundant functions and are divergent from group β Arabidopsis AUR3, in agreement with their subcellular localizations. We further show that AUR1 and AUR2 are crucial in regulating the orientation of formative cell divisions from early embryogenesis onward. An α Aurora double mutant (aur1-2 aur2-2) can be rescued by a chimeric AUR1 construct that is degraded following metaphase, suggesting that these kinases function in regulating the orientation of formative cell divisions prior to cytokinesis. These results give insight into division plane orientation and introduce a developmental function for group α Arabidopsis Aurora kinases.
RESULTS
The aur1-2 aur2-2 Double Mutant Is Affected in Lateral Root Formation
The orientation of formative divisions in plants likely depends on the polarization of cells (De Smet and Beeckman, 2011; Rasmussen et al., 2011). As Drosophila AURORA A is involved in establishing polarity and spindle orientation during formative divisions (Wirtz-Peitz et al., 2008; Johnston et al., 2009), we investigated the biological function of the Arabidopsis Auroras in this process via a genetic approach focusing on the α-group. We identified Arabidopsis T-DNA insertion mutants in AUR1 and AUR2, designated as aur1-1, aur1-2, aur1-3, aur2-1, and aur2-2 (Figure 1A; see Supplemental Figure 1A online). As single mutants showed no macroscopic phenotype, double mutants were generated. Combining aur1-1 with aur2-2 alleles lead to gametophytic lethality as no plants could be recovered in which either one of the gametes carried both mutations (n = 480; see Supplemental Figure 1B online). The aur2-1 allele caused overexpression of AUR2 and was not studied further (see Supplemental Figure 1C online). Combining the weaker aur1-2 allele with aur2-2 (see Supplemental Figures 1D to 1F online) resulted in viable double homozygous plants with short internodes and a bushy appearance (see Supplemental Figures 1G to 1J online). Although ~12% of the seeds aborted (n = 125/1008; see Supplemental Figure 1I online), the plants were fertile. Cell cycle–dependent HISTONE H3 phosphorylation, a previously reported function of Arabidopsis Aurora kinases (Demidov et al., 2005, 2009; Kawabe et al., 2005) was not impaired in the aur1-2 aur2-2 double mutant, yet the dose-dependent hypersensitivity to Aurora inhibitor II (Mortlock et al., 2005) shows that these plants are affected in Aurora-specific functions (see Supplemental Figure 2 online).
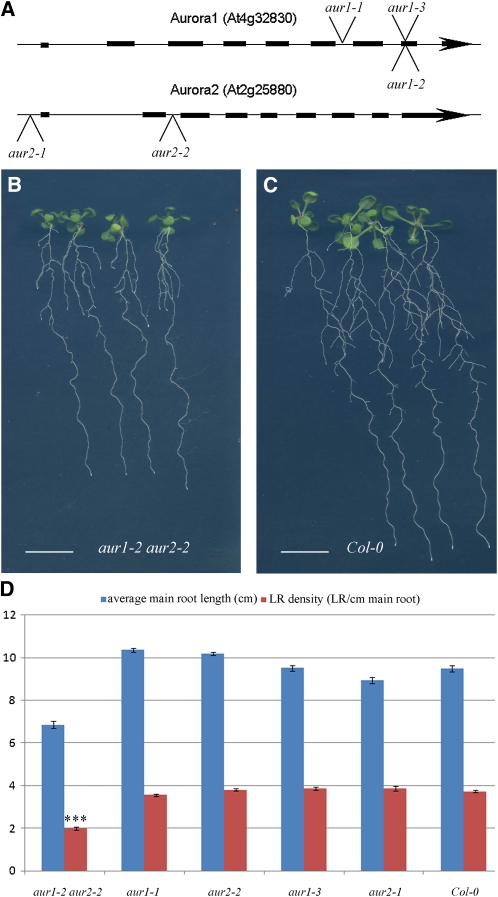
Figure 1.
The aur1-2 aur2-2 Double Mutant Shows Reduced Lateral Root Density.
(A) Gene models of Arabidopsis AUR1 and AUR2 with indications of T-DNA insertion lines analyzed. Introns are indicated by a line and exons by a black box.
(B) and (C) Representative images of the aur1-2 aur2-2 double mutant seedlings (B) grown for 12 d in continuous light showing a reduction in main root growth and lateral root density compared with wild-type Col-0 (C) seedlings.
(D) Quantification of average main root length and lateral root density between wild-type (Col-0, n = 36), several single Aurora T-DNA insertion lines (aur1-3, n = 27; aur2-1, n = 15; aur1-1, n = 57; aur2-2, n = 37), and the aur1-2 aur2-2 double mutant (n = 43). All single mutants show lateral root densities comparable to the wild type, while the aur1-2 aur2-2 mutant shows a statistically significant reduction in lateral root density (t test; triple asterisk; P < 0.0001).
Bars = 1 cm in (A) and (B), and error bars in (C) indicate se.
Twelve-day-old aur1-2 aur2-2 double mutant seedlings have an average main root length of ~72% of wild-type root length. Under these conditions, aur1-2 aur2-2 double mutants show a strong reduction in emerged lateral root density compared with the wild type and single mutants (Figures 1B to 1D). Arabidopsis AUR1 and AUR2 are expressed in the pericycle cells undergoing initial lateral root cell divisions (see Supplemental Figures 3C and 3D online), and detailed analysis of a translational fusion of AUR1 with β-glucuronidase showed expression in the pericycle nuclei before the first round of asymmetric divisions (see Supplemental Figure 4C online), in agreement with a function during the earliest stages of lateral root development.
The α and β Arabidopsis Aurora Kinases Are Functionally Divergent
To investigate whether reduced Aurora levels are causal to the observed mutant phenotypes, a complementation experiment was set up that also allowed us to assess functional redundancy among the Arabidopsis Auroras. Restoring the expression of either AUR1 or AUR2 using genomic constructs tagged with green fluorescent protein (GFP) (genomic fusions) complemented both the lateral root density and the bushy phenotype of aur1-2 aur2-2. However, introducing an extra genomic copy in addition to the wild-type AUR3 copy present or enhancing the expression of AUR3 from the functional AUR1 promoter did not (Figure 2; see Supplemental Figures 5A to 5H online). These results point to redundancy within and functional diversification between both groups of Aurora kinases. Subcellular localizations of functional α group Auroras in the aur1-2 aur2-2 background were highly similar with both proteins accumulating at the prophase spindle, mitotic microtubules, and the forming cell plate, whereas the genomic fusion of AUR3 accumulated at pericentromeric regions prior to cell division, marked the metaphase chromosomes, and reentered the reformed daughter nuclei without associating with the cell plate (see Supplemental Figure 3 online). The localization of the three Aurora kinases in Arabidopsis root meristem cells confirms previously reported localizations of these kinases in tobacco Bright Yellow-2 cells (Demidov et al., 2005; Kawabe et al., 2005), and the differential subcellular localization throughout cell division of both groups underlines their functional diversification.
