The orphan small-molecule glycosyltransferase UGT76B1 is a novel player in plant pathogen defense that represses the salicylic acid pathway, yet enhances the jasmonic acid pathway. A nontargeted metabolome approach led to the identification of its substrate, 2-hydroxy-3-methyl-pentanoic acid, which itself was able to induce resistance against Pseudomonas syringae infection.
Abstract
Plants coordinate and tightly regulate pathogen defense by the mostly antagonistic salicylate (SA)- and jasmonate (JA)-mediated signaling pathways. Here, we show that the previously uncharacterized glucosyltransferase UGT76B1 is a novel player in this SA-JA signaling crosstalk. UGT76B1 was selected as the top stress-induced isoform among all 122 members of the Arabidopsis thaliana UGT family. Loss of UGT76B1 function leads to enhanced resistance to the biotrophic pathogen Pseudomonas syringae and accelerated senescence but increased susceptibility toward necrotrophic Alternaria brassicicola. This is accompanied by constitutively elevated SA levels and SA-related marker gene expression, whereas JA-dependent markers are repressed. Conversely, UGT76B1 overexpression has the opposite effect. Thus, UGT76B1 attenuates SA-dependent plant defense in the absence of infection, promotes the JA response, and delays senescence. The ugt76b1 phenotypes were SA dependent, whereas UGT76B1 overexpression indicated that this gene possibly also has a direct effect on the JA pathway. Nontargeted metabolomic analysis of UGT76B1 knockout and overexpression lines using ultra-high-resolution mass spectrometry and activity assays with the recombinant enzyme led to the ab initio identification of isoleucic acid (2-hydroxy-3-methyl-pentanoic acid) as a substrate of UGT76B1. Exogenously applied isoleucic acid increased resistance against P. syringae infection. These findings indicate a novel link between amino acid–related molecules and plant defense that is mediated by small-molecule glucosylation.
INTRODUCTION
Plants are sessile organisms and cannot escape adverse environmental cues. Therefore, they have evolved elaborate mechanisms to antagonize these stresses and to organize defense or tolerance. These measures involve a complex reprogramming of plant cells, which relies on major changes in gene expression, protein modification, and a range of different compounds active in defense and signaling. Several small-molecule hormones, such as salicylic acid (SA), jasmonic acid (JA), ethylene, and abscisic acid play crucial roles in regulating responses of plants to both biotic and abiotic stresses. These signaling pathways interact with each other in synergistic as well as antagonistic manners, enabling the plant to fine-tune its response to the stressor(s) encountered (Jones and Dangl, 2006; Koornneef and Pieterse, 2008). Constitutive production of signaling molecules and the concomitant expression of defense genes is energetically costly, and reallocation of resources toward defense seems to decrease plant overall fitness (Heil and Baldwin, 2002; Lorrain et al., 2003). Therefore, plants need a tight control of the defense response and its suppression in the absence of pathogen attack or other stresses (Heidel et al., 2004; Bolton, 2009). Mostly, SA- and JA-mediated signaling pathways are triggered when plants defend themselves against pathogens. Although concerted actions of both pathways have been reported, they usually act in an antagonistic manner via mutual repression (Glazebrook, 2005; Jones and Dangl, 2006; Spoel et al., 2007; Koornneef et al., 2008; Vlot et al., 2009). Whereas biotrophic pathogens (bacteria, fungi, and viruses) are mostly combatted by the SA pathway, the opposite prioritization of defense signaling is mobilized to fight necrotrophic pathogens (bacteria and fungi) and herbivores.
Arabidopsis thaliana genetics has defined a plethora of genes involved in both SA and JA signaling and their interplay. A number of mutants resulted in enhanced susceptibility to biotrophic pathogens and suppression of SA responses and could therefore be used to define the crucial steps in SA signaling. These include components of the mitogen-activated protein kinase signaling pathway, such as ERD1, MPK3, and MPK6; genes related to SA biosynthesis, such as ICS1/SID2, PAD4, and EDS1; central downstream regulators of SA signaling, such as NPR1; as well as WRKY and TGA transcription factors. Induction of these transcription factors eventually leads to the activation of SA-responsive genes, including PR genes that are involved in defense responses. Similarly, mutations in genes such as JAR1, COI1, and JIN1, which define different steps in JA signaling, negatively affect the JA pathway (Kazan and Manners, 2008). Resistance toward necrotrophic pathogens is reduced in the corresponding mutants concomitant with the abolished induction of marker genes, like the defensin PDF1.2. By contrast, several gain-of-resistance Arabidopsis mutants, such as mlo, mpk4, wrky, acd, lsd, hrl1, hlm1, or dnd, show constitutive defense responses in the absence of (biotrophic) pathogen attack, which affects pathogen perception and response or leads to primed defense (Greenberg et al., 1994; Petersen et al., 2000; Devadas et al., 2002; Balagué et al., 2003; Lorrain et al., 2003; Consonni et al., 2006; Journot-Catalino et al., 2006; Genger et al., 2008). Other interesting classes of mutants with enhanced resistance affecting various steps in signal transduction are the cpr mutants, named after the CONSTITUTIVE ACTIVATION OF PR genes, and several suppressors of npr1 mutants, such as ssi and sni. These mutants are usually characterized by transcriptional activation of PR genes and constitutive accumulation of SA (Bowling et al., 1994; Li et al., 1999; Shah et al., 1999; Gou et al., 2009). In addition, several of the mutants resistant to biotrophic pathogens exhibit retarded growth and/or accelerated senescence. Notably, developmental senescence is at least in part regulated by an SA-dependent pathway (Buchanan-Wollaston et al., 2005).
