This work describes a chloroplast-to-nucleus retrograde signaling pathway involving SAL1, a chloroplast and mitochondrial enzyme that degrades the phosphonucleotide 3′-phosphoadenosine 5′-phosphate (PAP). In the absence of SAL1 or in response to drought and high light, PAP accumulates and can move between the chloroplast and the nucleus, where it inhibits exoribonucleases, thereby inducing stress-responsive genes.
Abstract
Compartmentation of the eukaryotic cell requires a complex set of subcellular messages, including multiple retrograde signals from the chloroplast and mitochondria to the nucleus, to regulate gene expression. Here, we propose that one such signal is a phosphonucleotide (3′-phosphoadenosine 5′-phosphate [PAP]), which accumulates in Arabidopsis thaliana in response to drought and high light (HL) stress and that the enzyme SAL1 regulates its levels by dephosphorylating PAP to AMP. SAL1 accumulates in chloroplasts and mitochondria but not in the cytosol. sal1 mutants accumulate 20-fold more PAP without a marked change in inositol phosphate levels, demonstrating that PAP is a primary in vivo substrate. Significantly, transgenic targeting of SAL1 to either the nucleus or chloroplast of sal1 mutants lowers the total PAP levels and expression of the HL-inducible ASCORBATE PEROXIDASE2 gene. This indicates that PAP must be able to move between cellular compartments. The mode of action for PAP could be inhibition of 5′ to 3′ exoribonucleases (XRNs), as SAL1 and the nuclear XRNs modulate the expression of a similar subset of HL and drought-inducible genes, sal1 mutants accumulate XRN substrates, and PAP can inhibit yeast (Saccharomyces cerevisiae) XRNs. We propose a SAL1-PAP retrograde pathway that can alter nuclear gene expression during HL and drought stress.
INTRODUCTION
The evolution of the eukaryotic cell necessitated the development of signaling between compartments or organelles to coordinate cell differentiation, development, and acclimation to altered environmental stimuli. In plants, the transcriptional and developmental program of the chloroplast is tightly integrated with the nuclear program (Vranová et al., 2002; Nott et al., 2006; Pogson et al., 2008; Kleine et al., 2009; Pfannschmidt, 2010). This is required because chloroplast multiprotein complexes, such as ribosomes and the photosystems, are mosaics of subunits transcribed from both the plastid and nuclear genomes. Thus, coexpression from both genomes is essential to enable coordinated assembly and maintenance of photosynthesis. For example, if chloroplasts become damaged, they initiate retrograde signals that are sent to the nucleus to preclude unnecessary transcription of nuclear-encoded proteins that are targeted to the chloroplast (Bradbeer et al., 1979). A range of signals and pathways have been proposed and actively debated (Pogson et al., 2008; Kleine et al., 2009; Pfannschmidt, 2010). However, no chemical signal has been reported that moves directly from the chloroplast to the nucleus via the cytosol to regulate gene expression nor has a protein been reported that directly regulates the levels of such a compound.
There is evidence for multiple retrograde pathways; indeed, given the complexity and number of different metabolic reactions undertaken within the plastid, that is to be expected (Pogson et al., 2008; Pfannschmidt, 2010). Retrograde signals can be divided into two classes: those related to chloroplast and photosystem biogenesis (biogenic control) and those related to the operation of the chloroplast in response to changing environmental stimuli (operational control) (Pogson et al., 2008). Forward genetic screens have identified several protein components of biogenic control signaling pathways. Examples of biogenic control mutants include the snowy cotyledon (Albrecht et al., 2010) and genomes uncoupled (gun) mutants (Susek et al., 1993; Larkin et al., 2003; Strand et al., 2003; Koussevitzky et al., 2007; Ruckle et al., 2007). With respect to biogenic control, a tetrapyrrole was proposed to move from the chloroplast to cytosol where it was hypothesized it would interact with cytosolic targets such as HSP90 (Strand et al., 2003; Kindgren et al., 2011). However, this has been actively debated by other groups (Mochizuki et al., 2008; Moulin et al., 2008). Recently, another tetrapyrrole, heme, was proposed as a putative plastid biogenic signal in plants, but no changes in heme levels or evidence for heme movement from the chloroplast were reported nor were cytosolic/nuclear signaling partners described (Woodson et al., 2011). Although there is evidence that tetrapyrroles trigger retrograde signaling in plants, what the actual pathways are remains an open question (Pfannschmidt, 2010).
With respect to operational control or chloroplast-nuclear signaling in response to environmental stimuli, considerable detail is understood about the initiation of signaling cascades in the chloroplast and transcriptional changes in the nucleus, but the intervening steps are largely unknown. Environmental stresses that perturb photosynthesis, such as high light (HL) and drought, induce reactive oxygen species (ROS), changes in redox state of plastoquinone, and changes in abscisic acid (ABA) concentration that are implicated in the HL response pathways (Karpinski et al., 1999; Vranová et al., 2002; Nott et al., 2006; Rossel et al., 2006; Lee et al., 2007; Pogson et al., 2008; Van Breusegem et al., 2008; Foyer and Noctor, 2009; Galvez-Valdivieso et al., 2009; Kleine et al., 2009; Pfannschmidt et al., 2009; Wilson et al., 2009). In the nucleus, HL alters the expression of ~700 genes (Rossel et al., 2007), including APX2 (Karpinski et al., 1999; Rossel et al., 2006) and EARLY LIGHT INDUCIBLE PROTEIN2 (ELIP2) (Harari-Steinberg et al., 2001; Kimura et al., 2003). A number of transcription factors are induced by HL, including DROUGHT RESPONSE BINDING 2A (DREB2A) and ZAT10; the latter can regulate the expression of 18% of the HL transcriptome, including APX2 (Rossel et al., 2007). Other HL-inducible genes, such as ELIP2, are regulated by cryptochromes (Kleine et al., 2007).
To identify steps between initiation of the signal and perception in the nucleus, screens for altered gene expression during oxidative stress have identified a series of mutations, including executer1 and 2, regulator of APX2, and altered APX2 expression8 (alx8) (Ball et al., 2004; Wagner et al., 2004; Rossel et al., 2006; Wilson et al., 2009). Yet, the actual retrograde signals regulated by the multiple biogenic and operational pathways still remain unknown.
The alx8 mutant exhibits constitutive upregulation of 25% of the HL-regulated transcriptome, including ZAT10, DREB2A, ELIP2, and APX2, along with hyperexpression of these transcripts upon HL stress (Rossel et al., 2006; Wilson et al., 2009). Indeed, as 70% of HL-inducible genes are also upregulated by drought (Kimura et al., 2002), it was not surprising that the alx8 mutant is also drought tolerant, surviving water deprivation up to 50% longer than wild-type plants. These phenotypes are caused by a lesion in the SAL1/ALX8/FRY1 gene and implicate SAL1 as a component of HL and drought stress signaling networks (Wilson et al., 2009; Hirsch et al., 2011).
SAL1 is a phosphatase that hydrolyzes a phosphate group from both phosphonucleotides and inositol polyphosphates in vitro (Quintero et al., 1996; Xiong et al., 2001). Inositol 1,4,5-trisphosphate (IP3) is viewed as one of the most logical targets for SAL1 in vivo (Xiong et al., 2001; Zhang et al., 2011). However, other findings using mutants and transgenic plants suggest SAL1 may be degrading 3′-phosphoadenosine 5′-phosphosulfate (PAPS) (Rodríguez et al., 2010) or 3′-phosphoadenosine 5′-phosphate (PAP) (Gy et al., 2007; Kim and von Arnim, 2009). Moreover, the enzymatic activity of recombinant SAL1 is similar for both phosphoadenosines (Gil-Mascarell et al., 1999), but the phosphatase activity against IP3 is only 4% of that against PAP (Xiong et al., 2001). The in vivo substrate is not resolved, as a recent article proposed for IP3 (Zhang et al., 2011). SAL1 is involved in many cellular processes, and identification of its primary substrates is required to better understand the mode of action of this phosphatase.
PAP is produced from PAPS during sulphation reactions catalyzed by cytosolic sulfotransferases (Klein and Papenbrock, 2004). Although PAP was originally viewed as a byproduct with no physiological function in plants, it can inhibit the activity of the two yeast (Saccharomyces cerevisiae) 5′ to 3′ exoribonucleases (XRNs), thereby altering RNA catabolism (Dichtl et al., 1997). Treatment of yeast with lithium (Li+), a strong inhibitor of the yeast SAL1 homolog (Sc-SAL1), results in an increase in PAP (Murguía et al., 1996) sufficient to inhibit XRNs, resulting in the accumulation of transcripts targeted by Xrn1 in yeast (Dichtl et al., 1997; van Dijk et al., 2011). Moreover, a PAP concentration of 0.1 mM inhibits the in vitro activity of the two yeast XRNs by 40 to 65% (Dichtl et al., 1997). SAL1 has recently been linked to several developmental and morphological processes in plants (Wilson et al., 2009; Robles et al., 2010; Rodríguez et al., 2010; Zhang et al., 2011); interestingly, Arabidopsis thaliana xrn mutants have a similar leaf and root morphology to that of sal1 mutants (Gy et al., 2007; Hirsch et al., 2011).
Although SAL1 functions in stress signaling and other fundamental plant processes, the subcellular localization, the in vivo substrate, and the mode of action of SAL1 are either unknown or debated. For example, the SAL1 protein has been reported to be localized in the chloroplast (Rodríguez et al., 2010), cytosol (Zhang et al., 2011), and nucleus (Kim and von Arnim, 2009) by different techniques. Consequently, it is critical to resolve its cellular location, to identify the in vivo substrates, and to investigate how the accumulation of its substrates in the cell might function in cellular signaling. More significantly, there is no indication in the literature whether PAP could act as a retrograde signal linking organelle status with nuclear gene expression. Indeed, there is no report of PAP measurements in plants, which precluded the study of its role in planta.
In this study, we show that SAL1 accumulates in both the chloroplasts and the mitochondria and provide evidence that PAP levels are modulated by SAL1. We propose that PAP functions as a mobile signal that alters RNA metabolism by inhibiting XRNs to affect stress and developmental gene expression and that the chloroplastic SAL1 protein can prevent its action by degrading PAP in the chloroplastic compartment.
RESULTS
SAL1 Expression Correlates Spatially with Responses to HL Stress
We previously showed that the lack of the SAL1 protein in the alx8 mutant promotes constitutive APX2:LUCIFERASE (LUC) expression in the vascular tissue (Rossel et al., 2006; Wilson et al., 2009). Here, we investigated the expression pattern of SAL1 in mature leaf tissue by reporter gene analyses. Expression of green fluorescent protein (GFP) driven by the SAL1 promoter (pSAL1:SAL1:GFP) was stronger in the vascular tissue than in the mesophyll tissue of the leaf (Figure 1A). The vascular tissue is the primary site of production of H2O2, especially after HL stress (Fryer et al., 2003) (Figure 1B). By contrast, the HL induction of H2O2 in alx8 vascular tissue was much lower, and the total H2O2 foliar level of plants grown under normal conditions was half of that in the wild type (Figure 1C). Thus, SAL1 expression colocalizes with APX2:LUC activity, with the loss of SAL1 leading to increased APX2 expression (Rossel et al., 2006; Wilson et al., 2009) and reduced H2O2 levels in the vasculature. Furthermore, it demonstrates that the elevated expression of APX2 and other HL-regulated genes is not due to elevated levels of H2O2.
Figure 1.
SAL1 Colocalizes in the Vascular Tissue with H2O2.
(A) Stable expression of the SAL1-GFP fusion protein in Arabidopsis plants. Gene expression was driven by a genomic sequence containing the endogenous SAL1 promoter, which was fused to GFP (pSAL1:SAL1:GFP). Top panel: 8-d leaf showing a strong GFP signal in the vascular tissue and a more moderate one in the mesophyll tissues. The bottom left image is GFP channel, middle image is chlorophyll channel, and right image shows the overlay from a 2-week-old leaf.