Figure 2.
Complementation of the aur1-2 aur2-2 Double Mutant.
(A) Lateral root density experiment (11 d after sowing) comparing the aur1-2 aur2-2 double mutant (n = 24), the aur1-2 aur2-2 double mutant transformed with genomic fusions of the three Arabidopsis Aurora kinases (AUR1, n = 44; AUR2, n = 47; AUR3, n = 51), and the wild type (Col-0, n = 29). Restoring expression of either AUR1 or AUR2 restores lateral root densities to wild-type levels, while introducing another copy of AUR3 does not (t test; triple asterisk; P < 0.0001).
(B) Representative seedlings used for the quantification in (A) showing the rescue of the mutant phenotype by AUR1 and AUR2. Bar = 1 cm.
(C) Lateral root density experiment (11 d after sowing) comparing the aur1-2 aur2-2 double mutant (n = 64), the aur1-2 aur2-2 double mutant transformed with either the AUR1 (n = 77) or the AUR3 (n = 80) open reading frame fused to GFP expressed from the AUR1 promoter, and the wild type (Col-0, n = 28). Expressing AUR3 in the expression domain of AUR1 is not sufficient to rescue the aur1-2 aur2-2 phenotype (t test; triple asterisk; P < 0.0001), while expressing AUR1 fused to GFP from the same promoter returns lateral root density to wild-type values.
(D) Quantitative PCR analysis of AUR3 expression in the wild type (Col-0), the aur1-2 aur2-2 double mutant, and the aur1-2 aur2-2 double mutant expressing AUR3 from the promoter of AUR1 showing strongly enhanced expression of AUR3.
Error bars represent se ([A] and [C]) and sd (D).
Defective Lateral Root Outgrowth in the Double Mutant Is Caused by Aberrant Positioning of Division Planes
To test whether the decrease in lateral root density observed in the aur1-2 aur2-2 mutant is caused by reduced initiation of lateral root primordia or by defects in lateral root emergence, we calculated the average density of emerged and nonemerged lateral root primordia for wild-type (n = 20) and aur1-2 aur2-2 plants (n = 40) expressing pCYCB1;1:GUS (for β-glucuronidase). Whereas the total number of lateral roots and primordia did not statistically differ between the wild type and the aur1-2 aur2-2 double mutant, the latter showed significantly more nonemerged primordia compared with the wild type (t test; P < 0.0001). This indicates that aur1-2 aur2-2 seedlings are affected in primordium development and/or outgrowth rather than lateral root founder cell establishment (Figure 3A).
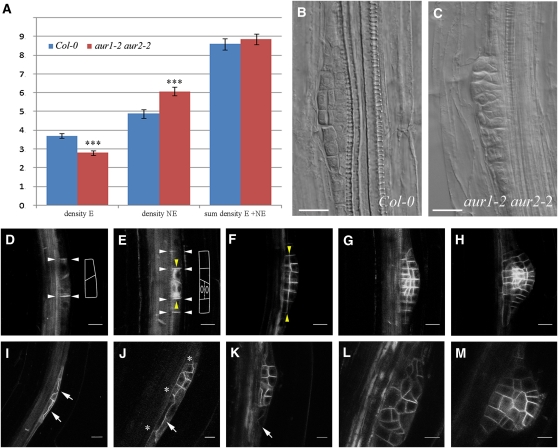
Figure 3.
The aur1-2 aur2-2 Double Mutant Shows Defects in Orienting Initial Formative Divisions during Lateral Root Primordium Formation.
(A) Analysis of emerged (E), nonemerged (NE), and total lateral root densities in wild-type (Col-0; n = 20) and aur1-2 aur2-2 double mutant seedlings (n = 40) measured by quantification of the proCycB1;1:GUS signal. The aur1-2 aur2-2 double mutant shows a clear reduction in emerged laterals and an increase in nonemerged laterals (t test; triple asterisk, P < 0.0001) compared with wild-type plants, while the total number of lateral root primordia does not statistically differ from the wild type.
(B) Wild-type (Col-0) stage III lateral root primordium.
(C) Multilayered lateral root primordium from the aur1-2 aur2-2 double mutant. Absence of patterning in the double mutant primordia is likely to interfere with the development and subsequent emergence of the lateral roots.
(D) to (M) Representative confocal images and traces for clarity of different stages of lateral root formation in the wild type (Col-0; [D] to [H]) and the aur1-2 aur2-2 double mutant ([I] to [M]) using proPIN1:PIN1-GFP as plasma membrane marker. White and yellow arrowheads indicate respective anticlinal and periclinal divisions. The aur1-2 aur2-2 double mutant shows defects in initial formative divisions, which are often periclinal instead of anticlinal ([I] to [K], arrows) and contain bifurcated cell plates ([J], asterisks). These aberrant divisions lead to unstructured lateral root primordia ([L] and [M]).
Bars = 10 μm in (B) to (H) and 20 μm in (I) to (M).
Lateral root primordia consist of a highly ordered layered pattern that is the result of controlled 90° switches between anticlinal and periclinal divisions and that has enabled the classification of successive developmental stages (Malamy and Benfey, 1997). When aur1-2 aur2-2 lateral root primordia were examined closely, it became apparent that these primordia showed defective patterning. In contrast with wild-type primordia, lateral root primordia in the aur1-2 aur2-2 mutant did not show a layered pattern and appeared to be built up out of random divisions, making it impossible to relate them to a defined developmental stage (cf. Figures 3B with 3C). This patterning phenotype was also rescued by reintroducing genomic fusions of either AUR1-GFP or AUR2-GFP in the aur1-2 aur2-2 double mutant background (see Supplemental Figure 6 online).