It has been shown that some of the genes mentioned above (such as those encoding MPK4, WRKY transcription factors, and NPR1) exert opposite effects on the SA and JA pathways. Thus, they are integral to the SA-JA crosstalk (Koornneef and Pieterse, 2008; Vlot et al., 2009). Interestingly, two aminotransferase mutants, agd2 and ald1, have an opposite influence on pathogen susceptibility, which points toward a possible involvement of amino acid–related molecules in the regulation of defense (Song et al., 2004). Although the existence of several SA and JA amino acid conjugates is known, the direct involvement of amino acids in defense has been shown only in the case of JA-Ile, which is the major, bioactive form of jasmonate (Staswick and Tiryaki, 2004; Fonseca et al., 2009).
Plant secondary metabolite UDP-dependent glycosyltransferases (UGTs) catalyze the transfer of a carbohydrate from an activated donor sugar onto small molecule acceptors by the formation of a glycosidic bond (Mackenzie et al., 1997; Li et al., 2001). Glycosylation changes the stability and/or solubility of the aglyca, and it may even create a higher diversity due to differential and multiple conjugations. These reactions are an important feature of the biosynthesis of many secondary metabolites and in many cases of the regulation of the activity of signaling molecules and defense compounds. They may include detoxification and compartmentation of endogenous compounds and xenobiotics (Jones and Vogt, 2001). One hundred and twenty-two different UGT isoforms exist in Arabidopsis, which represent 0.5% of all annotated genes in this species (Ross et al., 2001; Bowles et al., 2005; Gachon et al., 2005). Analyses of recombinant UGT proteins led to the identification of UGTs with in vitro activity toward several endogenous compounds, like auxin (Jackson et al., 2001), abscisic acid (Lim et al., 2005), flavonoids (Jones et al., 2003), lignin precursors, hydroxybenzoic acids (Lim et al., 2002), and thiohydroximate (Grubb et al., 2004), as well as toward xenobiotics (Messner et al., 2003; Brazier-Hicks and Edwards, 2005). However, these activities could be confirmed in vivo only in a few cases, possibly due to the broad substrate acceptance of some UGT enzymes in vitro or to a limited substrate availability in vivo (Jones and Vogt, 2001; Gachon et al., 2005; Bowles et al., 2006). So far, there is in vivo evidence that flavonoids, SA, indole-3-acetic acid, glucosinolates, and brassinosteroids function as endogenous substrates of UGT enzymes in Arabidopsis (Jackson et al., 2002; Grubb et al., 2004; Poppenberger et al., 2005; Tohge et al., 2005; Dean and Delaney, 2008; Song et al., 2008; Yonekura-Sakakibara et al., 2008). The substrates of the vast majority of UGT isoforms, however, have not been identified, and these isoforms thus remain orphan glycosyltransferases. An approach to gauge the impact of UGTs on plant defense irrespective of the knowledge of their substrates was also undertaken. The expression of various candidate UGT genes was altered (e.g., ugt73b3 and ugt73b5 knockout mutants were generated; UGT74F2 was overexpressed), and a decrease in resistance to pathogen infection indicated a role for these isoforms in defense (Langlois-Meurinne et al., 2005; Song et al., 2008).
In this project, we scanned public expression databases for stress-responsiveness of UGT genes and found that the otherwise uncharacterized UGT76B1 was the member of this family that exhibited the greatest induction in response to stress. UGT76B1 was broadly upregulated by both abiotic and biotic cues. Furthermore, it was one of only three UGT genes (with UGT72B1 and UGT75B1) that was induced by both SA and JA (methyl jasmonate) application. ugt76b1 knockout lines exhibited enhanced resistance toward Pseudomonas syringae infection, yet higher susceptibility toward necrotrophic Alternaria brassicicola, and they progressed earlier into senescence. By contrast, UGT76B1 overexpression resulted in the opposite phenotypes. Using a nontargeted metabolomic approach based on ultra-high-resolution Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS), we could pinpoint isoleucic acid (ILA) as an endogenous substrate of UGT76B1. Exogenously applied ILA itself activated SA-dependent marker gene expression and enhanced resistance toward infection with avirulent P. syringae. Collectively, these findings indicate that UGT76B1 and ILA are novel players in SA- and JA-mediated responses. UGT76B1 acts as a negative regulator of SA-dependent plant defense in the absence of pathogens, promotes the JA response, and negatively influences the onset of senescence.
RESULTS
A Subset of Genes Accounts for the Majority of Stress-Dependent Transcriptional Inductions within the UGT Family
To analyze the distribution of transcriptional responses to exogenous stresses within the Arabidopsis UGT genes, we examined public expression data of plants exposed to several abiotic and biotic stress cues. A total of 112 probe sets on the ATH1 microarray represent 105 UGT genes by gene-specific probes, while seven hybridize to two highly related genes each. Normalized expression data from Columbia (Col) wild-type leaves or seedlings were retrieved from the BAR database (bbc.botany.utoronto.ca; Toufighi et al., 2005). A large group of UGT genes was induced in one or several experiments, but stress responsiveness was not equally distributed across the genes analyzed. In both abiotic and biotic stress experiments, a clear clustering of stress-dependent induction was observed (Figure 1; see Methods). UGT76B1 was the top stress-induced UGT, being highly responsive to abiotic cues such as UV-B irradiation, osmotic, oxidative, drought, or wounding stresses as well as to both biotrophic and necrotrophic pathogens. Since the UGT76B subfamily only contains this unique member, UGT76B1 may have an important and specific role in plant stress responses.