(B) Visualization of H2O2 in Col-0 and alx8 leaves after 1 h HL treatment using DAB. H2O2 accumulation is visualized as a dark, brown precipitate.
(C) Quantification of leaf H2O2 in 6-week-old, soil-grown Col-0 and alx8. After extraction and incubation with the Amplex Red, the amount of H2O2 was quantified against a standard curve and normalized to the FW. The mean and sd are shown. Asterisk indicates significant difference relative to Col-0 (t test, P < 0.05, n = 5).
PAP Accumulates in sal1 Mutants and during Drought Stress
In vitro, SAL1 has a dual phosphatase activity against both polyphosphoinositols, such as IP3 (Xiong et al., 2001; Zhang et al., 2011), and 3′(2′),5′-biphosphate nucleotides, such as PAP or PAPS (Quintero et al., 1996). PAPS and IP3 have been reported to be in vivo substrates (Xiong et al., 2001; Rodríguez et al., 2010; Zhang et al., 2011); PAP has been suggested as a substrate based on work with transgenic plants (Kim and von Arnim, 2009; Chen and Xiong, 2010; Hirsch et al., 2011). We used two different approaches to investigate if the SAL1 phosphatase activity against PAP regulated the expression of APX2 and, thus, ROS levels. First, we tested the hypothesis that accumulation of PAP could upregulate the APX2 promoter and induce the alx8 phenotype by feeding PAP to Columbia-0 (Col-0) plants transformed with the reporter gene APX2:LUC. However, no significant change in APX2:LUC activity was observed when feeding increasing concentrations of PAP to 7-d-old plants via the roots under either low light (LL) or HL (see Supplemental Figure 1 online). Failed root substrate uptake or impaired vasculature transport, import into leaf cells, or perception are possible explanations for the negative results of the feeding experiment.
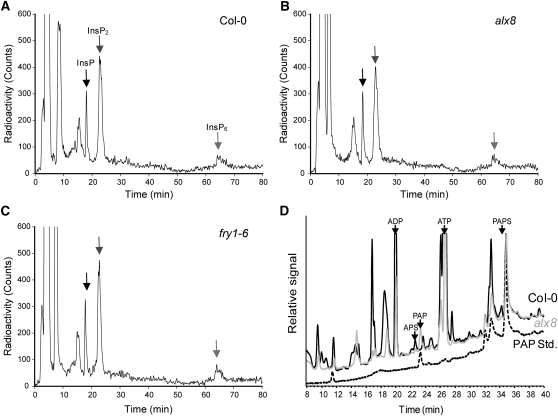
Next, we directly measured inositol phosphates (IPs) in the sal1 mutants alx8, fry1-6, and fry1-1. The alx8 mutant harbors a point mutation in SAL1 that renders the recombinant protein enzymatically inactive (Wilson et al., 2009), whereas the fry1-6 allele is a T-DNA insertion mutant (SALK_020882). Both mutants, in the Col-0 background, lacked detectable SAL1 protein and showed very similar rosette morphology (see Supplemental Figure 2 online). The third mutant, fry1-1, previously described in the C24 background, was reported to have increased IP3 (Xiong et al., 2001), and increased IP3 by 1.5- to 2.0-fold was recently reported in another sal1 mutant (Zhang et al., 2011), as measured by displacement bioassays. It was not revealed whether other IP compounds could be affected. To test this, we grew young seedlings in the presence of radiolabeled myo-[2-3H]inositol and showed that all the IP pools were similar in Col-0, alx8, fry1-6 (Figures 2A to 2C), and fry1-1 (see Supplemental Figure 3 online).
Figure 2.
PAP, and Not IPs, Accumulates in alx8.
(A) to (C) Analyses of phosphoinositols in different genotypes. Five to six 10-d-old Col-0 (A), alx8 (B), or fry1-6 (C) seedlings were labeled with myo-[2-3H]inositol for 72 h and IPs extracted in HCl prior to separation by HPLC. Chromatograms show representative profile of phosphoinositols of one of two independent experiments. The intensity and periodicity of the peaks corresponding to inositol mono-, bis-, and hexakisphosphate (arrows) are similar in all genotypes.
(D) Isolation and identification of phosphoadenosine nucleotides. Metabolites were extracted from leaves of 30-d-old plants and adenosines fluorescently labeled by derivatization. PAPS, APS, and PAP in wild-type Col-0 (black) and alx8 (gray) were identified by coelution with external PAP standard (dashed black line). A typical chromatogram is shown. Note that approximately fourfold less extract of alx8 than Col-0 was injected in this experiment. Quantification was undertaken using a standard curve (see Supplemental Figure 4 online).
Finally, we investigated whether PAP, or PAPS, is the in vivo substrate of SAL1 and developed a highly sensitive and specific fluorescence labeling–based HPLC method for quantification of these nucleotides (Figure 2D; see Supplemental Figure 4 online). Using this technique, we could clearly quantify PAP and PAPS based on the chromatograms of plant extracts with adenosines derivatized to enable their detection by fluorescence spectroscopy. We found that PAP accumulated 20-fold in three sal1 mutants, alx8 (Table 1), fry1-6, and fou8 (see Supplemental Figure 5 online), compared with wild-type plants. There was also a significant but minor increase in PAPS in alx8 and no significant changes in adenosine 5′-phosphosulfate (APS), the sole precursor of PAPS, and an intermediate in the synthesis of Cys. Nor was there any change in GSH, the storage form of Cys. The specific and substantial increase of PAP in sal1 mutants provides the first direct evidence that SAL1 has in vivo nucleotidase activity preferentially against PAP.
Table 1.
Quantification of Nucleotide Phosphates and Glutathione-Related Metabolites
| PAP Metabolism |
Glutathione Metabolism |
|||||
| Germplasm | APS | PAPS | PAP | Cys | γ-EC | GSH |
| Col-0 | 4.2 ± 1.4 | 1.1 ± 0.1 | 0.6 ± 0.2 | 4.8 ± 1.0 | 2.7 ± 0.6 | 176 ± 48 |
| alx8 | 3.8 ± 0.2 | 1.8 ± 0.1 | 11.8 ± 1.4 | 8.1 ± 3.4 | 4.5 ± 1.6 | 197 ± 62 |
| ns | P < 10−4 | P < 10−4 | P < 0.05 | P < 0.05 | ns | |
Metabolite concentrations were determined by HPLC for 30-d-old plants. γ-EC, l-γ-glutamylcysteine. Values are the concentration in pmol/mg FW ± sd (n = 4). Individual P numbers compared to Col-0 after t test analyses assuming two-tailed and two-sample unequal variance are indicated. ns, not significant.
The correlation between higher PAP levels in alx8 and drought tolerance led us to investigate a role for this metabolite during abiotic stress responses. Thus, we analyzed PAP levels in response to drought and HL in wild-type plants. PAP levels increased 30-fold in leaves of drought-stressed wild-type plants, coincident with a substantial decrease in plant relative water content (RWC) after 7 to 11 d of drought (Figure 3). Analysis of variance (ANOVA) two-factor analyses indicated strong interaction between day and genotype for RWC and PAP, being significantly higher for alx8 relative to Col-0 (P < 0.001). This increase did not occur in the early phase of drought and was observed only when there was a decline in RWC. A similar trend was observed for alx8, but it was delayed, again with PAP only rising as RWC declined. Similarly, exposure of Col-0 plants to HL for just 1 h resulted in significantly (P < 0.005) higher PAP levels than in plants kept at LL (0.9 ± 0.2 versus 0.6 ± 0.2 pmol of PAP/mg fresh weight [FW], respectively), although this increase was much smaller than that observed during drought. Taken together, these results revealed that the level of the sulfur-related metabolite PAP was elevated in mutants lacking SAL1 and increased in response to at least two abiotic stresses.
Figure 3.
PAP Accumulates during Drought in Arabidopsis.
(A) Correlation between RWC of plants and PAP concentration on a dry weight (DW) basis ± sd (n > 8) during drought. The day in drought is indicated in italics. Measurements performed as in Figure 2 and Supplemental Figure 4 online. Data were fitted to exponential curves, and results are shown in the table (R2, correlation coefficient). ANOVA two-factor analyses indicated a highly significant difference for day × genotype for RWC and PAP (P < 0.005).
(B) Images of representative plants harvested for the PAP measurements performed in (A).
SAL1 Localizes to Both Chloroplast and Mitochondria
Contrasting results have been obtained regarding the cellular localization of SAL1 (Kim and von Arnim, 2009; Rodríguez et al., 2010; Zhang et al., 2011). To resolve this debate, we used three different methods to investigate SAL1 location. First, a full-length SAL1 fused at the C terminus to GFP accumulated in both chloroplasts and mitochondria of transiently transformed Arabidopsis cells (Figure 4A). The chloroplasts and mitochondria were visualized by red fluorescent protein (RFP) fused to either the small subunit (SSU) of ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) transit peptide or to the targeting domain of the ALTERNATIVE OXIDASE1 (AOX1), respectively. Second, to verify that this expression pattern was not an artifact due to the use of transitory expression systems, we generated stable transgenic lines. SAL1:GFP fusion protein driven by the native promoter showed SAL1 in both organelles in mesophyll protoplasts isolated from stably transformed pSAL1:SAL1:GFP plants (Figure 4B). The chloroplasts were visualized by chlorophyll fluorescence and the mitochondria by MitoRed. Effectively all compartmentalized GFP could be attributed to either mitochondria or chloroplasts, not nuclei. Third, we developed a new chloroplast and cytosolic fractionation method (see Supplemental Figure 6 online and Methods) that allowed us to detect SAL1 unequivocally as an ~38-kD band in purified chloroplast and mitochondria fractions of Col-0 leaves by immunological methods (Figure 4C). The molecular mass of this band matches that of the recombinant protein lacking the deduced 54–amino acid chloroplast transit peptide (Wilson et al., 2009). The relative purity of the fractions was demonstrated by probing with antibodies against chloroplastic Lhcb, mitochondrial TOM40, and cytosolic UGPase. UGPase was enriched in the cytosolic fraction, and although there was a band in the chloroplast fraction, it is of lower molecular mass than UGPase, and as it is the same size as Rubisco, it is likely that this is a cross-reaction to Rubisco. Significantly, neither Lhcb, TOM40, nor the unprocessed or mature forms of SAL1 were detected in the cytosolic fraction.
Figure 4.
SAL1 Accumulates in the Chloroplasts and Mitochondria of Arabidopsis.
(A) Transient expression of 35S:SAL1:GFP in Arabidopsis cells. The full-length cDNA encoding SAL1 was fused in frame with GFP and cotransformed into Arabidopsis cells with either mitochondrial-targeted RFP (AOX-RFP) or plastid-targeted RFP (SSU-RFP).
(B) Stable expression of the SAL1:SAL1:GFP fusion in Arabidopsis leaf mesophyll protoplasts. The endogenous SAL1 promoter was fused to the SAL1 genomic sequence (pSAL1:SAL1:GFP) and transformed into Col-0 plants. The same construct transformed into sal1 mutants complemented all analyzed sal1 phenotypes (Hirsch et al., 2011). The SAL1:GFP pattern was identical for several independent transgenic lines in the Col-0 background. White arrows indicate mitochondria. Bars = 12 μm.
(C) Chloroplastic and cytosolic fractions were isolated from Col-0 protoplasts, while mitochondria were purified from seedlings using free-flow electrophoresis. Five micrograms of total protein was loaded per sample and subjected to immunoblots with polyclonal antibodies against SAL1 (Wilson et al., 2009) and other cellular markers.
(D) Detection of PAP in isolated Col-0 chloroplasts. Col-0 chloroplasts isolated using a Percoll gradient and assayed for PAP. The PAP peak for both the chloroplast extract and the PAP standard is indicated with an arrow.