To analyze the earliest lateral root divisions in vivo, we used proPIN1:PIN1-GFP (Benková et al., 2003) to visualize the plasma membrane in the aur1-2 aur2-2 mutants (Figures 3I to 3M and 4; see Supplemental Figure 7 online). Lateral root formation starts with the migration of nuclei in two neighboring pericycle cells toward the common cell wall (De Rybel et al., 2010), followed by two rounds of anticlinal asymmetric divisions, creating a primordium of two and four cells centered on a usually skewed pericycle cell wall (Figures 3D and 3E, white arrowheads). The central cells of this early primordium will then shift their division plane by 90° and divide periclinally (Figure 3E, yellow arrowheads). These initial rounds of division are followed by additional divisions, creating a layered dome-shaped primordium (Figures 3F to 3H). aur1-2 aur2-2 mutants are defective in orienting the initial asymmetric cell divisions during lateral root primordium formation, although the preceding nuclear migration appears unaffected (Figure 3I). Aberrant first (Figure 4) and second (Figure 3J; see Supplemental Figure 7 online) asymmetric cell divisions the aur1-2 aur2-2 mutant lead to lateral root primordia with a reduced number of first layer cells (Figure 3K). Aberrant divisions include anchoring defects (Figure 4; t = 0 min) and aberrantly expanding cell plates (see Supplemental Figure 7 online). Aberrantly expanding plates are either periclinal from the beginning or initiate as anticlinal and shift to periclinal, resulting in an S-shaped cell plate (Figure 4; see Supplemental Figure 7 online). During these aberrantly oriented divisions, the expanding cell plate often bifurcates (Figures 3J and 4). The observed shift in cell plate orientation, bifurcated cell plates, and anchoring defects are likely a consequence of the aberrant guidance of these cell plates. Due to defective initial formative divisions, subsequent divisions also lack organization, causing the formation of a randomly patterned primordium (Figures 3L and 3M) that likely fails to grow out.
Figure 4.
Time Lapse of LR Formation in the aur1-2 aur2 Double Mutant Expressing proPIN1:PIN1-GFP.
Representative lateral root primordium starting with two aberrant and bifurcated first asymmetric divisions. Subsequent divisions occur but are randomized. The division marked by the white arrows starts out as anticlinal and then shifts to periclinal, making an S-shaped cell plate (e.g., image at 2 h 48 min). Also, the second round of asymmetric anticlinal divisions is substituted by periclinal divisions (yellow arrows) with bifurcated cell plates (asterisks). Bar = 10 μm.
We reasoned that the aberrantly oriented formative lateral root divisions could be caused by either altered division plane determination early in mitosis or by cell plate–associated functions of the α Auroras. To discriminate between these two possibilities, we designed a rescue construct targeting AUR1 for degradation prior to cytokinesis by fusing the coding sequence of AUR1 to the destruction box (Dbox) sequence of Arabidopsis CYCLINB1;1 and EGFP (Figure 5A). This Dbox sequence was previously reported to be sufficient to cause stimulus-dependent degradation of heterologous proteins (Colón-Carmona et al., 1999). In contrast with the cell plate association of the AUR1-GFP fusion protein (Figure 5B), AUR1-DboxGFP labeled the forming spindle until anaphase, after which it disappeared due to degradation (Figures 5C to 5C’’). However, expression of this construct rescued the aur1-2 aur2-2 phenotype (Figures 5D to 5G; see Supplemental Figures 5I to 5K online). These results indicate that the presence of AUR1 at the forming cell plate during cytokinesis does not significantly contribute to the observed lateral root phenotype and that Arabidopsis α group Auroras function in division plane orientation prior to cytokinesis.
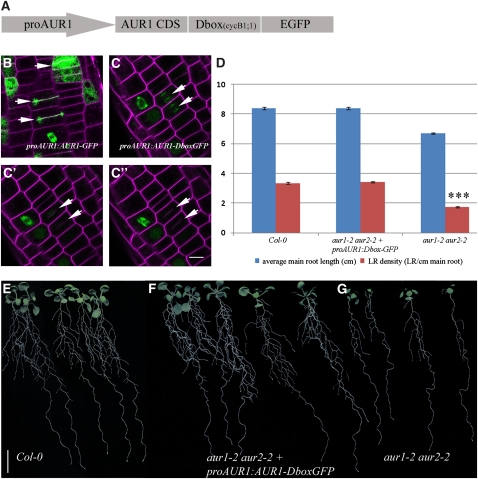
Figure 5.
Aurora Functions before Cytokinesis.
(A) Schematic representation of the Dbox construct used to assess complementation of the aur1-2 aur2-2 double mutant.
(B) Localization of proAUR1:AUR1-GFP in Arabidopsis root cells stained with FM4-64 showing AUR1 accumulation at the forming cell plate (arrows).
(C) to (C’’) Time-lapse image series of aur1-2 aur2-2 double mutant root cells expressing the Dbox construct depicted in (A) and stained with FM4-64. Modulation of AUR1 expression by the Dbox of CycB1;1 abolishes the accumulation of AUR1-GFP at the forming cell plate (arrows).
(D) Average main root length and lateral root density comparison between the aur1-2 aur2-2 double mutant (n = 82), the wild type (Col-0, n = 36), and the aur1-2 aur2-2 double mutant expressing Dbox-tagged AUR1-GFP (n = 74). Expression of AUR1-Dbox-GFP restores main root length and lateral root density to wild-type levels and is highly statistically different from the lateral root density in the aur1-2 aur2-2 double mutant (t test; triple asterisk; P < 0.0001).
(E) to (G) Representative seedlings grown for 13 d in continuous light showing the reduced lateral root density of the aur1-2 aur2-2 double mutant and average lateral root density of Dbox-tagged AUR1, which is comparable to the wild type (Col-0).
Bar = 10 μm in (B) to (C’’) and 1 cm in (E) to (G). Error bars in (D) indicate se.
The Aurora Double Mutant Shows Defects in Orienting Formative Cell Division throughout Development
To determine whether the defect in formative division plane orientation of the aur1-2 aur2-2 mutant was confined to lateral root development, we examined other types of formative divisions. Next to lateral root primordia, AUR1 expression was observed in meristematic root and shoot tissues, during embryogenesis, and in the undifferentiated cells of the stomatal lineage (see Supplemental Figures 4A to 4G online). aur1-2 aur2-2 embryos until the octant stage contained aberrant divisions in >40% of embryos analyzed (n = 74/242), while in wild-type plants, deviating divisions were found only in ~5% of embryos analyzed (n = 9/187). Deviations in division plane orientation in the aur1-2 aur2-2 mutant occurred both in the embryo proper and suspensor cells and ranged from oblique to a 90° shift (Figures 6A to 6D). At later stages of embryo development (Figures 6E to 6H), aberrant divisions were apparent at the basal pole where the root meristem is initiated, which translates into seedlings without a main root in 6% of germinated aur1-2 aur2-2 mutants (n = 28/466; Figure 6I). Seedlings that developed a main root displayed misoriented meristematic cell divisions, for instance, in the endodermis layer (Figure 6K) and showed ectopic division planes around the quiescent center (QC) in 80% (n = 83/106) of analyzed aur1-2 aur2-2 seedlings without abolishing QC cell fate (Figures 6J to 6L). These aberrant divisions likely affect root meristem activity, resulting in the shorter main roots observed in aur1-2 aur2-2 double mutants (Figures 1B to 1D). Stomatal development itself did not appear to be impaired in the aur1-2 aur2-2 double mutant. Nevertheless, aberrant epidermal divisions leading to the formation of small cells that are linked to defective breaking of asymmetry (Dong et al., 2009) were observed frequently (Figures 6M to 6O, arrows; see Supplemental Figure 8 online, brackets). These results suggest that AUR1 and AUR2 play an important role in orienting the division plane of formative cell divisions throughout plant development.