Figure 1.
Stress-Responsive Expression of UGT Genes in Arabidopsis Leaves and Seedlings Based on Affymetrix ATH1 Microarray Data.
The distribution of maximal inductions to abiotic and biotic stress factors among all UGT members is shown. Significant inductions equal or higher than twofold (dark gray) and higher than 1.5-fold (light gray) are indicated. Genes are sorted from highest to lowest abiotic and biotic stress inducibility using mutual ranking (see Methods). The top stress-induced gene, UGT76B1, which was used for further analysis, is indicated with an arrow. A1, osmotic; A2, salt; A3, UV-B; A4, oxidative; A5, wounding; A6, drought; A7, cold; A8, heat; B1, P. syringae pv tomato DC3000 (virulent); B2, P. syringae pv phaseolicula (avirulent); B3, Phytophthora infestans; B4, Botrytis cinerea; B5, P. syringae pv maculicola ES4326 (virulent); B6, Erysiphe orontii.
ugt76b1 Knockout and UGT76B1 Overexpression Lines
To study whether UGT76B1 has any function in plant stress responses and to determine how this might affect the plant, we obtained Arabidopsis loss-of-function mutants and generated constitutive overexpression lines of UGT76B1. Two T-DNA insertion lines, SAIL_1171A11 and GT_5_11976, in two different genetic backgrounds (Col-0 and Landsberg erecta [Ler]) were characterized as ugt76b1-1 and ugt76b1-2 knockout mutants, respectively (see Methods; see Supplemental Figure 1 online). Arabidopsis lines overexpressing UGT76B1 under the control of 35S-derived constitutive promoters were generated, and two homozygous lines with single insertions were selected (see Methods). Both lines, UGT76B1-OE-5 and UGT76B1-OE-7, showed a significantly higher transcript level compared with the wild type (see Supplemental Figure 1 online).
UGT76B1 Expression Affects Onset of Senescence
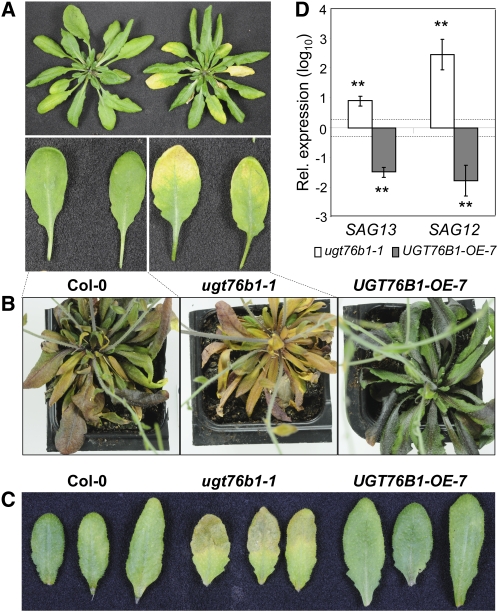
UGT76B1-OE-7 and ugt76b1 knockout lines were examined for morphological or developmental phenotypes associated with a change in UGT76B1 expression. All genotypes germinated at the same time. No obvious morphological differences were found in lines with altered UGT76B1 expression compared with the wild type, except for a tendency for smaller rosettes of the ugt76b1 knockout lines and for enlarged rosettes in the case of UGT76B1-OE-7 (see Supplemental Figure 2 online). However, mutant and overexpression lines showed a clearly altered onset of developmental as well as dark-induced senescence. The knockout plants developed yellowing of leaves 6 weeks after germination, while the wild type did not yet show any signs of senescence (Figure 2A; see Supplemental Figure 3A online). After 9 weeks, ugt76b1-1 was completely senescent, the wild type only started to show the first signs of leaf yellowing, and the overexpression line still showed mostly dark-green leaves (Figure 2B).
Figure 2.
Senescence Phenotypes of ugt76b1 Knockout and UGT76B1 Overexpression Lines.
(A) Natural senescence in 6.5-week-old Col-0 and ugt76b1-1 mutant plants.
(B) Natural senescence in 9-week-old Col-0, ugt76b1-1, and UGT76B1-OE-7 plants.
(C) Dark-induced senescence in Col-0, ugt76b1-1, and UGT76B1-OE-7 plants. Excised leaves from 5-week-old plants were kept in water and darkness for 5 d. The second knockout line, ugt76b1-2, showed the same senescence-related phenotype (see Supplemental Figures 3A and 3B online).
(D) Relative quantification of senescence-associated marker genes SAG13 and SAG12 in 7-week-old wild-type, ugt76b1-1, and UGT76B1-OE-7 plants (leaves 7 to 9). Transcript levels were normalized to UBIQUITIN5 and S16 transcripts; levels relative to Col-0 plants are displayed. Arithmetic means and standard errors from log10-transformed data of two experiments (each based on three independent replicates) were calculated. Asterisks indicate significance of the difference to the wild-type line; **P value < 0.01. The dashed lines indicate a twofold change.