We attempted to assay PAP from the cytosolic fractions used for the protein purification, but we observed that PAP is labile in tissue extracts, and as a consequence, it was not detected after the lengthy purification procedure. However, analyses of phosphonucleotides from Col-0 chloroplasts prepared by a different, more rapid protocol identified a peak that matched that of the PAP standard, suggesting that PAP is present in the chloroplasts of Arabidopsis (Figure 4D). All this evidence supports the conclusion that SAL1 accumulates in mitochondria and chloroplasts and that PAP can be detected in chloroplasts.
Complementation of sal1 Mutants Demonstrates That PAP Regulates Nuclear Gene Expression
Having demonstrated that SAL1 is required for the catabolism of PAP, we sought to investigate whether PAP could act as a mobile signal within the cell, capable of entering the nucleus and regulating gene expression. Due to the aforementioned technical difficulties of measuring adenosines in different cellular compartments, we used a genetic approach to test the hypothesis of PAP movement by the analysis of plant lines with SAL1 targeted to different subcellular compartments kindly provided by Kim and von Arnim (2009) and Rodríguez et al. (2010). In these earlier studies, the authors reported on partial complementation of morphological phenotypes, but neither PAP, nuclear gene expression, retrograde signaling, nor stress tolerance was measured in these studies.
First, does the chloroplast-localized SAL1 regulate PAP concentration? Given the unexpected dual targeting of SAL1 to the chloroplast and mitochondria, it was necessary to determine if chloroplast-localized SAL1 can modulate PAP levels. Targeting the yeast SAL1 (Sc-SAL1) to the chloroplast using the transit peptide of the Rubisco SSU resulted in a significant (P < 0.01) lowering of PAP levels and APX2 mRNA accumulation, demonstrating that chloroplast localization of SAL1 functions in regulating PAP content and APX2 mRNA levels (Figures 5A and 5B).
Figure 5.
PAP Levels Can Be Lowered by Nuclear or Chloroplastic Targeting of SAL1.
(A) Schematic representations of the SAL1 gene, mutant alleles, and constructs used to transform the corresponding genotypes. White boxes represent the mature protein, while the gray ones represent the transit peptides. The reported cellular location is indicated (C, chloroplast; M, mitochondria, N, nucleus; −, no SAL1 protein). Bar graphs represent the average PAP concentrations from leaf tissue in pmol/mg FW ± sd (n = 5). Asterisk indicates significant difference relative to Col-0, and “°” indicates significant difference to alx8. ns, not significant (t test; P < 0.01).
(B) Correlation between PAP levels and APX2 expression. The PAP concentration of 26-d-old plants was plotted versus the logarithm of the fold change of APX2 mRNA relative to Col-0. Four biological replicates were run for Col-0, alx8, and fou8 and six for fou8+SSU:ScSAL1. (A) and (B) show results from two different experiments. For PAP, asterisk indicates significant difference relative to Col-0, and “°” indicates significant difference to alx8 or fou8. ns, not significant (t test; P < 0.01). For APX2 mRNA, “#” indicates significant difference relative to Col-0, and “^” indicates significant difference to alx8 or fou8. ns, not significant (t test; P < 0.01).
(C) Schematic representation of the effect of SAL1 cellular localization on PAP levels and APX2 expression summarizing the results presented in Table 2 and (A) and (B). Shaded compartments indicate the location of SAL1 protein in each of the germplasms.
Second, can PAP move between cellular compartments? Targeting of SAL1 to the nucleus resulted in complete complementation of PAP levels (Figure 5A), APX2 mRNA abundance in LL and drought-stressed leaves, ELIP2 mRNA abundance in LL- and HL-treated leaves, and the viability of plants in response to terminal drought (Table 2). The combined results of the targeting experiments indicate that the degree of complementation of PAP levels is somewhat proportional to the degree of complementation of APX2 expression (Table 2, Figure 5). More importantly, they show that PAP can be catabolized by either nuclear or chloroplastic targeting of SAL1, demonstrating that PAP can move between subcellular compartments.
Table 2.
Survival of and Expression Levels of Stress-Inducible Genes in Col-0, alx8 Mutants, and fry1-6 Complemented with SAL1 Directed to the Cytosolic-Nuclear Compartment
| Genotype | Col-0 | fry1-6 | fry1-6 + trSAL1 | alx8 |
| SAL1 proteina | Endogenous | − | Truncated | − |
| SAL1 location | C, M | − | N | − |
| Survival (days)b | ||||
| Experiment 1 | 7.7 ± 0.4 (7) | 11.0 ± 0.4 (7) | 8.1 ± 0.2 (14) | 11.1 ± 0.3 (7) |
| Experiment 2 | 13.4 ± 0.2 (9) | 17.0 ± 0.3 (6) | 11.8 ± 0.2 (9) | 15.2 ± 0.2 (8) |
| APX2 mRNAc | ||||
| Control | 1 ± 0 | 661.7 ± 133.2 | 0.5 ± 0 | 313 ± 15.2 |
| Drought | 5.9 ± 1.3 | 501 ± 28.6 | 6.5 ± 1.9 | 274 ± 33.1 |
| ELIP2 mRNA | ||||
| LL | 1 ± 0.2 | 87 ± 63 | 1.5 ± 0.6 | 109 ± 19 |
| HL | 347 ± 93 | 3490 ± 993 | 711 ± 140 | 4191 ± 1336 |
The SAL1 protein lacking the chloroplastic transit peptide (trSAL1) accumulates in the nucleus (N) (Kim and von Arnim, 2009). C, chloroplast; M, mitochondria; −, absent.
Survival time, or the number of days plants remain viable, during drought was calculated as described by Woo et al. (2008) from measurements of the maximum efficiency of photosystem II (FvFm) using chlorophyll fluorescence. Values represent the average of days ± se; numbers of replicates are indicated in parentheses. Two independent experiments were conducted at different times.
The expression levels (fold change) of APX2 in plants grown in control conditions or after 9 d of drought were measured by qRT-PCR. ELIP2 message was quantified in the same manner but for plants after 1 h of LL or HL (1500 μmole m−2 s−1). The values represent the relative mRNA levels compared to the Col-0 control ± se (n = 3).
SAL1 and Nuclear XRNs Coregulate a Large Subset of Genes
Given that PAP levels are elevated in sal1 mutants, and PAP is known to inhibit the activity of the yeast XRNs (Dichtl et al., 1997; van Dijk et al., 2011), we hypothesized that PAP could regulate the expression of stress-responsive genes via attenuation of XRN-mediated RNA catabolism. Although the Arabidopsis XRNs are less well characterized than their S. cerevisiae counterparts, they play key roles in multiple RNA processing pathways and as posttranscriptional gene silencing suppressors (Kastenmayer and Green, 2000; Souret et al., 2004; Gy et al., 2007; Zakrzewska-Placzek et al., 2010). The XRNs belong to a small gene family in Arabidopsis with three members. XRN2 and XRN3 are homologs of Xrn2p/Rat1p and are nuclear localized. By contrast, the cytosolic XRN4 is a functional homolog of S. cerevisiae Xrn1p (Kastenmayer and Green, 2000). Identified substrates of XRN2 and XRN3 are excised hairpin loops that form part of precursor MIRNA transcripts, which also accumulate in fry1-6 (Gy et al., 2007). On the other hand, XRN4 is involved in mRNA decay by degrading the resulting 3′ cleavage products of microRNA (miRNA) targets (Souret et al., 2004).
Analysis of the alx8 transcriptome revealed that transcripts for 19 genes that are confirmed targets of miRNAs were increased by at least threefold (see Supplemental Table 1 online). As probes for Affymetrix arrays are biased toward the 3′ region of genes, we tested whether this reflected increased abundance of full-length mRNA or accumulation of cleaved transcripts due to inhibition of XRN4 by increased PAP levels. We undertook quantitative real-time PCR (qRT-PCR) of four miRNA targets using primers specific for 3′ sequences or spanning the miRNA cleavage site (see Supplemental Figure 7 online). The uncleaved transcripts did not increase; rather, the 3′ cleavage products for ATHB15, PHB, MYB33, and REV accumulated to higher levels in both sal1 mutants compared with the wild type, with MYB33 showing the highest increase (sixfold). The accumulation of 3′ cleavage products in alx8 that should otherwise be degraded by XRNs is consistent with XRNs being inhibited in SAL1 mutants.
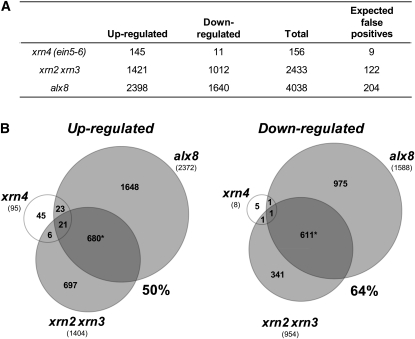
To determine if XRNs and SAL1-PAP regulate a common set of genes, we undertook gene expression profiling in the PAP-accumulating alx8 mutant, xrn2 xrn3 double mutant, and xrn4 (ein5-6, ethylene-insensitive5; Gregory et al., 2008). We used Affymetrix GeneChip Arabidopsis ATH1 genome arrays to analyze global changes in transcript abundance between Col-0 and the mutant genotypes. Whole rosettes of seedlings at the 10 true leaf stage of development were used for analysis of each genotype. We found that for alx8 and xrn2 xrn3, there were 4038 and 2433 transcripts, respectively, that showed a significant change in transcript abundance (>1.5-fold, false discovery rate [FDR] corrected at P < 0.05) relative to Col-0 (Figure 6A; see Supplemental Data Set 1online). By contrast, only 156 transcripts were significantly altered in xrn4 compared with Col-0. This low level of transcriptome change in xrn4 is consistent with previous transcriptome profiling data of xrn4 (Souret et al., 2004; German et al., 2008; Gregory et al., 2008).
Figure 6.
SAL1 and Nuclear XRNs Regulate a Large Subset of Genes.
(A) Summary table of transcriptome changes in xrn4, xrn2 xrn3, and alx8 mutants. Total number of genes whose transcripts were significantly different in abundance by >1.5-fold in each mutant compared with Col-0 after FDR correction at PPDE (>P) > 0.95 (95% confidence interval). Expected false positives at this FDR cutoff level are also indicated.
(B) Venn diagrams showing the overlap of changes in gene expression relative to Col-0 (>1.5-fold) between alx8, xrn2 xrn3, and xrn4. Numbers in the Venn diagrams indicate transcripts that are significantly (PPDE [>P] > 0.95) up- or downregulated in the mutant genotype compared with Col-0. The percentage of genes in xrn2 xrn3 that are regulated in the same manner as in alx8 is given. The number of genes that significantly change excluding antagonistic changes is given in parentheses (see Supplemental Figure 8 online). Asterisk indicates significantly (P < 0.02) more transcripts overlapping than expected by chance according to a χ2 test.
More importantly, there was a large and significant overlap between the alx8 and xrn2 xrn3 transcript profiles (Figure 6B). Of the 1404 genes upregulated in xrn2 xrn3 (relative to Col-0), 50% (680 transcripts) were also upregulated in alx8, which is a significantly greater overlap than would be expected by random chance (P < 0.05), while only 14 transcripts showed an antagonistic response (i.e., were downregulated in alx8), which is statistically fewer than would be expected by chance (P < 0.05; see Supplemental Figure 8 online). Similarly, in the downregulated transcript set, there was a significant overlap of 64% (611 of 954 transcripts) between the xrn2 xrn3 transcriptome and that of alx8, again with lower levels of antagonistic change than would be expected by random chance (24 transcripts; P < 0.05; see Supplemental Figure 8 online). Additionally, coexpression of four highly upregulated transcripts in the alx8 microarray (Wilson et al., 2009) in alx8 and xrn2 xrn3 was confirmed by qRT-PCR (see Supplemental Table 2 online). Furthermore, the fold change of all genes upregulated by more than fivefold was also comparable for xrn2 xrn3 and alx8 (Table 3). Transcripts encoding transferases, transporters, hormone-related transcription factors, and starch synthase were coexpressed in both alx8 and xrn2 xrn3 to the same extent (Table 3).
Table 3.