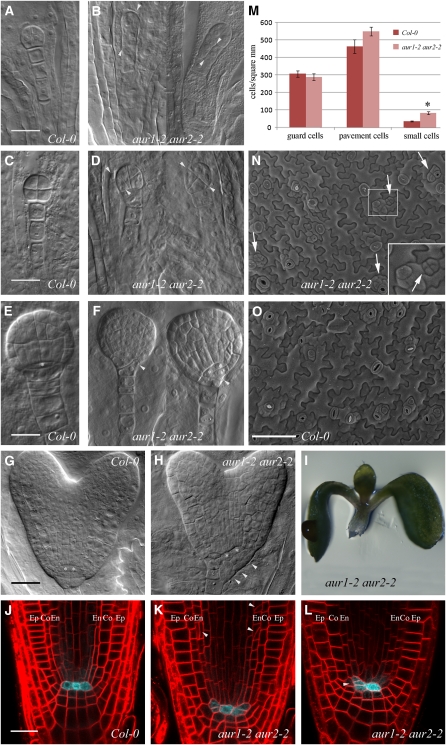
Figure 6.
aur1-2 aur2-2 Double Mutants Are Affected in Formative Divisions throughout Development.
(A) to (H) Defective orientation of formative divisions during embryogenesis.
(A) to (D) Wild-type (Col-0; [A] and [C]) and aur1-2 aur2-2 embryos ([B] and [D]) showing aberrant orientations (arrowheads) of early divisions.
(E) to (H) Wild-type (Col-0; [E] and [G]) and aur1-2 aur2-2 embryos ([F] and [H]) with altered division orientations of the hypophysis (arrowheads). Asterisks mark QC cells.
(I) Representative seedling germinating without a main root, which is observed in 6% of the germinated aur1-2 aur2-2 seedlings (n = 28/466) and likely the result of accumulated defective formative divisions of the hypophysis.
(J) to (L) FM4-64–stained main root meristems of the wild type (Col-0; [J]) and aur1-2 aur2-2 mutants ([K] and [L]) expressing the QC25:CFP marker. aur1-2 aur2-2 mutants maintain QC identity but show aberrant formative divisions of the cortex and endodermis initials (arrowheads in [L]), leading to altered organization of cells surrounding the QC. Co, cortex; En, endodermis layer; Ep, epidermis.
(M) Quantification of wild-type (Col-0) and aur1-2 aur2-2 epidermal cells from 8-d-old cotyledons showing defective cell divisions in the stomatal lineage. aur1-2 aur2-2 mutants contain comparable densities of stomatal and pavement cells but produce significantly more small cells than the wild type (t test; asterisk, P < 0.05).
(N) and (O) Representative scanning electron microscopy overview image of 8-d-old aur1-2 aur2-2 mutant (N) and wild-type (Col-0; [O]) cotyledons. Aberrant orientations of cell division leading to the production of small cells in the aur1-2 aur2-2 mutant are indicated by arrows and shown in the inset in (N).
Bars = 20 μm in (A) to (L) and 100 μm in (N) and (O). Error bars in (M) indicate sd.
DISCUSSION
Although Aurora kinases are conserved throughout the eukaryotic kingdom, their functions have somehow evolved and diversified. Vertebrates evolved two subclades of Aurora kinase, A and B, with separate functions and interacting proteins (Carmena et al., 2009). Yeasts contain a single Aurora kinase resembling B-type Aurora (Bohnert et al., 2009; Nakajima et al., 2009), while the Dictyostelium discoideum (DdAurora) and Asterina pectinifera (ApAurora) Aurora kinases have properties of both A- and B-type Auroras (Li et al., 2008; Abe et al., 2010). Arabidopsis, like vertebrates, also contains three Aurora kinases that can be clustered into two groups that do not appear to resemble the A and B type (Demidov et al., 2005; Kawabe et al., 2005), highlighting the diversification of Aurora kinases throughout the eukaryotic kingdom.
To date, research on Arabidopsis Aurora kinases has focused mainly on HISTONE H3 phosphorylation and chromosome segregation without making functional discriminations among the different members. Cell cycle–dependent phosphorylation of HISTONE H3 is performed by all three Arabidopsis Aurora kinases (Demidov et al., 2005, 2009; Kawabe et al., 2005; Kurihara et al., 2006, 2008). Therefore, the activity of AUR3, together with residual activity of α Auroras, might explain why no differential immunostaining was observed between Arabidopsis wild-type and aur1-2 aur2-2 mutant plants using an antibody against phosphorylated HISTONE H3.
The data presented here, the differential localization of both groups of Aurora kinases in Bright Yellow-2 cells (Demidov et al., 2005) and Arabidopsis seedlings (this work), and in silico modeling (Vos et al., 2008) nevertheless point toward diverged functions for both groups as the specificity of these kinases is largely determined by their interactors (Eyers et al., 2005; Hans et al., 2009). Both members of group α likely have redundant functions, as the aur1-2 aur2-2 mutant is equally complemented by reintroducing genomic fusions of AUR1 or AUR2. By contrast, AUR3, the single member of the group β, is unable to overcome the lack of group α Auroras, independent of the level or domain of expression (Menges et al., 2003; Demidov et al., 2005), proving functional divergence of the groups for certain functions.
Chemical treatment of wild-type plants with high doses of Aurora kinase inhibitors will also likely target AUR3 and potentially even nonrelated kinases, making it difficult to attribute potential effects to reduced AUR1 and AUR2 activities. Therefore, we have taken a genetic approach to unravel the function of the group α Aurora kinases and were able to identify a developmental function for this subclade through the identification of a viable double mutant. This viability likely depends on a combination of the expression of a C-terminally truncated AUR1 and/or a partially functional AUR2 lacking the first two exons. This follows from the facts that (1) stronger allelic combinations (aur1-1 aur2-2) resulted in gametophytic lethality, (2) no changes in cell cycle–dependent HISTONE H3 phosphorylation could be observed in the double mutant, and (3) these mutant plants exhibit hypersensitivity to Aurora kinase inhibitor II. The latter property could provide an easy and powerful system to screen for specific Aurora kinase inhibitors, a research field that has developed actively over the last couple of years (Pérez Fidalgo et al., 2009).