Similar visible differences were found when we analyzed dark-induced senescence in detached leaves (Figure 2C; see Supplemental Figure 3B online).
To confirm that the leaf yellowing of ugt76b1-1 is due to an accelerated onset of developmentally induced senescence, we monitored the expression of two marker genes. SAG13 is induced during early senescence, whereas SAG12 is specifically activated during the later stages of developmentally regulated senescence, when the leaves start to show yellowing (Weaver et al., 1998). Both senescence marker genes SAG12 and SAG13 were more strongly induced in ugt76b1-1 knockout plants than in the wild type, whereas expression was much lower in UGT76B1-OE-7 (Figure 2D).
UGT76B1 Overexpression and Loss of Function Alter Pathogen Susceptibility in an Opposite Manner
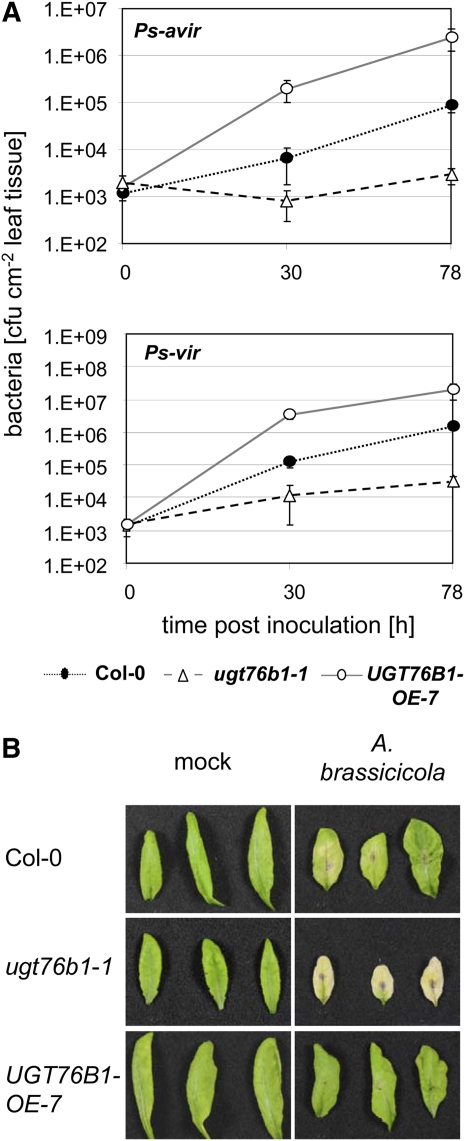
As changing UGT76B1 expression had no influence on plant resistance to abiotic stressors like UV irradiation and salt, we tested whether alterations in UGT76B1 expression would affect the susceptibility of the plant to biotrophic pathogens. Whole leaves of ugt76b1-1, UGT76B1-OE-7, and Col-0 were inoculated with 5 × 105 colony-forming units (cfu) mL−1 avirulent P. syringae strain D3000 AvrRpt2 (Ps-avir). The bacteria showed the typical proliferation of Ps-avir in Col-0 30 and 78 h after inoculation. In the knockout plant, nearly no bacterial growth was observed, pointing to a significantly reduced susceptibility, whereas in the overexpression line, the bacterial population strongly increased, indicating a reduced resistance (Figure 3A). The second knockout line, ugt76b1-2, showed the same pathogen resistance–related phenotype (see Supplemental Figure 3C online). Similar results were obtained with virulent P. syringae DC3000 (Ps-vir, Figure 3). In both cases, UGT76B1 expression negatively correlated with plant resistance to the biotrophic pathogen P. syringae.
Figure 3.
Pathogen Susceptibility Is Affected in Opposite Directions in UGT76B1 Overexpression and Loss-of Function Lines.
(A) Bacterial growth of avirulent (avir) and virulent (vir) P. syringae (Ps) in Arabidopsis leaves of wild-type, ugt76b1-1, and UGT76B1-OE-7 plants. Leaves were infiltrated with an inoculum of 5 × 105 cfu mL−1 of Ps-avir (top graph) and Ps-vir (bottom graph). Bacteria (cfu cm−2) were quantified 30 and 78 h after inoculation. The graphs represent the means and standard deviations of three replicates.
(B) Enhanced/decreased resistance of UGT76B1-OE-7/ugt76b1-1 lines to A. brassicicola. Four-week-old plants were infected with 7.5 × 103 spores (see Methods). Photographs were taken 2 weeks after infection.
The experiments were repeated with similar results.
We also analyzed the susceptibility of mutant and overexpression lines toward the necrotrophic fungus A. brassicicola. Here, exactly the opposite effect was observed: ugt76b1-1 was less resistant to infection, whereas UGT76B1 overexpression led to enhanced resistance (Figure 3B). Thus, UGT76B1 expression directly correlated with resistance against the necrotrophic infection.