A Subset of Coregulated Genes in xrn2 xrn3 and alx8 That Are Fivefold Up- or Downregulated Compared to Col-0
|
xrn2 xrn3 versus Col |
alx8 versus Col |
||||
| AGIa | Gene Description | F.C.b | P value | F.C. | P value |
| AT3G61630 | CYTOKININ RESPONSE FACTOR6 | 109.3 | 1.48E-05 | 47.5 | 6.95E-07 |
| AT2G04050 | MATE efflux family protein | 84.4 | 4.83E-07 | 19.5 | 6.95E-07 |
| AT1G05680 | UDP-glucuronosyl family protein | 44.5 | 4.17E-08 | 7.4 | 6.95E-07 |
| AT2G41730 | Unknown protein | 22.5 | 6.41E-10 | 10.4 | 6.95E-07 |
| AT1G61800 | Glc-6-phosphate transporter | 19.4 | 4.92E-10 | 12.1 | 6.95E-07 |
| AT2G04040 | ATDTX1 transporter | 18.1 | 3.07E-04 | 4.5 | 6.95E-07 |
| AT2G21640 | Unknown protein | 16.7 | 1.13E-06 | 6.4 | 6.95E-07 |
| AT3G53980 | Lipid transfer protein family protein | 16.0 | 4.58E-06 | 15.8 | 6.95E-07 |
| AT5G08030 | Glycerophosphoryl diester phosphodiesterase | 15.0 | 3.00E-07 | 14.5 | 6.95E-07 |
| AT2G40230 | Transferase family protein | 11.4 | 1.09E-09 | 11.8 | 6.95E-07 |
| AT1G75580 | Auxin-responsive protein, putative | 9.2 | 6.14E-08 | 12.3 | 6.95E-07 |
| AT5G25350 | EIN3 BINDING F BOX PROTEIN2 | 7.5 | 2.42E-08 | 6.9 | 6.95E-07 |
| AT5G27660 | Ser-type peptidase/trypsin | 6.9 | 1.67E-05 | 51.9 | 6.95E-07 |
| AT1G56150 | Auxin-responsive family protein | 5.7 | 5.21E-09 | 6.5 | 6.95E-07 |
| AT1G32900 | Starch synthase, putative | 5.1 | 9.05E-07 | 5.1 | 6.95E-07 |
| AT2G22810 | 1-Aminocyclopropane-1-carboxylate synthase | −5.1 | 8.47E-05 | −5.8 | 6.95E-07 |
| AT4G25490 | CBF1 transcription factor | −5.3 | 5.71E-06 | −15.3 | 6.95E-07 |
| AT5G26200 | Mitochondrial substrate carrier family protein | −5.4 | 1.89E-07 | −18.5 | 6.95E-07 |
| AT5G18060 | Auxin-responsive protein, putative | −5.5 | 5.30E-07 | −8.9 | 6.95E-07 |
| AT5G05250 | Similar to unknown protein (TAIR: AT3G56360.1) | −6.2 | 1.87E-03 | −15.5 | 6.95E-07 |
| AT2G17880 | DNAJ heat shock protein, putative | −6.5 | 3.31E-04 | −15.4 | 6.95E-07 |
| AT2G37950 | Zinc finger (C3HC4-type RING finger) family | −8.6 | 2.96E-05 | −17.9 | 6.95E-07 |
| AT2G26710 | BAS1 (PHYB SUPPRESSOR1) | −9.0 | 3.17E-07 | −10.8 | 6.95E-07 |
| AT2G44130 | Kelch repeat–containing F-box family protein | −9.1 | 2.92E-07 | −9.9 | 6.95E-07 |
| AT5G54610 | ANKYRIN; protein binding | −9.6 | 9.07E-05 | −7.6 | 6.95E-07 |
| AT1G78450 | SOUL heme binding family protein | −14.9 | 3.98E-09 | −12.5 | 6.95E-07 |
AGI, Arabidopsis Genome Initiative.
F.C., fold change compared to Col-0.
See Supplemental Data Set 1 online for the complete gene list.
Finally, comparison of all coexpressed genes up- or downregulated by more than threefold against a series of microarray experiments (Hruz et al., 2008) revealed a high degree of coexpression under HL, ABA, drought stress, and a combined moderate HL and mild drought on a plant with a defective mitochondrial stress-inducible protein, AOX1A (Giraud et al., 2008) (Figure 7). This same set of genes was not differentially expressed in LL or under different light quality and wavelengths, nor was the set similarly coexpressed upon treatment with the plastid translational inhibitor, lincomycin, that suppresses GUN-regulated genes. Both H2O2, known to induce some HL-responsive genes, and the mitochondrial respiratory complex I inhibitor, rotenone (Clifton et al., 2005; Garmier et al., 2008), resulted in increased expression of some of the upregulated genes, but the converse was observed for the downregulated genes. This suggests the coexpressed set of SAL1- and XRN2 XRN3–regulated genes respond to specific organelle signals, such as HL, but not translational inhibitors, such as lincomycin.
Figure 7.
Heat Map of Genes Coregulated in both alx8 and xrn2 xrn3 Mutants.
Heat map of genes upregulated (A) and downregulated (B) threefold or more in both genotypes compared with their regulation in response to abiotic stress and chemical treatments (Hruz et al., 2008).
Nuclear XRNs Regulate the Induction of HL and Drought Stress Genes
To investigate further the potential role of PAP in stress signal transduction pathways, we focused on the expression of the model chloroplast stress-responsive genes ELIP2 and APX2 in the xrn mutants. Our analysis of xrn4/ein5-6 transcriptome data (see Supplemental Data Set 1 online; Souret et al., 2004; German et al., 2008; Gregory et al., 2008) revealed that neither ELIP2 nor APX2 is upregulated in the xrn4 mutant. Thus, we focused on XRN2 and XRN3.
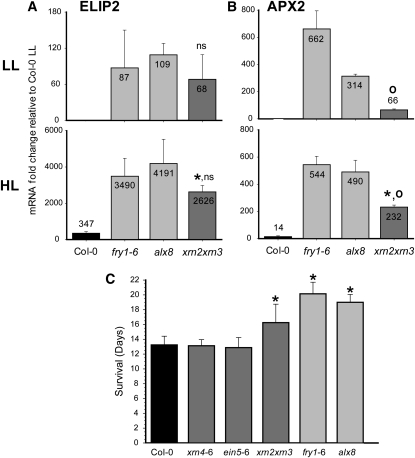
We aimed to mimic PAP inhibition of the nuclear XRNs by comparing the induction of stress-responsive genes between xrn2 xrn3 and the sal1 mutants under both LL and HL conditions. Indeed, mRNA levels of ELIP2 were remarkably similar in alx8, fry1-6, and xrn2 xrn3 mutants for nonstressed leaves, with APX2 expression being induced in all three mutants but less so in xrn2 xrn3 (Figure 8). Overall, all the mutants exhibited around 100-fold higher levels of ELIP2 and 60- to 600-fold increased APX2 compared with the wild type. Under HL stress, ELIP2 was hyperinduced to the same extent (by ~3000-fold) in alx8, fry1-6, and xrn2 xrn3, and again this was not substantially different between the three mutants, yet it was 10-fold higher than the induction in Col-0 (Figure 8A). The results were comparable for APX2, with expression changes in xrn2 xrn3 being of a similar order of magnitude to the alx8 and fry1-6 mutants, although attenuated (Figure 8B). This is consistent with the observations that APX2 is regulated by multiple signaling pathways in response to HL stress, including H2O2, ABA, glutathione metabolism, and plastoquinone redox state (Karpinski et al., 1997; Fryer et al., 2003; Ball et al., 2004; Rossel et al., 2006; Pogson et al., 2008; Galvez-Valdivieso et al., 2009) as well as PAP (Figure 5).
Figure 8.
Light-Induced Gene Regulation and Drought Tolerance Is Similar in sal1 and xrn2 xrn3 Double Mutants.
(A) Expression levels of ELIP2 after LL and HL.
(B) Expression levels of APX2 after LL and HL.
For both (A) and (B), the transcript levels were quantified by real-time PCR for both alx8 and xrn2 xrn3 mutants plants grown under standard growth conditions and after 1 h of HL stress (~1500 μmol m−2 s−1). The bars represent the average of the fold change compared with that of the wild type ± sd (n = 3). For xrn2 xrn3, “°” and ns indicate significant or no significant difference, respectively, relative to sal1 mutants (t test; P < 0.05). For HL, asterisk indicates significant difference relative to Col-0 HL (t test; P < 0.05).
(C) Survival time of plants during drought calculated as described by Woo et al. (2008). Periodic measurements of the maximum efficiency of photosystem II (Fv/Fm) using chlorophyll fluorescence were recorded during drought and used to calculate plant survival. Bar graphs represent the average survival time as measured in days ± sd (n > 7). Asterisk indicates significant difference relative to Col-0 (t test; P < 0.001).
The results presented above clearly indicate that nuclear XRNs are negative regulators of stress-inducible genes. Significantly, a triple mutant in both cytosolic and nuclear XRNs was better able to survive drought when compared with the wild type in a soil-based experiment (Hirsch et al., 2011), indicating that XRNs are regulators of the drought response. To elucidate whether cytosolic or nuclear XRNs mediate the drought response, we investigated the degree of drought tolerance of nuclear xrn2 xrn3 and sal1 mutants, and we also included a second xrn4 single mutant, ein5-6 (Olmedo et al., 2006). Both sal1 mutants survived the drought almost 50% longer compared with the wild type as previously shown (Wilson et al., 2009) (Figure 8C). Interestingly, plants with impaired nuclear XRN activity (i.e., the xrn2 xrn3 mutant), but not those in which the cytosolic homolog was mutated (i.e., xrn4), survived longer than the wild type but not as long as the sal1 mutants.
Taken together, these results suggest that PAP can stimulate gene expression by repressing the activity of the nuclear XRNs. That is, XRN2 and XRN3 negatively regulate the expression of the stress-responsive genes APX2 and ELIP2 and the drought response, most likely via the SAL1-PAP signaling pathway.
DISCUSSION
PAP Is a Primary in Vivo Substrate of SAL1 and Accumulates in Response to HL and Drought Stress
Chloroplasts and mitochondria can be viewed as environmental sensors mediating cellular responses to external stimuli that result in short- and long-term acclimation responses, ranging from induction of stress-responsive genes to changes in leaf thickness and petiole length. Likewise, SAL1 has been linked to stress responses (Rossel et al., 2006; Wilson et al., 2009) and many developmental processes (Kim and von Arnim, 2009; Robles et al., 2010; Wilson et al., 2009; Rodríguez et al., 2010; Hirsch et al., 2011; Zhang et al., 2011), demonstrating that the enzyme influences multiple biological processes. This requires either multiple enzymatic activities or that the substrate of SAL1 can initiate multiple responses. Thus, understanding the enzymatic activity of SAL1 in vivo will shed light on many different fields of plant biology.