Surprisingly, although Arabidopsis Auroras are expressed strongly in dividing cells (Demidov et al., 2005) and although a transcriptional fusion of AUR1 accumulates highly in the root apical meristem and the central cylinder, proliferative cell division orientations in the main root are hardly affected in aur1-2 aur2-2 mutants. Rather, the observed defects in embryonal and pericycle divisions and divisions surrounding the QC point to a prominent role for α Auroras in orienting formative divisions, when correct changes in the orientation of cell divisions are most crucial. Lateral root primordium formation is highly sensitive toward division plane orientation defects, as tightly regulated switches in orientation are essential to produce the layered dome-shaped primordium able to penetrate the several layers of root cells during emergence (Swarup et al., 2008; Péret et al., 2009). The occurrence of small cotyledon epidermal cells in the aur1-2 aur2-2 mutant is also in agreement with a function for α Auroras in orienting formative divisions, as defective formative stomatal lineage divisions lead to the formation of small cells via additional rounds of cell division (Dong et al., 2009).
However, the specific defects observed in the aur1-2 aur2-2 mutant also can be explained by a higher stringency of formative over proliferative divisions to regulate their division plane or by the fact that residual truncated expression of AUR1 and/or AUR2 in the aur1-2 aur2-2 mutant is sufficient to orient proliferative divisions. In light of this, the stronger effects on lateral root and embryonal divisions compared with main root proliferative divisions in the aur1-2 aur2-2 mutant could be a consequence of the strict temporal requirement to switch the division plane orientation in these fast-occurring divisions.
Although we cannot completely exclude chromosome separation defects and aneuploidy causing the observed defects, the specificity of the defects, the fertility of the aur1-2 aur2-2 mutant plants, and the repetitive rounds of random divisions occurring during lateral root primordium formation in the double mutant argue against this. Together, the data provided favor a role for Arabidopsis AUR1 and AUR2 in formative division plane orientation, a process that we are only beginning to unravel in plants (Dong et al., 2009).
Drosophila Aurora A has been shown to indirectly function in orienting asymmetric mitotic divisions by affecting spindle orientation (Johnston et al., 2009). In contrast with animal cells, land plant cells determine their somatic division plane much earlier in mitosis. The first visible sign of division plane determination consists of the construction of the preprophaseband (PPB), which encircles the premitotic nucleus. The PPB aids oriented bipolar spindle formation and positions positive and negative markers that prolong the determination of the division plane throughout mitosis and guide the growing cytokinetic cell plate (Chan et al., 2005; Ambrose and Cyr, 2008; Müller et al., 2009; Van Damme, 2009). Therefore, ectopically positioned cell plates either result from aberrant division plane determination or from altered centrifugal growth of the cell plate (Van Damme, 2009). Cell plate growth is not impaired in the double mutant, and rescue does not require increased AUR1 protein levels during cytokinesis, arguing for a function of α Auroras in regulating cell division orientation earlier during mitosis. It remains to be seen if formative and proliferative cell division demarcation share common mechanisms and if the defects observed in the aur1-2 aur2-2 mutant are linked with PPB formation and/or known division plane markers (Vanstraelen et al., 2006; Walker et al., 2007; Azimzadeh et al., 2008; Xu et al., 2008; Dong et al., 2009; Wright et al., 2009; Spinner et al., 2010) To our knowledge, no division plane markers have so far been reported to function in formative root divisions in Arabidopsis, and functional Aurora-GFP fusions do not associate with the PPB in Arabidopsis roots. One future task will be to clarify the mechanism by which these kinases affect cell division orientation.
METHODS
T-DNA Insertion Lines in AUR1 (At4g32830) and AUR2 (At2g25880)
For AUR1, two SALK lines (SALK_031697, aur1-2; SALK_112121, aur1-3) were mapped to the same insertion site on exon 8 by sequencing the T-DNA–specific PCR fragments (primers are given in Supplemental Table 1 online), and a Gabi-Kat line (GK225FO7, aur1-1) was mapped to intron 6. For AUR2, a Gabi-Kat line (GK403B02, aur2-2) was identified to disrupt the second intron, whereas a Wisconsin DsLox line (WsDsLx368B03, aur2-1) mapped to the promoter region.
Plant Growth and Transformation
Arabidopsis thaliana ecotype Columbia-0 (Col-0) plants were grown under standard growth conditions in continuous light on vertical plates containing half-strength Murashige and Skoog (MS) medium supplemented with 8 g/L plant tissue culture agar and 1% Suc. All Aurora single and double mutants were genotyped using the primers listed in Supplemental Table 1 online. Wild-type Col-0 and aur1-2 aur2-2 double mutant plants were transformed using the floral dip protocol (Clough and Bent, 1998). proCycB1;1-GUS, proPIN1:PIN1-GFP, and proQC25:CFP (for cyan fluorescent protein) used in this study were described previously (Ferreira et al., 1994; Benková et al., 2003; Sabatini et al., 2003). The basl-2 mutant (Dong et al., 2009) was kindly provided by Dominique Bergmann.
Lateral Root Density Measurements
Seeds were sown and vernalized at 4°C for 3 d prior to transfer to growing conditions. Plants were grown on vertical plates for 12 to 13 d in continuous light conditions. Synchronization of germination was scored after 2 d in the light, and late germinating seedlings were excluded from further analysis. Emerged lateral roots were counted manually using a Leica S4E stereomicroscope. Plates were scanned using an Epson perfection V700 photo scanner at 300 dpi, and main root length was analyzed from these scans using the ImageJ software package (http://rsbweb.nih.gov/ij/). Col-0 and aur1-2 aur2-2 double mutants expressing proCYCB1:1-GUS were grown in continuous light for 10 d. Main root length was measured from the scanned plates, and GUS staining was performed on individual seedlings as described by Péret et al. (2007). Seedlings were subsequently mounted between slide and cover slip, and early primordia were scored on an Olympus BX51 light microscope. Very young primordia were taken into account only when multiple cell divisions could be observed to avoid including proliferative pericycle divisions.