Defense Marker Gene Expression Is Constitutively Altered in UGT76B1-OE and ugt76b1 Lines
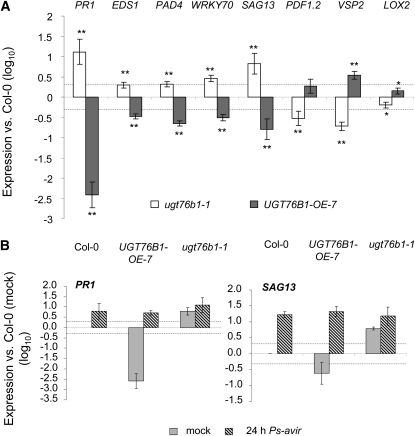
Since the differential effects of UGT76B1 overexpression and loss of function on either biotrophic or necrotrophic pathogen infections pointed to an altered plant defense in these lines, we analyzed several marker genes diagnostic for the antagonistic SA- and JA-dependent pathways using relative quantification by real-time quantitative RT-PCR (qRT-PCR). PAD4 and EDS1 act upstream from SA biosynthesis but are also induced by SA (Rustérucci et al., 2001). PR1 is a pathogen- and SA-responsive gene, which is a well-established marker gene for the defense responses of Arabidopsis against P. syringae (Uknes et al., 1992). SAG13 is an early senescence marker, which is also induced by several stress factors and SA (Weaver et al., 1998). WRKY70 encodes a transcription factor and is an important regulator in the interplay of SA- and JA-related plant defense responses (Li et al., 2004). PDF1.2 and VSP2 are marker genes frequently used to monitor JA responses (Pieterse et al., 2009), whereas LOX2, which is involved in JA biosynthesis, is activated by a positive feedback loop (Sasaki et al., 2001).
Changing UGT76B1 expression had a strong effect on the transcript level of these defense-related genes (Figure 4A). PR1, PAD4, EDS1, WRKY70, and SAG13 were induced in leaves of 5-week-old untreated ugt76b1 knockout plants compared with the wild type. By contrast, JA-responsive genes PDF1.2 and VSP2 as well as LOX2 were downregulated. UGT76B1-OE-7 showed the opposite regulation for all measured genes. PR1, PAD4, EDS1, WRKY70, and SAG13 were downregulated, whereas VSP2 and LOX2 were upregulated. The upregulation of PDF1.2 in UGT76B1-OE-7 was more variable in different experiments. To exclude an age-dependent effect on PR1 expression (Kus et al., 2002), PR1 and SAG13 were analyzed also in 3-week-old plants. Again, both genes showed a similar, opposite regulation in knockout and overexpression lines (see Supplemental Figure 4 online).
Figure 4.
Defense Marker Gene Expression in ugt76b1-1 and UGT76B1-OE-7 Plants before and after Pathogen Infection.
(A) Gene expression of PR1, EDS1, PAD4, WRKY70, SAG13, PDF1.2, VSP2, and LOX2 in 5-week-old ugt76b1-1 and UGT76B1-OE-7 measured by qRT-PCR. Expression levels were normalized to UBIQUITIN5 and S16 transcripts; levels relative to Col-0 plants are displayed. Arithmetic means and standard errors from log10-transformed data of two experiments each consisting of three independent replicates were calculated using ANOVA. Asterisks indicate significance of the difference to the wild-type line; **P value < 0.01 and *P value < 0.05.
(B) Transcript levels of PR1 and SAG13 in 5-week-old wild-type plants 24 h after infection (5 × 105 cfu mL−1 Ps-avir) measured by qRT-PCR. Values are relative to expression 24 h after mock treatment and log10 transformed. Bars represent the mean and standard deviation of three replicates.
The dashed, horizontal lines indicate a twofold change.
To analyze whether the overexpression line was still able to induce SA-dependent defense after P. syringae challenge, although it was compromised by the strong constitutive repression of this pathway, we quantified the transcription of PR1 and SAG13 in wild-type, mutant, and UGT76B1-overexpressing plants after bacterial inoculation. In wild-type plants, PR1 and SAG13 were induced 24 h after infection with Ps-avir to similar levels as those constitutively expressed in the ugt76b1 loss-of-function mutant. In the overexpression line, transcripts of both PR1 and SAG13 reached similar levels as in wild-type plants 24 h after pathogen challenge. Thus, the potential to perceive the pathogen and eventually activate the SA signaling pathway was retained in the UGT76B1 overexpression line.
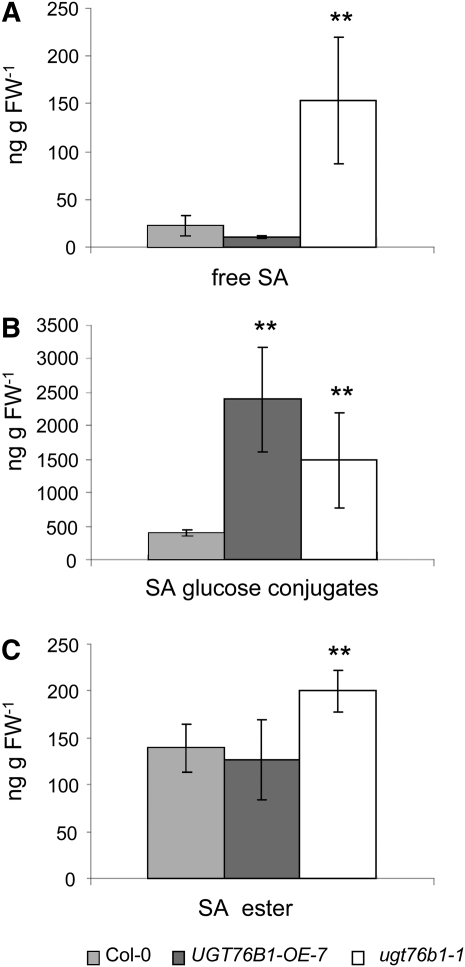
Endogenous Levels of Free and Conjugated SA Are Elevated in ugt76b1
EDS1 and PAD4 are essential regulators of basal resistance and are known to regulate the accumulation of the signaling molecule SA (Zhou et al., 1998; Rustérucci et al., 2001). In addition, several mutants with constitutive transcriptional activation of PR genes are known to have increased levels of SA and its glucosides (Silva et al., 1999; Balagué et al., 2003; Gou et al., 2009). We therefore assessed whether the high level of PR1 expression in ugt76b1-1 plants was correlated with higher endogenous SA levels (Figure 5). Indeed, ugt76b1-1 showed a considerably higher basal level of SA and its glucosides than wild-type plants in the absence of any inducer. By contrast, the overexpression line contained a slightly repressed, yet not significantly altered, amount of free SA compared with wild-type plants but also higher levels of the SA conjugates. The SA ester level did not significantly change in overexpression lines but was slightly increased in the knockout mutant (Figure 5).