We addressed this issue by in vivo measurements of proposed substrates. The results presented here clearly support the view that PAP is a major in vivo substrate of the phosphatase SAL1 and not PAPS or IPs, as previously proposed (Xiong et al., 2001; Rodríguez et al., 2010; Zhang et al., 2011). First, a distinctive peak corresponding to PAP was detected using HPLC coupled to fluorescence detection (Figures 2D; see Supplemental Figure 4 online). This method is more sensitive and specific than the absorbance detection used by Rodríguez et al. (2010), which could explain the lack of specific signal for PAP in that work, especially as PAP is very labile. Second, we were able to show that PAP increased by ~20-fold, whereas PAPS increased by just 1.6-fold (Table 1) and IPs either did not change (Figure 2) or increased by 1.5-fold to twofold (Zhang et al., 2011), which may be direct catalysis or an indirect effect, as many metabolites change in sal1 mutants (Wilson et al., 2009). In this study, we measured sulfur metabolites (Cys, γ-EC, and GSH), adenosines (APS, PAPS, PAPS, SAM, Ade, AMP, ADP, and ATP), and inositols, none of which changed significantly or to the same degree as PAP. Thus, we consider that an indirect change in glutathione metabolism is not a consequence of the SAL1 mutation; thus, the increase in APX2 mRNA in alx8 is not a consequence of glutathione metabolism–mediated signaling as proposed for rax1 (Ball et al., 2004). Furthermore, in a prior study, we measured the alx8 metabolome using gas chromatography–mass spectrometry and of the metabolites that changed, such as carbohydrates and polyamines, and none could be readily viewed as likely substrates of a nucleotide phosphatase (Wilson et al., 2009). Also, the expression of the PAP-specific phosphatase AHL enzyme that lacks IP3 activity complements sal1 mutants (Kim and von Arnim, 2009; Hirsch et al., 2011). This is also consistent with IP3 not being important for drought tolerance in Arabidopsis (Perera et al., 2008). There is also a reported in vitro preference of the recombinant SAL1 enzyme for PAP (Gil-Mascarell et al., 1999; Xiong et al., 2001). Additionally, the alx8 point mutation results in a recombinant SAL1 protein that cannot dephosphorylate PAP (Wilson et al., 2009). Thus, while we cannot preclude another as yet unknown enzymatic activity for SAL1, we conclude PAP is a primary substrate of SAL1 in vivo.
If the basis of the altered response to HL and drought in alx8 is mediated by increased PAP, then it might be expected that this metabolite should increase during stress in wild-type plants. Indeed, PAP increased 30-fold after 7 d of drought (Figure 3) and increased significantly within 1 h of HL stress. Interestingly, there was a tight correlation between leaf water status and PAP levels in both Col-0 and alx8 plants.
SAL1 Localizes to Both Chloroplasts and Mitochondria
Critical to understanding the function of PAP is determining the subcellular localization of the enzyme that regulates its levels, SAL1. Conflicting reports have suggested that SAL1 fusions are targeted to nuclei (Kim and von Arnim, 2009), cytosol in the roots (Zhang et al., 2011), or chloroplasts of onion epidermal peels (Rodríguez et al., 2010). The reported nuclear localization of SAL1 (Kim and von Arnim, 2009) likely reflects the authors’ use of a truncated SAL1 gene that lacked the transit peptide. To define the subcellular location of a protein, it is necessary to use multiple techniques, including in vivo analyses in the species of interest (Millar et al., 2009). Both stable and transient transformation lead to accumulation of SAL1:GFP in the chloroplast and mitochondria (Figures 4A and 4B), and the SAL1 protein was unequivocally detected in the purified chloroplastic and mitochondrial fractions of Col-0 leaves (Figure 4C). Significantly, no SAL1 protein was detected in the cytosolic fraction, and effectively all SAL1:GFP fluorescence could be attributed to either chloroplasts or mitochondria, not nuclei (Figure 4).
The finding of SAL1 in the chloroplast is consistent with the chloroplastic localization of isoenzymes for the synthesis of APS and PAPS (Mugford et al., 2009) and the detection of SAL1 by chloroplast proteomic analysis (Peltier et al., 2006; Olinares et al., 2010). Thus, using three different approaches, we demonstrated that SAL1 is a dual-localized protein found in chloroplasts and mitochondria, not the cytosol or nucleus. In addition, SAL1 inactivation results in a 20-fold increase in PAP levels.
The detection of PAP in isolated chloroplasts demonstrates it can accumulate in this organelle. Interestingly, it is believed that PAPS is largely synthesized in the plastid but that its conversion to PAP occurs in the cytosol. PAP could move back into the chloroplast via an unknown PAP/PAPS antiporter. This same proposed, but yet to be identified, transporter would allow its exit from the organelle back to the cytosol (Klein and Papenbrock, 2004; Mugford et al., 2009). An alternative to the PAP/PAPS antiporter is that movement is promoted by chloroplast damage during extreme stress. However, even photobleached cells contain viable and intact chloroplasts, as do sal1 mutants (Wilson et al., 2009). Rather than membrane damage per se enabling movement, it is more likely that stress can modulate the transport of proteins or signaling molecules, such as PAP. Regardless of the mechanism of transport, the lowering of PAP to near-wild-type levels by targeting Sc-SAL1 to the chloroplast demonstrated that PAP can move from the site of synthesis in the cytosol to the chloroplast.
The observation that SAL1 was also found in mitochondria was unexpected and raises the question as to its role and enzymatic activity in that organelle. Whereas it is beyond the scope of this study, it is worth noting that the partial complementation observed in sal1 mutants by Sc-SAL1 targeted to the chloroplast could reflect a reduced expression or activity of the yeast enzyme in transgenic Arabidopsis or a mitochondrial role for SAL1. Additionally, knockouts of a nuclear gene, AOX1A, encoding a mitochondrial protein used to study mitochondrial retrograde signaling (Giraud et al., 2008) resulted in similar coexpression of genes in response to a moderate drought and light as those coexpressed in alx8 and xrn2 xrn3, and a possible, but weaker, correlation was observed for the mitochondrial electron transporter chain inhibitor, rotenone (Figure 7).
Evidence for a SAL-PAP Retrograde Pathway
Two studies have reported on the complementation of the morphological phenotypes of sal1 mutants by targeting SAL1 to the nucleus (Kim and von Arnim, 2009) and Sc-SAL1 to the chloroplast (Rodríguez et al., 2010). However, the significance of these findings with respect to PAP acting as a retrograde signal was not considered by the authors; rather, they concluded that their constructs demonstrated the location of SAL1. PAP levels, chloroplast-specific responses, and drought responses were not measured. Furthermore, as mentioned above, the differing reported localizations of SAL1 have prevented any systematic analysis.
In this study, we demonstrated that total leaf PAP pools can be significantly lowered by targeting Sc-SAL1 exclusively to the chloroplast (Figure 5) and that induction of the nuclear gene APX2, which is routinely used to study HL and drought stress–induced retrograde signaling, was lowered when PAP was lowered by chloroplastic SAL1 complementation.
There are several lines of evidence that support the notion that PAP can move between cellular compartments, as shown in Figure 9. First and most compelling is that nuclear targeting of SAL1 results in the full complementation of sal1 mutant phenotypes, including total leaf PAP levels, APX2 expression in LL and drought, ELIP2 expression in LL and HL, and drought tolerance (Figure 5, Table 2). Second, sal1 and xrn2 xrn3 double mutants show a very similar molecular and morphological phenotype, suggesting that PAP accumulation can inhibit XRN function as originally proved in yeast (Dichtl et al., 1997) and suggested to occur in plants (Gy et al., 2007). Indeed, it is reasonable to assume that once in the cytosol, PAP would diffuse freely through the nuclear pore as do other nucleotides. Thus, degradation of PAP pools in either the chloroplast, mitochondria, or nucleus have the potential to restore the wild-type phenotype at the molecular level. This could be interpreted as PAP being able to move between cellular compartments.
Figure 9.
Proposed Model for a SAL1-PAP Retrograde Signaling Pathway.
PAP levels are negatively regulated by the chloroplastic SAL1 phosphatase (Figures 2 to 5). Upon environmental stresses, such as HL and drought, PAP levels increase (Figure 3). PAP can move between cellular compartments as evidenced by the complementation studies (Figure 5). Elevated PAP levels likely inhibit XRNs in the cytosol and nucleus. Nuclear XRN inhibition causes similar changes in expression to sal1 mutants, such as ELIP2 and APX2, and a degree of drought tolerance (Figures 6 to 8, Table 3).
[See online article for color version of this figure.]
Given the effectiveness of nuclear complementation, it begs the question as to why SAL1 is targeted to the organelles and not the nucleus. One speculative option is that this provides the potential to regulate PAP content in the cell in response to environmental stimuli. How, for instance, are HL and drought regulating PAP accumulation and/or transport? The observed increase in PAP levels during drought in alx8 plants lacking SAL1 suggests that PAP pools may be regulated at least in part by increased biosynthesis of PAP rather than a decrease in catabolism. However, whether SAL1 activity is directly regulated by HL and drought is not known, nor is it known if the stress-mediated PAP accumulation precedes ABA accumulation or not. Additionally, it would be worth investigating whether there could be regulation of PAP movement between compartments. Given the relatively small but significant change in PAP in response to HL, it might be that relocation of PAP within the cell also contributes to the response.
Does the proposed retrograde pathway operate in parallel or series with other proposed retrograde pathways? The gene expression profiles of other retrograde mutants pertain to the specific signals being studied, such as the repression of Lhcb mRNA in response to lincomycin and the chloroplast bleaching herbicide, NFZ, in the GUN signaling cascade. Lhcb mRNA does not change in alx8, and few of the coexpressed alx8 and xrn genes are induced or repressed by lincomycin (Figure 7; see Supplemental Data Set 1 online). Given the low levels of hydrogen peroxide in sal1 mutants (Figure 1), it is unlikely that the SAL1-PAP pathway is epistatic to or regulated by H2O2. With respect to ABA, the increase in PAP during drought and the induction of drought and ABA-responsive genes in sal1 and xrn mutants would suggest some interaction (Figures 3 and 7). Both drought and light alter chloroplasts and initiate gene expression changes, as demonstrated by the initial isolation of the alx8 mutant that results in changed expression of chloroplast proteins (ELIP2) and cytosolic proteins (APX2) and confers cellular tolerance to stress (e.g., plasma membrane damage) (see Wilson et al., 2009) and lowers ROS (Figure 1). That is, while drought is a general stress on a cell, a component of the drought response can be viewed as chloroplast specific, namely, inhibition of photosynthesis, leading to elevated ROS. Thus, it is not unexpected that 70% of HL genes are drought inducible. However, that does not preclude other drought response pathways operating independently or concurrently with the proposed SAL1-PAP pathway.
Evidence for XRNs Being Targets of the SAL1-PAP Pathway
A key question is, what are the targets of PAP? PAP is an adenosine phosphate and thus can bind irreversibly to yeast XRNs, inhibiting their activity (Dichtl et al., 1997). Thus, we investigated whether xrn knockouts would phenocopy elevated PAP levels.
Whereas the single xrn mutants show a wild-type phenotype, there is a strikingly similar expression levels of 56% of transcripts altered in xrn2 xrn3 with those in alx8, similar drought tolerance of the xrn2 xrn3 (Figures 6 to 8, Tables 2 and 3), and similar altered leaf morphology (Gy et al., 2007). All of this suggests that XRN2 and XRN3 are negative regulators of stress gene expression and may function in the SAL1-PAP pathway. xrn4 is also likely to be inhibited by elevated PAP; here, we show alx8 alters the abundance of the 3′ cleavage products of four miRNA targets (see Supplemental Figure 7 online), in addition to those reported for xrn4-5 (Souret et al., 2004) and the sal1 mutants fry1-4 and fry1-5 (Gy et al., 2007). Based on gene expression analysis (Figures 6 and 7) and xrn4 phenotypes, it is unlikely that inhibition of XRN4 accounts for the majority of the phenotypes observed in alx8, as the majority of the transcript changes, altered morphology, and drought tolerance better correlate with the xrn2 xrn3 double mutant. With respect to the drought tolerance, UDP-glucoronosyl/UDP-glucosyl transferase family protein (AT1G05680) is significantly upregulated in both sal1 and xrn2 xrn3 mutants (see Supplemental Data Set 1 online), and increasing its levels can cause drought tolerance (Tognetti et al., 2010). However, xrn2 xrn3 plants are not as tolerant as alx8 plants, and ~40% of the alx8 transcriptome changes are not found in the xrn2 xrn3 arrays. Although it remains to be directly demonstrated that PAP inhibits the activity of plant XRNs, the weight of evidence presented here and by Gy et al. (2007) is in favor of this interaction. Whether there are other targets for PAP or processes altered by SAL1 is the subject of investigation.