Cloning of Constructs
All constructs were made using single and multiple Gateway recombination reactions (Invitrogen). Cloning of coding sequence constructs of AUR1 and AUR3 in pDONR207 have been described before (Van Damme et al., 2004). Genomic fusions with EGFP were made by amplifying genomic clones (promoter until the last codon) of AUR1, AUR2, and AUR3 using the primers listed in the Supplemental Table 1 online. PCR products were separated and purified from gel using the high pure PCR purification kit (Roche) and cloned into pDONR221 by BP reaction and to pB7FWG,0 by LR reaction to allow expression of Aurora-GFP fusions at endogenous levels. The proAUR1:GUS-AUR1, proAUR1:AUR1-GFP, and proAUR1:AUR3-GFP clones were made by recombining proAUR1 in pDONRP4P1R with the GUS open reading frame in pDONR221 and AUR1 in pDONRP2P3R, by combining proAUR1 with AUR1 in pDONR207 and EGFP in pDONRP2P3R, or by combining proAUR1 with EGFP in pDONR221 and AUR1 in pDONRP2P3R into pB7m34GW (AUR1-GFP and AUR3-GFP) or pK7m34GW (GUS-AUR1) (Karimi et al., 2005, 2007). The Dbox-GFP clone was made by amplifying the first 116 amino acids of Arabidopsis Cyclin B1;1 (At4g37490) in pDONR221 (Boruc et al., 2010) according to what has been described previously (Colón-Carmona et al., 1999), using a forward primer with an attB2 site and a reverse primer containing the first 20 bp of EGFP. EGFP was then amplified from pDONRP2P3R-EGFP with a forward primer containing the last 20 bp of CycB1;1 and a reverse EGFP primer attached to an attB3 site. Both the Dbox-amplified sequence and the EGFP amplified sequence were stitched together by a sewing PCR reaction, and the product (Dbox-EGFP) was cloned into pDONRP2P3R.
Inhibitor Treatments
Aurora kinase inhibitor II (Calbiochem) was dissolved in DMSO to a 20 mM stock solution and added to half-strength MS plates at 10 and 20 μM final concentration. Arabidopsis seedlings were germinated on inhibitor-containing plates or on control plates supplied with an equal amount of DMSO. Main root length was scored after 6 d in continuous light.
Confocal Microscopy
Image acquisition was obtained with a FluoView1000 inverted confocal microscope (Olympus) equipped with a water-corrected ×60 objective (numerical aperture of 1.2) using 488-nm laser excitation and a spectral detection bandwidth of 500 to 530 nm for EGFP and 559-nm laser excitation together with a spectral detection bandwidth of 570 to 670 nm for FM4-64 detection, with a Zeiss 710 inverted confocal microscope with the ZEN 2009 software package and equipped with ×40 and ×63 water-corrected objectives (numerical aperture of 1.2). EGFP was visualized using 488-nm laser excitation and 500- to 530-nm spectral detection; FM4-64 was visualized using 458-nm laser excitation and 592- to 754-nm spectral detection, and propidium iodide was visualized using 514-nm laser excitation and 566- to 649-nm spectral detection.
Lateral root development time-lapse microscopy was done by mounting Arabidopsis seedlings in a chambered cover glass system (Lab-Tek). Seedlings were covered with a slice of half-strength MS medium to immobilize and prevent dehydration. Early lateral root primordium development was followed over time using the time-lapse acquisition tool of the Olympus FV1000 or Zeiss 710 confocal microscopes using a ×60 (Olympus) or ×63 (Zeiss) water-corrected lens.
Quantitative RT-PCR
RNA was prepared from young whole seedlings grown on vertical plates. RNA was extracted using the RNeasy mini kit (Qiagen), and cDNA was made using the iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR reactions were performed using the Platinum SYBR Green qPCR Supermix-UDG kit (Invitrogen) on a Bio-Rad iCycler, and cycle threshold values were analyzed using the Qbase software package (Hellemans et al., 2007). Primers against EEF1A4 were used as normalization genes in all quantitative PCR experiments.
Embryo Analysis
Embryo development was examined in several plants that were confirmed as double mutants by genotyping PCR. Plants that were genotyped as wild-type from the same mother plant served as internal controls. Embryos were mounted in Hoyer’s medium (Bougourd et al., 2000) and visualized after clearing on an Olympus BX51 light microscope. GUS staining of proAUR1:GUS-AUR1–expressing embryos was performed as described by Péret et al. (2007).
List of Primers Used in This Study
All primers used in this study can be found in Supplemental Table 1 online.
Indirect Immunofluorescence
Preparation of chromosomes and immunostaining was performed as described by Manzanero et al. (2000). Rabbit antibodies against histone H3S10ph (Upstate) were diluted 1:400 in PBS. After 12 h of incubation at 4°C and subsequent washing, slides were incubated with rhodamine-conjugated anti-rabbit IgG (Dianova) diluted 1:200. After final washes, preparations were mounted in antifade containing 4',6-diamidino-2-phenylindole as counterstain. Immunofluorescence was recorded with an Olympus BX61 microscope equipped with an ORCA-ER charge-coupled device camera (Hamamatsu). All images were collected in gray scale and pseudocolored with Adobe Photoshop.
Epidermal Cell Measurements
Cotyledons from 8-d-old plants (grown on half-strength MS medium without added sugar under a 16/8 photoperiod) were detached and adhered on the adaxial side to a flat surface covered with double-sided sticky tape. Dental resin Genie VPS light body (Sultan Healthcare) was applied to the abaxial surface. The dental resin mold was filled with nail polish to create a cast that was imaged using a Hitachi TM-1000 scanning electron microscope. Cells were counted using the ImageJ software package (http://rsbweb.nih.gov/ij/). Cells were classified as guard cells, pavement cells, or small cells; the latter defined as being smaller than a guard cell, with round to rectangular shape and without any protrusions as defined by Dong et al. (2009). Visualization of adaxial cotyledon epidermal patterning by fluorescence microscopy was done by dipping seedlings (3 d after germination) in 1 μg/μL propidium iodide (Sigma-Aldrich) followed by mounting seedlings in water between slide and cover slip.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or databases under the following accession numbers: AUR1, At4g32830; AUR2, At2g25880; AUR3, At2g45490.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Quantitative PCR Analysis of T-DNA Insertion Lines in AUR1 and AUR2 and Macroscopic Phenotype of the aur1-2 aur2-2 Double Mutant.
Supplemental Figure 2. Cell Cycle–Dependent HISTONE H3 Phosphorylation and Sensitivity to Aurora Kinase Inhibitor of the aur1-2 aur2-2 Double Mutant.
Supplemental Figure 3. Localization of Genomic Aurora-GFP Constructs in Arabidopsis Root Cells.