Figure 5.
SA and Conjugated SA Levels in 5-Week-Old Seedlings of the Wild Type, ugt76b1-1, and UGT76B1-OE-7.
Values represent the means and standard deviations obtained from five replicates. Asterisks indicate significance of the difference to the wild-type line; **P value < 0.01. The experiment was repeated with similar results. Free SA (A), SA Glc conjugates (B), and SA ester (C). FW, fresh weight.
Dependence of UGT76B1-Related Responses on SA and JA Pathways
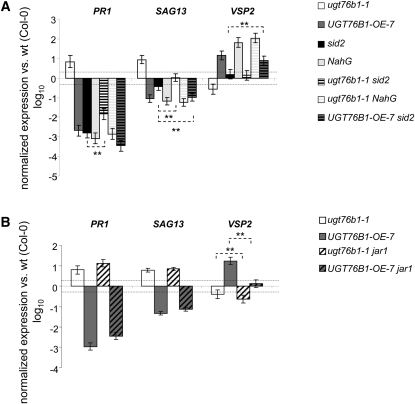
UGT76B1 overexpression and loss of function antagonistically affected SA- and JA-dependent responses. To analyze how these responses were integrated into the corresponding pathways, ugt76b1-1 and UGT76B1-OE-7 were introgressed in sid2 and NahG as well as in jar1 lines. SID2 is responsible for stress-induced SA biosynthesis; consequently, its loss of function leads to an impaired SA-dependent defense (Nawrath and Métraux, 1999). The NahG line, by contrast, leads to an almost complete loss of SA, including basal levels, due to the hydrolytic activity of the bacterial transgene NahG (Gaffney et al., 1993). On the other hand, loss of JAR1 blocks the JA pathway at the formation of the bioactive JA-Ile conjugate (Staswick and Tiryaki, 2004).
The ugt76b1-dependent induction of the SA marker genes PR1 and SAG13 was reverted (or at least eliminated) after introgression into the sid2 and NahG backgrounds, indicating its dependence on SA in both cases (Figure 6A). However, PR1, and, to a lesser extent, SAG13 were still significantly increased in ugt76b1 sid2 compared with sid2, similar to their induction in ugt76b1-1 compared with the wild type. Thus, the ugt76b1-dependent activation was at least partially functional in the sid2 background, which still contains basal SA levels. However, the activation completely relied on total SA levels, since in ugt76b1 NahG plants, PR1 and SAG13 were fully suppressed like in the NahG line alone (Figure 6A). The early senescence phenotype as well as the reduced rosette size of ugt76b1-1 was completely abolished by sid2, in agreement with the expression of the marker gene SAG13, which was identical for the wild type and ugt76b1 sid2. Lack of JAR1 did not influence these phenotypes (Figure 6B; see Supplemental Figure 5 online).
Figure 6.
SA and JA Marker Gene Expression in UGT76B1 Overexpression and Knockout Lines after Introgression into sid2, NahG, and jar1.
(A) Gene expression in sid2 and NahG introgressed lines. Wt, wild type.
(B) Gene expression in jar1 introgressed lines.
PR1, SAG13, and VSP2 transcripts were quantified in 4-week-old ugt76b1-1, UGT76B1-OE-7, and the crossed lines by qRT-PCR. Expression levels were normalized to UBIQUITIN5 and S16 transcripts; levels relative to Col-0 plants are displayed. Arithmetic means and standard errors from log10-transformed data of at least four independent replicates from two separate experiments were calculated using ANOVA. Asterisks indicate significance of the difference between the two bars indicated by the ends of the dotted line; **P value < 0.01.
The ugt76b1-dependent suppression of the JA marker VSP2 was fully dependent on both SID2-related and total SA levels, since in both ugt76b1 sid2 and ugt76b1 NahG the expression of VSP2 was reverted to the level of the sid2 and NahG lines, respectively (Figure 6A).
The combination of UGT76B1-OE-7 and sid2 indicated that the transcriptional effects of the ectopic expression of UGT76B1 did not rely on the suppression of SID2. VSP2 induction in relation to the wild type was basically unchanged with and without introgression of sid2 and also significant in UGT76B1-OE sid2 with respect to sid2 alone. The UGT76B1-OE–dependent suppression of PR1 and SAG13 was also retained in the sid2 background (Figure 6A).