XRN-mediated gene regulation does not consist just of altered mRNA degradation as for MYB33 (see Supplemental Figure 7 online) but also includes elevated transcription, as demonstrated by APX2:LUC (Rossel et al., 2006). We envisage at least two potential mechanisms of action: XRNs alter mRNA levels by altering small and/or cleaved RNA pools, or XRNs alter gene transcription by affecting transcription termination. With respect to the latter, XRNs alter the release of RNA polymerases from the gene, thereby affecting transcription in yeast and human (Kim et al., 2004; West et al., 2004). Regarding changes to gene silencing, XRNs may inhibit accumulation of small RNAs that target positive regulators of stress gene expression. Alternatively, inhibition of the XRNs by PAP could prevent the degradation of the uncapped RNA templates triggering post-transcriptional gene silencing (Gy et al., 2007) of genes that repress stress responses. The determination of the substrates and the function of the nuclear XRNs will be critical to elucidate the underlying gene regulatory mechanisms.
In this article, we provide evidence for a previously undiscovered retrograde mechanism, the SAL1-PAP pathway, which would rely on chloroplastic SAL1 enzyme–regulating PAP levels, thereby affecting its action on nuclear targets, most likely XRNs. We resolve the chloroplastic localization of SAL1 and provide the unreported finding that it also accumulates in mitochondria but not in the cytosol or nuclei and that a primary in vivo substrate is PAP. PAP accumulates as result of HL and drought (20-fold), and it correlates with upregulation of 25% of the HL transcriptome. PAP is known to inhibit yeast XRNs, and we show here that SAL1 and XRNs mediate accumulation of 3′ mRNA cleavage products and expression of a common set of genes. Based on this, together with our finding that PAP can be depleted by targeting SAL1 to chloroplasts or nuclei, we conclude that SAL1 and PAP function in one of the cellular retrograde signaling pathways.
METHODS
Plant Material and Growth of Plants
Plant growth and drought stress conditions were as previously described (Wilson et al., 2009). Seeds from fry1-6 (SALK_020882) overexpressing a truncated form of SAL1 (AT5G63980) cDNA (Kim and von Arnim, 2009) and the xrn2-1 xrn3-1 (xrn2 xrn3) double mutant (Gy et al., 2007) were donated by A.G. von Arnim (University of Tennessee). Seeds for fou8 and SSU:ScSAL1-complemented fou8 (Rodríguez et al., 2010) were kindly provided by E.E. Farmer (University of Lausanne). Survival time of plants during drought was calculated as described by Woo et al. (2008) from measurements of the maximum efficiency of photosystem II (FvFm) using chlorophyll fluorescence. xrn4 mutants were xrn4-6 (SALK_ 014209) and ein5-6 (Olmedo et al., 2006). All insertion mutants were confirmed by PCR. See the primers listed in Supplemental Table 3 online.
RNA Isolation and RT-PCR
Total RNA was extracted from ~50 mg of leaf tissue using the Spectrum Total RNA kit (Sigma-Aldrich). RNA was reversed transcribed into cDNA using the Roche Transcriptor first-strand cDNA synthesis kit (Roche Diagnostics) and oligo(dT) primers. Gene expression was analyzed on a Roche LightCycler480 using hydrolysis probes from the Universal Probe Library and applying the relative quantification method described by Pfaffl (2001). Samples were normalized against CYCLOPHILIN5 (ATCYP5, AT2G29960) or GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE C2 (GAPC2, AT1G13440). At least three biological replicates per genotype per experiment were sampled, and each sample was run in triplicate.
Global Transcript Analyses
Analysis of the changes in transcript abundance between Col-0, xrn4 (ein5), alx8, and xrn2 xrn3 seedlings was performed using Affymetrix GeneChip Arabidopsis ATH1 genome arrays. Whole rosettes from several seedlings at the 10 true leaf stage of development (synchronized for development to account for the slower growth rate of alx8), grown under a 16-h photoperiod, were pooled for each biological replicate. Col-0 and mutant tissue samples were collected in biological triplicate. For each replicate, total RNA was isolated from the leaves using the RNeasy plant mini protocol (Qiagen) and quality verified using a Bioanalyzer (Agilent Technologies), and spectrophotometric analysis was performed to determine the A260:A280 and A260:A230 ratios. Preparation of labeled copy RNA from 400 ng of total RNA (3′ IVT Express kit; Affymetrix), target hybridization, as well as washing, staining, and scanning of the arrays were performed exactly as described in the Affymetrix GeneChip expression analysis technical manual, using an Affymetrix GeneChip Hybridization Oven 640, an Affymetrix Fluidics Station 450, and an GeneChip Scanner 3000 7G at the appropriate steps.
Statistical Analysis
Data quality was assessed using GCOS 1.4 before CEL files were exported into AVADIS Prophetic (version 4.3; Strand Genomics) and Partek Genomics Suite software, version 6.3, for further analysis. MAS5 normalization algorithms were performed only to generate present/absent calls across the arrays. Probe sets that recorded absent calls over 11 or more of the gene chips analyzed were removed. Bacterial controls were also removed, resulting in a final data set of 16,022 probe identifiers. CEL files were also subjected to GC-content background Robust Multi-array Average normalization for computing fluorescence intensity values used in further analyses. Correlation plots were examined between all arrays using the scatterplot function in the Partek Genomics Suite, and in all cases r ≥ 0.98 (data not shown). The values of gene expression after normalization with GC-content background Robust Multi-array Average were analyzed to identify differentially expressed genes by a regularized t test based on a Bayesian statistical framework using the software program Cyber-T (Baldi and Long, 2001) (http://cybert.microarray.ics.uci.edu/). Cyber-T employs a mixture model-based method described by Allison et al. (2006) for the computation of the global false-positive and false-negative levels inherent in a DNA microarray experiment. To accurately control for FDR and minimize false positives within the differential expression analysis, posterior probability of differential expression PPDE(P) values and PPDE(>P) values were calculated, as a means to measure the true discovery rate (1 – FDR). Changes in transcript abundance were considered significant with a PPDE(>P) > 0.95 and a fold change >1.5-fold.
Overlaps in the transcript abundance responses for the different genotypes were plotted on Venn diagrams to determine statistically significant over- or underrepresentation in the overlap, compared with that which is expected by random chance, using a Pearson’s χ2 test for independence.
ANOVA and t test analyses were performed using Microsoft Excel.
Cell Fractionation
Arabidopsis thaliana ecotype Col-0 was grown in 0.5× Murashige and Skoog (MS) medium, 1% (w/v) Suc, and 0.7% (w/v) agar plates under LL (100 μmol photon m–2 s–1) with a 12-h photoperiod at 20 ± 2°C for 16 d. Approximately 13 g of seedlings were harvested in the morning before commencement of the light period. The tissue was vacuum infiltrated in 40 mL of digestion medium (1.5% [w/v] cellulase, 0.4% [w/v] macerozyme, 0.5 M Suc, 20 mM KCl, 10 mM CaCl2, and 20 mM MES) for 30 min and incubated for an additional 3 h. All incubation steps were performed in the dark. After incubation, the mixture was filtered through two layers of miracloth presoaked in floating medium (Gardeström and Wigge, 1988) (0.5 M Suc, 1 mM MgCl2, and 5 mM HEPES, pH 7.0). Protoplasts were released by passing the eluant through the slurry several times, pressing it with a spatula after the last elution, and collecting it in a glass beaker in ice. All subsequent steps were performed at 4°C. The dark-green flow-through was divided into 10-mL aliquots in four 30-mL Corex tubes and topped with 5 mL of floating medium II (FMII; 0.4 M Suc, 0.1 M sorbitol, 1 mM MgCl2, and 5 mM HEPES, pH 7.0) and 3 mL of floating medium III (FMIII; 0.5 M sorbitol, 1 mM MgCl2, and 5 mM HEPES, pH 7.0). This was performed very carefully to avoid disrupting the interfaces between the different solutions. The tubes were centrifuged at 250g for 5 min at 4°C in a swing-out rotor (brake off). Intact protoplasts were recovered from the FMIII/FMII interface and the chloroplast-containing pellet further processed (see below). The intact protoplasts were disrupted by six strokes in a prechilled 10-mL Wheaton Potter-Elvehjem tissue grinder. An ~8-mL sample was transferred to a 15-mL Corex tube, a 500 μL 85% (v/v) Percoll cushion was layered at the bottom with a long glass Pasteur pipette, and a 1000-μL layer of FMIII was added on the top. The gradient was then centrifuged at 2000g for 10 min at 4°C in a swing-out rotor (brake off). Contaminating protoplasts settled in the FMIII/sample interface, and the remainder of the chloroplasts were on top of the 85% (v/v) Percoll cushion. The middle phase, containing the cytosolic fraction, was carefully removed and centrifuged at 13,000g for 30 min at 4°C in a fixed-angle rotor to remove any contaminating organelles. The supernatant was further concentrated with 5-kD Ultrafree centrifugal filter devices (Millipore).
For the purification of chloroplasts, the pellets after the first centrifugation step were resuspended in a final volume of 10 mL of chloroplast buffer (50 mM HEPES-KOH, pH 8.0, 5 mM EDTA, 5 mM EGTA, 330 mM sorbitol, 5 mM Cys, and 5 mM ascorbic acid) and cleared by centrifugation at 250g for 5 min at 4°C in a swing-out rotor. The pellets were gently resuspended in a minimum volume (~3 mL) of the same buffer, loaded onto a 45/85% (v/v) Percoll gradient (Aronsson and Jarvis, 2002), and spun at 3000g 15 min at 4°C in a swing-out rotor (brake off). The intact chloroplasts were recovered from the 45/85% (v/v) interface, washed once with 10 volumes of chloroplast buffer, and spun at 800g for 15 min in a swing-out rotor at 4°C. The clean, intact chloroplasts were resuspended in 500 μL of chloroplast buffer.
Mitochondria were purified by free-flow electrophoresis as described by Eubel et al. (2007).
Protein Gel Electrophoresis and Immunoblotting
Total protein from tissue and cell fractions were extracted in 10% (w/v) tricarboxylic acid in cold acetone. Protein gel electrophoresis was performed as described (Wilson et al., 2009). Immunoblotting was performed using the SNAPid system (Millipore) and antibodies against SAL1 (1:1000; Wilson et al., 2009), Lhcb2 (1:1000; Agrisera AS01-003), TOM40 (1:5000; Carrie et al., 2009), and UGPase (1:500; Agrisera AS05-086).
GFP Fusion for Subcellular Localization
The full-length cDNA of SAL1 was cloned as a C-terminal GFP fusion by Gateway cloning under the control of a 35S promoter (Murcha et al., 2007; Carrie et al., 2009). Primers used were as follows: SAL1FOR, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCunderline>ATG/underline>underline>ATGTCTATAAATTGTTTTCGAA/underline>-3′, and SAL1REV, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCCACCTCCGGATCCunderline>GAGAGCTGAA/underline>underline>GCTTTCTCTTGC/underline>-3′, where underlined sections correspond to SAL1 and the rest for homologous recombination. The PCR product was cloned into pDonr201 (Invitrogen) and then into pDest/pGem/CGFP (Carrie et al., 2009). The plastid localization control, Rubisco SSU, and the mitochondrial control, AOX1, were cloned as C-terminal RFP fusions using the same system. SAL1-GFP along with either one of the control constructs was cotransformed into Arabidopsis cell suspension as previously described (Thirkettle-Watts et al., 2003; Carrie et al., 2009). Localization of GFP and RFP expression was conducted using an Olympus BX61 fluorescence microscope and imaged using the CellR imaging software as previously described (Carrie et al., 2007; Murcha et al., 2007).
In Vivo SAL1:GFP Visualization
The SAL1 genomic fragment (1960 bp) and an additional 753-bp upstream region was PCR cloned by standard molecular techniques in the Wassilewskija accession. Primers used were as follows: FRY1promF, 5′-CACCGTTGGAGATTATCTTCTGTAGG-3′, and FRY1endR, 5′-GAGAGCTGAAGCTTTCTCTTGC-3′, which amplified a product of 2713 bp (753+1960). After sequencing in pENTR/D-TOPO, an LR clonase reaction was used to clone the genomic fragment in the binary vector pGWB4 (Nakagawa et al., 2007) and transformed into Arabidopsis Col-0 by simplified floral dip method (Logemann et al., 2006). Primary transformants were selected in medium containing 50 μg/L hygromycin. Their progeny were screened for GFP expression in standard in vitro growth conditions.