Supplemental Figure 4. Expression Pattern of AUR1.
Supplemental Figure 5. Complementation of the aur1-2 aur2-2 Double Mutant Bushy Phenotype.
Supplemental Figure 6. Rescue of the Lateral Root Patterning Defect by Genomic Fusions of AUR1-GFP and AUR2-GFP in the aur1-2 aur2-2 Double Mutant Background.
Supplemental Figure 7. Time-Lapse Images of a Developing aur1-2 aur2-2 Lateral Root Primordium.
Supplemental Figure 8. Comparison between the Small Cells Occurring in the basl-2 and the aur1-2 aur2-2 Mutant Backgrounds.
Supplemental Table 1. List of Primers Used in This Study.
Acknowledgments
We thank Dominique Bergmann for providing basl-2 seeds. This work was supported by grants from the Interuniversity Attraction Poles Programme (IUAP VI/33), initiated by the Belgian State, Science Policy Office, and the Research Foundation-Flanders (G.0065.08). D.V.D., I.D.S., and W.G. are postdoctoral fellows of the Research Foundation-Flanders. I.D.S. is supported by a Biotechnology and Biological Science Research Council David Phillips Fellowship (BB_BB/H022457/1). D.D. and A.H. are supported by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereiche 648). G.G. was supported by the Belgian Science Policy Office for a postdoctoral fellowship for Non-European Union Researchers and the Research Foundation-Flanders (G.0065.08) and is now a Career Investigator of the Consejo Nacional de Investigaciones Científicas y Técnicas. B.D.R. was the recipient of a predoctoral fellowship of the Special Research Fund of Ghent University.
AUTHOR CONTRIBUTIONS
D.V.D., I.D.S., A.H., T.B., and E.R. designed the research. D.V.D., B.D.R., G.G., D.D., and W.G. performed the research. D.V.D., B.D.R., G.G., D.D., W.G., I.D.S., A.H., T.B., and E.R. wrote the article.
References
- Abe Y., Okumura E., Hosoya T., Hirota T., Kishimoto T. (2010). A single starfish Aurora kinase performs the combined functions of Aurora-A and Aurora-B in human cells. J. Cell Sci. 123: 3978–3988 [DOI] [PubMed] [Google Scholar]
- Ambrose J.C., Cyr R. (2008). Mitotic spindle organization by the preprophase band. Mol. Plant 1: 950–960 [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Nacry P., Christodoulidou A., Drevensek S., Camilleri C., Amiour N., Parcy F., Pastuglia M., Bouchez D. (2008). Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 20: 2146–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr A.R., Gergely F. (2007). Aurora-A: The maker and breaker of spindle poles. J. Cell Sci. 120: 2987–2996 [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Besson S., Dumais J. (2011). Universal rule for the symmetric division of plant cells. Proc. Natl. Acad. Sci. USA 108: 6294–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert K.A., Chen J.-S., Clifford D.M., Vander Kooi C.W., Gould K.L. (2009). A link between aurora kinase and Clp1/Cdc14 regulation uncovered by the identification of a fission yeast borealin-like protein. Mol. Biol. Cell 20: 3646–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J., Mylle E., Duda M., De Clercq R., Rombauts S., Geelen D., Hilson P., Inzé D., Van Damme D., Russinova E. (2010). Systematic localization of the Arabidopsis core cell cycle proteins reveals novel cell division complexes. Plant Physiol. 152: 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougourd S., Marrison J., Haseloff J. (2000). Technical advance: An aniline blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. Plant J. 24: 543–550 [DOI] [PubMed] [Google Scholar]
- Carmena M., Ruchaud S., Earnshaw W.C. (2009). Making the Auroras glow: Regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 21: 796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Calder G., Fox S., Lloyd C. (2005). Localization of the microtubule end binding protein EB1 reveals alternative pathways of spindle development in Arabidopsis suspension cells. Plant Cell 17: 1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Demidov D., Hesse S., Tewes A., Rutten T., Fuchs J., Ashtiyani R.K., Lein S., Fischer A., Reuter G., Houben A. (2009). Aurora1 phosphorylation activity on histone H3 and its cross-talk with other post-translational histone modifications in Arabidopsis. Plant J. 59: 221–230 [DOI] [PubMed] [Google Scholar]
- Demidov D., Van Damme D., Geelen D., Blattner F.R., Houben A. (2005). Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell 17: 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., et al. (2010). A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- De Smet I., Beeckman T. (2011). Asymmetric cell division in land plants and algae: The driving force for differentiation. Nat. Rev. Mol. Cell Biol. 12: 177–188 [DOI] [PubMed] [Google Scholar]
- Dong J., MacAlister C.A., Bergmann D.C. (2009). BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyers P.A., Churchill M.E.A., Maller J.L. (2005). The Aurora A and Aurora B protein kinases: A single amino acid difference controls intrinsic activity and activation by TPX2. Cell Cycle 4: 784–789 [DOI] [PubMed] [Google Scholar]
- Ferreira P.C.G., Hemerly A.S., Engler J.D., van Montagu M., Engler G., Inzé D. (1994). Developmental expression of the Arabidopsis cyclin gene cyc1At. Plant Cell 6: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller B.G., Lampson M.A., Foley E.A., Rosasco-Nitcher S., Le K.V., Tobelmann P., Brautigan D.L., Stukenberg P.T., Kapoor T.M. (2008). Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature 453: 1132–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. (2008). Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9: 355–366 [DOI] [PubMed] [Google Scholar]
- Hans F., Skoufias D.A., Dimitrov S., Margolis R.L. (2009). Molecular distinctions between Aurora A and B: A single residue change transforms Aurora A into correctly localized and functional Aurora B. Mol. Biol. Cell 20: 3491–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8: R19.1–R19.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer A., Berdnik D., Wirtz-Peitz F., Žigman M., Schleiffer A., Knoblich J.A. (2006). Mitotic activation of the kinase Aurora-A requires its binding partner Bora. Dev. Cell 11: 147–157 [DOI] [PubMed] [Google Scholar]
- Johnston C.A., Hirono K., Prehoda K.E., Doe C.Q. (2009). Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell 138: 1150–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Bleys A., Vanderhaeghen R., Hilson P. (2007). Building blocks for plant gene assembly. Plant Physiol. 145: 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kawabe A., Matsunaga S., Nakagawa K., Kurihara D., Yoneda A., Hasezawa S., Uchiyama S., Fukui K. (2005). Characterization of plant Aurora kinases during mitosis. Plant Mol. Biol. 58: 1–13 [DOI] [PubMed] [Google Scholar]