The induction and repression of PR1 and SAG13 in ugt76b1-1 and UGT76B1-OE-7, respectively, were maintained after crossing both lines with jar1 and were thus independent from the related formation of JA-Ile. On the other hand, VSP2 induction in the overexpression line, which was not affected by sid2 (see above), was abolished by the introgression of jar1 (Figure 6B). Therefore, ectopic UGT76B1 expression acted in a JAR1-dependent, but SID2-independent, manner to activate the JA pathway. Nevertheless, UGT76B1 overexpression was able to revert the reduced growth phenotype of jar1 plants (see Supplemental Figure 5 online).
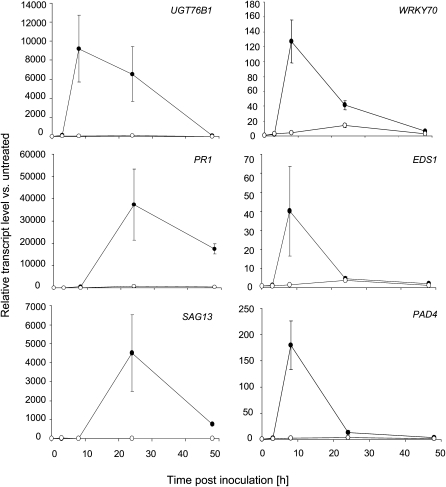
UGT76B1 Is Induced Early after Pathogen Infection before PR1
To determine at which time point after pathogen infection UGT76B1 transcription was activated, we analyzed the kinetics of UGT76B1 expression after pathogen infection compared with other defense marker genes known to be induced at early or late phases during the defense response. Figure 7 shows the time course of UGT76B1, SAG13, WRKY70, EDS1, PAD4, and PR1 expression during the incompatible interaction of wild-type plants with Ps-avir. PR1 and SAG13 were highly induced 24 h after pathogen inoculation. UGT76B1 as well as WRKY70, EDS1, and PAD4 preceded the upregulation of PR1 and SAG13.
Figure 7.
qRT-PCR Expression Profiles of UGT76B1, WRKY70, EDS1, and PAD4 Induction after Infection with Avirulent P. syringae.
Transcript levels were quantified at the indicated time points after inoculation with Ps-avir (closed circles) and mock (10 mM MgCl2; open circles) treatment. The transcript level (relative expression) was normalized to the transcript abundance of UBIQUITIN5 and S16 genes. Values correspond to the mean and standard deviation of triplicates. The experiment was repeated with similar results.
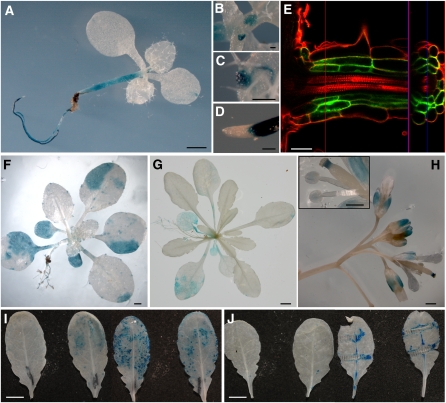
Spatial Expression Pattern of UGT76B1
To analyze the expression of UGT76B1 in different plant organs and at different developmental stages, transgenic lines carrying a UGT76B1pro:GFP-GUS (for green fluorescent protein and β-glucuronidase) construct were produced (see Methods). Plants in different developmental stages (8, 17, 28, and 36 d) showed consistent GUS activity among two independent transgenic lines. UGT76B1 was expressed throughout the roots, except in root tips. Stronger expression was found in young roots and in lateral roots (Figures 8A and 8D). Confocal laser scanning microscopy of lateral roots from the same promoter:GFP-GUS lines revealed that UGT76B1 was mainly expressed in the root cortex and endodermis (Figure 8E). GUS staining of aerial plant parts showed UGT76B1 expression in very young leaves (Figure 8B), hydathodes (Figures 8C and 8F), sepals, and style (Figure 8H). Expression in mature leaves of young plants (17 d) was patchy (Figure 8F). In 4-week-old plants, expression in leaves was reduced (Figure 8G). GUS staining also showed induction of UGT76B1 expression after P. syringae inoculation and wounding (Figures 8I and 8J).
Figure 8.
Localization of UGT76B1 Expression Using UGT76B1pro:GUS-GFP Lines.
Transgenic plants harboring UGT76B1pro:GUS-GFP constructs were stained for GUS activity in different developmental stages ([A] to [D] and [F] to [J]) or examined for GFP fluorescence by confocal microscopy (E) (see Methods). Results were consistent among at least two independent transgenic lines.
(A) to (D) Eight-day-old seedling (A) with leaf primordia (B), leaf hydatodes (C), and root tip (D).
(E) Lateral root of a 1-week-old seedling grown on an agar plate. Cell walls were counterstained with propidium iodide. The red and blue lines indicate the positions where the vertical (right part) and longitudinal (left part) optical cross sections were taken, respectively. Vertical and longitudinal projections are separated by the purple line.
(F) A 17-d-old plant.
(G) A 28-d-old plant.
(H) Inflorescence of a 36-d-old plant. The inset shows a magnification of the stigma and anthers.
(I) Two leaves of 5-week-old plants 8 h after mock treatment (left) or after inoculation with Ps-avir (right).
(J) Two leaves before (left) or 6 h after (right) mechanical wounding using a forceps.