Observations were made using either an upright DMR or a Leica SP2 AOBS inverted confocal microscope (Leica Microsystems). In the first case, a ×10 DRY objective lens (numerical aperture of 0.40) was mounted on an upright DMR microscope equipped with a mercury lamp. The GFP and chlorophyll signals were collected using a 515-nm-long pass filter (excitation with 450 to 490 nm). For the observations made with the Leica SP2 AOBS inverted confocal microscope, 2-week-old plants grown in soil in short-day conditions were used. A ×10 dry objective lens (numerical aperture of 0.40) was used for all observations. GFP and chlorophyll were excited with a 488-nm argon laser. The GFP signal was collected at 496 to 539 nm and chlorophyll at 676 to 718 nm (pinhole adjusted to 2.46). Following acquisition, brightness and contrast were adjusted using the LCS software.
Mesophyll protoplasts were prepared as described (Leonhardt et al., 2004). Protoplasts were incubated on ice with 400 nM MitoTracker Red CMXRos (Invitrogen) for 30 min, in the dark, before confocal observations. Observations were made using a Leica SP2 AOBS inverted confocal microscope (Leica Microsystems) equipped with an argon ion laser and a DPSS 561-nm laser. A ×63 water objective lens (numerical aperture of 1, 20) was used for all observations. GFP was excited at 488 nm and MitoTracker red at 561 nm. The GFP signal was collected at 500 to 530 nm, the MitoTracker Red signal was collected at 575 to 615 nm, and chlorophyll at 653 to 731 nm. Following acquisition, brightness and contrast were adjusted using the LCS software (identical parameters were applied to control and GFP images).
Quantification of Phosphoadenosines
Extraction of Adenosine Derivatives
Adenosine derivatives were extracted in HCl and quantified fluorometrically after specific derivatization of adenosine compounds with chloroacetaldehyde (CAA) based on a method previously described (Bürstenbinder et al., 2007). Briefly, 100 mg of tissue was ground in liquid nitrogen, extracted with 1 mL of 0.1 M HCl in ice for 15 min, and centrifuged twice at 16,000g at 4°C for 5 min. After clarification, 150 μL of the supernatant mixed with 770 μL of CP buffer [620 mM citric acid-1-hydrate and 760 mM (Na)2HPO4.2H2O, pH 4] was derivatized by adding 80 μL of 45% (v/v) chloroacetaldehyde for 10 min at 80°C. The sample was finally centrifuged at 16,000g for 45 min at 20°C before injection into the HPLC. The commercial standards used were as follows: adenosine (Sigma-Aldrich; A9251), ADP sodium salt (Sigma-Aldrich; A2754), AMP (Fluka; catalog number 1930), APS sodium salt (Sigma-Aldrich; A5508), ATP (Sigma-Aldrich; A-5394), PAP (Sigma-Aldrich; A5763), PAPS (Sigma-Aldrich; A1651), and S-(5′-adenosyl)-l-Met chloride (Sigma-Aldrich; A7007).
HPLC of Adenosine Derivatives
The analyses of adenosines was performed by reverse-phase HPLC on a Gemini-NX 5-μm C18 110A column (Phenomenex) connected to Waters 600E HPLC system and Waters 474 fluorescent detector (Waters). The gradient for separation of PAP was optimized as follows: equilibration of column for 0.2 min with 95% (v/v) of buffer A (5.7 mM [CH3(CH2)3]4N HSO4 and 30.5 mM KH2PO4, pH 5.8) and 5% (v/v) buffer B (67% [v/v] acetonitrile and 33% [v/v] buffer A), linear gradient for 53 min up to 50% (v/v) of buffer B, and re-equilibration for 7 min with 5% (v/v) buffer B. Chromatograms were recorded and processed with the Milenium32 software (Waters) and edited using Adobe Illustrator (Adobe).
Analysis of IPs
Feeding of Arabidopsis Seedlings with Radiolabeled myo-Inositol
Seeds were stratified and sown on 0.5× MS agar containing vitamins (Duchefa Biochemie) and germinated at 22°C in a Sanyo light cabinet under long-day (16 h light/8 h dark) conditions at a fluence rate of 120 μmol m−2 s−1 for 10 d. Five or six seedlings were subsequently placed in 0.4 mL of 0.5× MS agar basal salts (Duchefa) liquid media to which 1.11 mBq of myo-[2-3H]inositol (specific activity 752 GBq/mmol; PerkinElmer) and 0.1 mM unlabeled myo-inositol were added. Seedlings were labeled for 72 h. Seedlings were washed briefly in water, blotted dry, transferred to a conical Eppendorf tube, frozen in liquid N2, and ground with a plastic pestle. The sample was extracted, with further grinding, with 0.3 mL of 0.6 M HCl after addition of 10 μL of 1 mM phytic acid (Sigma-Aldrich; P8810) and left on ice for 30 min before centrifugation at 17,000g for 15 min at 4°C. The supernatant was diluted to 1 mL with water and applied directly to HPLC.
HPLC of IPs
IP extracts were analyzed on a 25 cm × 4.6-mm Partisphere SAX HPLC column (Whatman) with guard cartridge of the same material. Samples, 1 mL, were loaded and eluted with a gradient of phosphate, generated by mixing solvents from buffer reservoirs, A, water; B, 1.25 M NH4H2PO4, adjusted to pH 3.35 with H3PO4, according to the following schedule: time (min),% B; 0,0; 5,0; 65,100; 75,100. Radioactivity was estimated in a Canberra Packard A515 detector fitted with a 0.5-mL flow cell after admixture of Optima Flo AP scintillation fluid (Canberra Packard) at a flow rate of 2 mL min−1. The integration interval was 0.2 min. Data were exported from the Flo-One software (Canberra Packard) as ASCII files and redrawn in Delta Graph 4.0 (DeltaPoint).
Feeding of Arabidopsis Seedlings with PAP
Seven-day-old, plate-grown Col-0 seedlings were incubated with 100 μL of 0, 0.5, 1.0, and 2.0 mM PAP in 96-well plates. They were then incubated at LL (150 μmol m−2 s−1) or HL (1500 μmol m−2 s−1) for 50 min. After treatment, 1 mM luciferin was added and light emitted acquired overnight in a luminometer (FLUOStar OPTIMA).
H2O2 Detection and Quantification
Foliar H2O2 levels were visualized by 3,3′-diaminobenzidine (DAB) staining. Five-week-old plants grown hydroponically were transferred to a solution containing 25 mM DAB and incubated 1 h under HL (1000 μmol photons m−2 s−1). The leaves were then placed into ethanol to remove the chlorophyll and the presence of H2O2 visualized as a brown stain. Alternatively, leaf H2O2 was quantified in 6-week-old, soil-grown plants with the Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen). Approximately 100 mg of leaf tissue was flash frozen in liquid nitrogen, ground, and extracted with 500 μL of 20 mM K2HPO4, pH 6.5. The slurry was cleared by centrifugation at 16,000g for 15 min at 4°C. The supernatant was incubated with 0.1 mM Amplex Red reagent and 0.2 units mL−1 horseradish peroxidase at room temperature for 30 min in the dark (final reaction volume of 100 μL). Finally, absorbance was measured at 560 nm using a μQuant plate reader (BioTek Instruments) with Gen5 software, and the concentration of H2O2 was calculated using a standard curve (0.5, 1, 2, and 5 mM H2O2 range).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: ABI1 (At4g26080), APX2 (At3g09640), ATHB15 (At1g52150), ATCYP5 (AT2G29960), ELIP2 (At4g14690), GNAT (At2g39030), MYB33 (At5g06100), OST1 (At4g33950), PHB (At2g34710), REV (At5g60690), SAL1/FRY1 (At5g63980), SOT1 (At2g03760), T5C23 (At4g11710), VSP1 (At5g24780), XRN2 (At5g42540), XRN3 (At1g75660), and XRN4 (At1g54490).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. PAP Feeding Experiments
Supplemental Figure 2. The sal1 Mutants alx8 and fry1-6 Lack SAL1 Protein and Have Similar Rosette Morphology.
Supplemental Figure 3. Inositol Phosphate Profile in Col-0 and the sal1 Mutant fry1-1.
Supplemental Figure 4. Development of a Highly Sensitive Technique for Quantification of Phosphonucleotides.
Supplemental Figure 5. PAP, but Not PAPS, Accumulates in the fou8 Mutants.
Supplemental Figure 6. Development of a New Method for Simultaneous Isolation of Cytosolic and Chloroplastic Fractions.
Supplemental Figure 7. Accumulation of 3′ Cleavage Products in SAL1 Mutants.
Supplemental Figure 8. Antagonistic Changes in Global Gene Expression in alx8, xrn4, and xrn2 xrn3.
Supplemental Table 1. miRNA Targets Upregulated in alx8.
Supplemental Table 2. Genes Showing Similar Transcriptional Responses in Both alx8 and xrn2 xrn3 Mutants.
Supplemental Table 3. List of Primers.
Supplemental Data Set 1. Microarray Data for alx8, xrn2 xrn3, and xrn4 Mutants.
Acknowledgments
We thank Albrecht G. von Arnim (University of Tennessee) for providing seeds of fry1-6 (SALK_020882) overexpressing a truncated form of SAL1 and xrn2 xrn3 double mutants. Seeds of fou8 and fou8-complemented line were kindly provided by Edward Farmer (University of Lausanne). We thank Hayley Whitfield (Brearley lab) for plant care and technical assistance. Funding to Australian researchers was provided by the Australian Research Council Centre of Excellence in Plant Energy Biology (CE0561495).
AUTHOR CONTRIBUTIONS
G.M.E. performed cell fractionation and localization, adenosine derivative quantification, gene expression of complemented lines for the RT-PCR versus PAP correlations, immunoblots, and drought experiments. G.M.E. and B.J.P. planned and coordinated the experimental work, analyzed data, and wrote the manuscript. P.A.C. screened and characterized fry1-6:trSAL1 and xrn mutant lines, including drought testing, performed gene expression analyses, and assisted with PAP quantification and writing the manuscript. W.P. performed the feeding experiments with PAP, DAB staining, and H2O2 quantification. M.W. and R.H. developed the HPLC procedure to quantify PAP and also performed metabolite quantification. D.C. standardized the HPLC technique at Australian National University and helped to quantify PAP in chloroplastic fractions. C.C., E.G., and J.W. carried out the GFP fusion and microarray analyses. P.D., H.J., and E.M. prepared the pSAL1:SAL1:GFP construct, transformed the plants, and performed localization studies of the SAL1:GPF reporter gene in vivo. C.B. quantified the IPs in seedlings and seeds of different genotypes. All the authors discussed the results and commented on the manuscript.