- Kelly A.E., Funabiki H. (2009). Correcting aberrant kinetochore microtubule attachments: An Aurora B-centric view. Curr. Opin. Cell Biol. 21: 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D., Matsunaga S., Kawabe A., Fujimoto S., Noda M., Uchiyama S., Fukui K. (2006). Aurora kinase is required for chromosome segregation in tobacco BY-2 cells. Plant J. 48: 572–580 [DOI] [PubMed] [Google Scholar]
- Kurihara D., Matsunaga S., Uchiyama S., Fukui K. (2008). Live cell imaging reveals plant aurora kinase has dual roles during mitosis. Plant Cell Physiol. 49: 1256–1261 [DOI] [PubMed] [Google Scholar]
- Li H., Chen Q., Kaller M., Nellen W., Gräf R., De Lozanne A. (2008). Dictyostelium Aurora kinase has properties of both Aurora A and Aurora B kinases. Eukaryot. Cell 7: 894–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macůrek L., Lindqvist A., Lim D., Lampson M.A., Klompmaker R., Freire R., Clouin C., Taylor S.S., Yaffe M.B., Medema R.H. (2008). Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455: 119–123 [DOI] [PubMed] [Google Scholar]
- Malamy J.E., Benfey P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Manzanero S., Arana P., Puertas M.J., Houben A. (2000). The chromosomal distribution of phosphorylated histone H3 differs between plants and animals at meiosis. Chromosoma 109: 308–317 [DOI] [PubMed] [Google Scholar]
- Menges M., Hennig L., Gruissem W., Murray J.A.H. (2003). Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Mortlock A.A., Keen N.J., Jung F.H., Heron N.M., Foote K.M., Wilkinson R.W., Green S. (2005). Progress in the development of selective inhibitors of aurora kinases. Curr. Top. Med. Chem. 5: 807–821 [DOI] [PubMed] [Google Scholar]
- Müller S., Wright A.J., Smith L.G. (2009). Division plane control in plants: New players in the band. Trends Cell Biol. 19: 180–188 [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Tyers R.G., Wong C.C.L., Yates J.R., III, Drubin D.G., Barnes G. (2009). Nbl1p: A Borealin/Dasra/CSC-1-like protein essential for Aurora/Ipl1 complex function and integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 20: 1772–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H., Ohta N., Moon W., Matsuzaki F. (2009). Protein phosphatase 2A negatively regulates aPKC signaling by modulating phosphorylation of Par-6 in Drosophila neuroblast asymmetric divisions. J. Cell Sci. 122: 3242–3249 [DOI] [PubMed] [Google Scholar]
- Péret B., De Rybel B., Casimiro I., Benková E., Swarup R., Laplaze L., Beeckman T., Bennett M.J. (2009). Arabidopsis lateral root development: An emerging story. Trends Plant Sci. 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Péret B., Swarup R., Jansen L., Devos G., Auguy F., Collin M., Santi C., Hocher V., Franche C., Bogusz D., Bennett M., Laplaze L. (2007). Auxin influx activity is associated with Frankia infection during actinorhizal nodule formation in Casuarina glauca. Plant Physiol. 144: 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez Fidalgo J.A., Roda D., Roselló S., Rodríguez-Braun E., Cervantes A. (2009). Aurora kinase inhibitors: A new class of drugs targeting the regulatory mitotic system. Clin. Transl. Oncol. 11: 787–798 [DOI] [PubMed] [Google Scholar]
- Rasmussen C.G., Humphries J.A., Smith L.G. (2011). Determination of symmetric and asymmetric division planes in plant cells. Annu. Rev. Plant Biol. 62: 387–409 [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. (2007). Chromosomal passengers: Conducting cell division. Nat. Rev. Mol. Cell Biol. 8: 798–812 [DOI] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardon T., Peset I., Petrova B., Vernos I. (2008). Dissecting the role of Aurora A during spindle assembly. EMBO J. 27: 2567–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki A., Coppinger J.A., Jang C.Y., Yates J.R., Fang G. (2008). Bora and the kinase Aurora A cooperatively activate the kinase Plk1 and control mitotic entry. Science 320: 1655–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery S.D., Mancini M.A., Brinkley B.R., Hall R.M. (2009). Aurora-C kinase supports mitotic progression in the absence of Aurora-B. Cell Cycle 8: 2984–2994 [PubMed] [Google Scholar]
- Song S.J., Kim S.J., Song M.S., Lim D.-S. (2009). Aurora B-mediated phosphorylation of RASSF1A maintains proper cytokinesis by recruiting Syntaxin16 to the midzone and midbody. Cancer Res. 69: 8540–8544 [DOI] [PubMed] [Google Scholar]
- Spinner L., Pastuglia M., Belcram K., Pegoraro M., Goussot M., Bouchez D., Schaefer D.G. (2010). The function of TONNEAU1 in moss reveals ancient mechanisms of division plane specification and cell elongation in land plants. Development 137: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Swarup K., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Van Damme D. (2009). Division plane determination during plant somatic cytokinesis. Curr. Opin. Plant Biol. 12: 745–751 [DOI] [PubMed] [Google Scholar]
- Van Damme D., Bouget F.-Y., Van Poucke K., Inzé D., Geelen D. (2004). Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. Plant J. 40: 386–398 [DOI] [PubMed] [Google Scholar]
- Vanstraelen M., Van Damme D., De Rycke R., Mylle E., Inzé D., Geelen D. (2006). Cell cycle-dependent targeting of a kinesin at the plasma membrane demarcates the division site in plant cells. Curr. Biol. 16: 308–314 [DOI] [PubMed] [Google Scholar]
- Vos J.W., Pieuchot L., Evrard J.-L., Janski N., Bergdoll M., de Ronde D., Perez L.H., Sardon T., Vernos I., Schmit A.-C. (2008). The plant TPX2 protein regulates prospindle assembly before nuclear envelope breakdown. Plant Cell 20: 2783–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker K.L., Müller S., Moss D., Ehrhardt D.W., Smith L.G. (2007). Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr. Biol. 17: 1827–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Nishimura T., Knoblich J.A. (2008). Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell 135: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A.J., Gallagher K., Smith L.G. (2009). discordia1 and alternative discordia1 function redundantly at the cortical division site to promote preprophase band formation and orient division planes in maize. Plant Cell 21: 234–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.M., Zhao Q., Rodrigo-Peiris T., Brkljacic J., He C.S., Müller S., Meier I. (2008). RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc. Natl. Acad. Sci. USA 105: 18637–18642 [DOI] [PMC free article] [PubMed] [Google Scholar]