References
- Albrecht V., Simková K., Carrie C., Delannoy E., Giraud E., Whelan J., Small I.D., Apel K., Badger M.R., Pogson B.J. (2010). The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis. Plant Cell 22: 3423–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison D.B., Cui X., Page G.P., Sabripour M. (2006). Microarray data analysis: From disarray to consolidation and consensus. Nat. Rev. Genet. 7: 55–65 [DOI] [PubMed] [Google Scholar]
- Aronsson H., Jarvis P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529: 215–220 [DOI] [PubMed] [Google Scholar]
- Baldi P., Long A.D. (2001). A Bayesian framework for the analysis of microarray expression data: Regularized t -test and statistical inferences of gene changes. Bioinformatics 17: 509–519 [DOI] [PubMed] [Google Scholar]
- Ball L., Accotto G.-P., Bechtold U., Creissen G., Funck D., Jimenez A., Kular B., Leyland N., Mejia-Carranza J., Reynolds H., Karpinski S., Mullineaux P.M. (2004). Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16: 2448–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbeer J.W., Atkinson Y.E., Borner T., Hagemann R. (1979). Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesized RNA. Nature 279: 816–817 [Google Scholar]
- Bürstenbinder K., Rzewuski G., Wirtz M., Hell R., Sauter M. (2007). The role of methionine recycling for ethylene synthesis in Arabidopsis. Plant J. 49: 238–249 [DOI] [PubMed] [Google Scholar]
- Carrie C., Murcha M.W., Millar A.H., Smith S.M., Whelan J. (2007). Nine 3-ketoacyl-CoA thiolases (KATs) and acetoacetyl-CoA thiolases (ACATs) encoded by five genes in Arabidopsis thaliana are targeted either to peroxisomes or cytosol but not to mitochondria. Plant Mol. Biol. 63: 97–108 [DOI] [PubMed] [Google Scholar]
- Carrie C., Kühn K., Murcha M.W., Duncan O., Small I.D., O’Toole N., Whelan J. (2009). Approaches to defining dual-targeted proteins in Arabidopsis. Plant J. 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Chen H.A.O., Xiong L. (2010). The bifunctional abiotic stress signalling regulator and endogenous RNA silencing suppressor FIERY1 is required for lateral root formation. Plant Cell Environ. 33: 2180–2190 [DOI] [PubMed] [Google Scholar]
- Clifton R., Lister R., Parker K.L., Sappl P.G., Elhafez D., Millar A.H., Day D.A., Whelan J. (2005). Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Dichtl B., Stevens A., Tollervey D. (1997). Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 16: 7184–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H., Lee C.P., Kuo J., Meyer E.H., Taylor N.L., Millar A.H. (2007). Free-flow electrophoresis for purification of plant mitochondria by surface charge. Plant J. 52: 583–594 [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2009). Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11: 861–905 [DOI] [PubMed] [Google Scholar]
- Fryer M.J., Ball L., Oxborough K., Karpinski S., Mullineaux P.M., Baker N.R. (2003). Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J. 33: 691–705 [DOI] [PubMed] [Google Scholar]
- Galvez-Valdivieso G., Fryer M.J., Lawson T., Slattery K., Truman W., Smirnoff N., Asami T., Davies W.J., Jones A.M., Baker N.R., Mullineaux P.M. (2009). The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21: 2143–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardeström P., Wigge B. (1988). Influence of photorespiration on ATP/ADP ratios in the chloroplasts, mitochondria, and cytosol, studied by rapid fractionation of barley (Hordeum vulgare) protoplasts. Plant Physiol. 88: 69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmier M., Carroll A.J., Delannoy E., Vallet C., Day D.A., Small I.D., Millar A.H. (2008). Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol. 148: 1324–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German M.A., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Gil-Mascarell R., López-Coronado J.M., Bellés J.M., Serrano R., Rodríguez P.L. (1999). The Arabidopsis HAL2-like gene family includes a novel sodium-sensitive phosphatase. Plant J. 17: 373–383 [DOI] [PubMed] [Google Scholar]
- Giraud E., Ho L.H., Clifton R., Carroll A., Estavillo G., Tan Y.F., Howell K.A., Ivanova A., Pogson B.J., Millar A.H., Whelan J. (2008). The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory B.D., O’Malley R.C., Lister R., Urich M.A., Tonti-Filippini J., Chen H., Millar A.H., Ecker J.R. (2008). A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Gy I., Gasciolli V., Lauressergues D., Morel J.-B., Gombert J., Proux F., Proux C., Vaucheret H., Mallory A.C. (2007). Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19: 3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari-Steinberg O., Ohad I., Chamovitz D.A. (2001). Dissection of the light signal transduction pathways regulating the two early light-induced protein genes in Arabidopsis. Plant Physiol. 127: 986–997 [PMC free article] [PubMed] [Google Scholar]
- Hirsch J., et al. (2011). A novel fry1 allele reveals the existence of a mutant phenotype unrelated to 5′->3′ exoribonuclease (XRN) activities in Arabidopsis thaliana roots. PLoS ONE 6: e16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008: 420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S., Escobar C., Karpinska B., Creissen G., Mullineaux P.M. (1997). Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9: 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S., Reynolds H., Karpinska B., Wingsle G., Creissen G., Mullineaux P. (1999). Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284: 654–657 [DOI] [PubMed] [Google Scholar]
- Kastenmayer J.P., Green P.J. (2000). Novel features of the XRN-family in Arabidopsis: Evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl. Acad. Sci. USA 97: 13985–13990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.H., von Arnim A.G. (2009). FIERY1 regulates light-mediated repression of cell elongation and flowering time via its 3′(2′),5′-bisphosphate nucleotidase activity. Plant J. 58: 208–219 [DOI] [PubMed] [Google Scholar]
- Kim M., Krogan N.J., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S. (2004). The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522 [DOI] [PubMed] [Google Scholar]
- Kimura M., Manabe K., Abe T., Yoshida S., Matsui M., Yamamoto Y.Y. (2003). Analysis of hydrogen peroxide-independent expression of the high-light-inducible ELIP2 gene with the aid of the ELIP2 promoter-luciferase fusions. Photochem. Photobiol. 77: 668–674 [DOI] [PubMed] [Google Scholar]
- Kimura M., Yamamoto Y., Seki M., Sakurai T., Sato M., Shinozaki K., Manabe K., Matsui M. (2002). Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Plant Cell Physiol. 43: S159. [DOI] [PubMed] [Google Scholar]
- Kindgren P., Eriksson M.-J., Benedict C., Mohapatra A., Gough S.P., Hansson M., Kieselbach T., Strand A. (2011). A novel proteomic approach reveals a role for Mg-protoporphyrin IX in response to oxidative stress. Physiol. Plant. 141: 310–320 [DOI] [PubMed] [Google Scholar]
- Klein M., Papenbrock J. (2004). The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J. Exp. Bot. 55: 1809–1820 [DOI] [PubMed] [Google Scholar]
- Kleine T., Kindgren P., Benedict C., Hendrickson L., Strand A. (2007). Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T., Voigt C., Leister D. (2009). Plastid signalling to the nucleus: Messengers still lost in the mists? Trends Genet. 25: 185–192 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Larkin R.M., Alonso J.M., Ecker J.R., Chory J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Lee K.P., Kim C., Landgraf F., Apel K. (2007). EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N., Kwak J.M., Robert N., Waner D., Leonhardt G., Schroeder J.I. (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E., Birkenbihl R.P., Ulker B., Somssich I.E. (2006). An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.H., Carrie C., Pogson B., Whelan J. (2009). Exploring the function-location nexus: Using multiple lines of evidence in defining the subcellular location of plant proteins. Plant Cell 21: 1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Tanaka R., Tanaka A., Masuda T., Nagatani A. (2008). The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M., McCormac A.C., Terry M.J., Smith A.G. (2008). Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. USA 105: 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford S.G., et al. (2009). Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21: 910–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha M.W., Elhafez D., Lister R., Tonti-Filippini J., Baumgartner M., Philippar K., Carrie C., Mokranjac D., Soll J., Whelan J. (2007). Characterization of the preprotein and amino acid transporter gene family in Arabidopsis. Plant Physiol. 143: 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murguía J.R., Bellés J.M., Serrano R. (1996). The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J. Biol. Chem. 271: 29029–29033 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nott A., Jung H.-S., Koussevitzky S., Chory J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Olinares P.D.B., Ponnala L., van Wijk K.J. (2010). Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol. Cell. Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo G., Guo H., Gregory B.D., Nourizadeh S.D., Aguilar-Henonin L., Li H., An F., Guzman P., Ecker J.R. (2006). ETHYLENE-INSENSITIVE5 encodes a 5′—>3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl. Acad. Sci. USA 103: 13286–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J.-B., Cai Y., Sun Q., Zabrouskov V., Giacomelli L., Rudella A., Ytterberg A.J., Rutschow H., van Wijk K.J. (2006). The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics 5: 114–133 [DOI] [PubMed] [Google Scholar]
- Perera I.Y., Hung C.-Y., Moore C.D., Stevenson-Paulik J., Boss W.F. (2008). Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell 20: 2876–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T. (2010). Plastidial retrograde signalling—A true “plastid factor” or just metabolite signatures? Trends Plant Sci. 15: 427–435 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T., Bräutigam K., Wagner R., Dietzel L., Schröter Y., Steiner S., Nykytenko A. (2009). Potential regulation of gene expression in photosynthetic cells by redox and energy state: Approaches towards better understanding. Ann. Bot. (Lond.) 103: 599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B.J., Woo N.S., Förster B., Small I.D. (2008). Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Quintero F.J., Garciadeblás B., Rodríguez-Navarro A. (1996). The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 8: 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles P., Fleury D., Candela H., Cnops G., Alonso-Peral M.M., Anami S., Falcone A., Caldana C., Willmitzer L., Ponce M.R., Van Lijsebettens M., Micol J.L. (2010). The RON1/FRY1/SAL1 gene is required for leaf morphogenesis and venation patterning in Arabidopsis. Plant Physiol. 152: 1357–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez V.M., Chételat A., Majcherczyk P., Farmer E.E. (2010). Chloroplastic phosphoadenosine phosphosulfate metabolism regulates basal levels of the prohormone jasmonic acid in Arabidopsis leaves. Plant Physiol. 152: 1335–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel J.B., Walter P.B., Hendrickson L., Chow W.S., Poole A., Mullineaux P.M., Pogson B.J. (2006). A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ. 29: 269–281 [DOI] [PubMed] [Google Scholar]
- Rossel J.B., Wilson P.B., Hussain D., Woo N.S., Gordon M.J., Mewett O.P., Howell K.A., Whelan J., Kazan K., Pogson B.J. (2007). Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19: 4091–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle M.E., DeMarco S.M., Larkin R.M. (2007). Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret F.F., Kastenmayer J.P., Green P.J. (2004). AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15: 173–183 [DOI] [PubMed] [Google Scholar]
- Strand A., Asami T., Alonso J., Ecker J.R., Chory J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Susek R.E., Ausubel F.M., Chory J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Thirkettle-Watts D., McCabe T.C., Clifton R., Moore C., Finnegan P.M., Day D.A., Whelan J. (2003). Analysis of the alternative oxidase promoters from soybean. Plant Physiol. 133: 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti V.B., et al. (2010). Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F., Bailey-Serres J., Mittler R. (2008). Unraveling the tapestry of networks involving reactive oxygen species in plants. Plant Physiol. 147: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E.L., et al. (2011). XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475: 114–117 [DOI] [PubMed] [Google Scholar]
- Vranová E., Inzé D., Van Breusegem F. (2002). Signal transduction during oxidative stress. J. Exp. Bot. 53: 1227–1236 [PubMed] [Google Scholar]
- Wagner D., Przybyla D., Op den Camp R., Kim C., Landgraf F., Lee K.P., Würsch M., Laloi C., Nater M., Hideg E., Apel K. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- West S., Gromak N., Proudfoot N.J. (2004). Human 5′ —> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432: 522–525 [DOI] [PubMed] [Google Scholar]
- Wilson P.B., Estavillo G.M., Field K.J., Pornsiriwong W., Carroll A.J., Howell K.A., Woo N.S., Lake J.A., Smith S.M., Harvey Millar A., von Caemmerer S., Pogson B.J. (2009). The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 58: 299–317 [DOI] [PubMed] [Google Scholar]
- Woo N.S., Badger M.R., Pogson B.J. (2008). A rapid, non-invasive procedure for quantitative assessment of drought survival using chlorophyll fluorescence. Plant Methods 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J.D., Perez-Ruiz J.M., Chory J. (2011). Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L.M., Lee Bh, Ishitani M., Lee H., Zhang C.Q., Zhu J.K. (2001). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska-Placzek M., Souret F.F., Sobczyk G.J., Green P.J., Kufel J. (2010). Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res. 38: 4487–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2011). Inositol trisphosphate-induced Ca2+ signaling modulates auxin transport and PIN polarity. Dev. Cell 20: 855–866 [DOI] [PubMed] [Google Scholar]