Bars = 1 mm in (A) and (F), 0.1 mm in (B) to (D), 30 μm in (E), 0.5 cm in (G), (I), and (J), and 0.5 mm in (H).
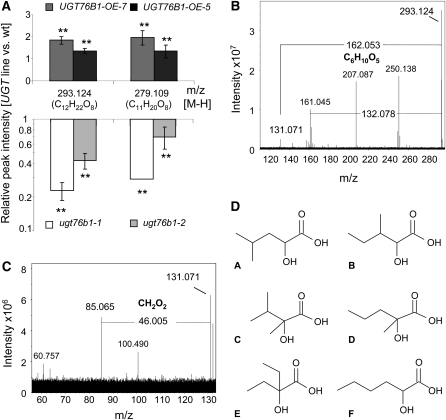
Nontargeted Metabolome Analysis Reveals Correlation between ILA Hexoside Formation and UGT76B1 Expression
Since there was neither an indication of the UGT76B1 substrate nor of the affected metabolic pathway, we embarked on a completely nontargeted strategy to obtain this information. An ultra-high-resolution 12 Tesla FT-ICR mass spectrometer run in the negative ionization mode was employed to compare the metabolic profile of UGT76B1-OE and ugt76b1 mutants with their respective wild type. Root material from plants grown in hydroponic culture was used as starting material for metabolite extraction because UGT76B1 was mainly expressed in roots and showed only lower expression in leaves under unstressed conditions. A stringent, combinatorial screening for metabolite changes was performed across the two independent knockout lines in two different wild-type backgrounds and both independent overexpression lines. By setting a P value cutoff smaller than 0.01 and by filtering for metabolites that showed consistent and opposite regulation in knockout and overexpression plants, only two metabolites were found whose accumulation was significantly and positively correlated with UGT76B1 expression. Both mass-to-charge ratio (m/z) peaks were repressed in the knockout and induced in the overexpression lines (Figure 9A; see Methods). In addition, both peaks were significantly enhanced compared with the wild type in leaf material of the UGT76B1 overexpression lines, although with an overall lower intensity than in roots (see Supplemental Figure 6 online).
Figure 9.
Nontargeted Metabolome Analysis of UGT76B1 Overexpression and ugt76b1 Knockout Lines.
(A) Metabolic changes found in roots of two independent knockout lines and two independent overexpression lines compared with the respective wild type (wt). Means and standard deviation of three independent biological replicates with two technical replicates each are displayed. m/z 279 was nearly undetectable and undetectable in ugt76b1-2 and ugt76b1-1, respectively. Therefore, a default value for the ugt76b1-1 peak was used for calculating the relative intensity (see Methods). Asterisks indicate significance of the difference to the wild type; **P value < 0.01. The predicted molecular formulae are indicated. The experiment was independently repeated with similar results.
(B) Fragmentation pattern of m/z 293. The loss of m/z 162 confirmed the presence of a hexosidic moiety. Other major peaks at m/z 207 and 250 could be unequivocally excluded as m/z 293–derived fragments; they were originating from electrical noise and from an N-containing contaminant, respectively. By contrast, m/z 161 was in agreement with a radical anion of deprotonated hexose, which was directly produced from m/z 293.
(C) Further in-cell fragmentation led to the elimination of CH2O2 (formic acid), which restricted the nature of the aglycon to α-hydroxy carboxylic acid isomers.
(D) Six possible isomeric molecular structures of the aglycon C6H12O3.
Due to the high accuracy in m/z determination, an exact molecular formula could be assigned to both peaks. Fragmentation studies further indicated that the molecule with m/z 293 was a glucoside (Figure 9B). No hexoside loss could be observed upon fragmentation of the second peak (m/z 279). Loss of the hexosidic moiety from m/z 293 led to a smaller compound with m/z 131. The molecular formula of this residual aglycon was C6H12O3. Further in-cell fragmentation of this molecule led to the loss of a formic acid (CH2O2) moiety and the formation of a second fragment, m/z 85. According to a previous study, this behavior indicated that the aglycon of m/z 293 was an α-hydroxy carboxylic acid with a free β-hydrogen (Bandu et al., 2006). Thus, six possible structures could be suggested for the aglycon m/z 131 (Figure 9C). Structures A, C, D, and F could be excluded because the fragmentation of the corresponding standard compounds gave rise to further fragments, which were not detected after fragmentation of the unknown aglycon from the plant extract (m/z 131) (see Supplemental Figure 7 online). Both compounds B and E gave the same fragmentation pattern as the unknown plant peak and therefore constituted possible candidate structures of the aglycon.
In Vitro Activity of Recombinant UGT76B1 toward ILA
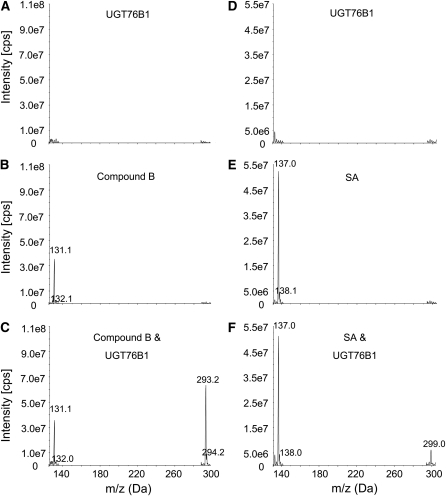
To further elucidate the structure of m/z 131, compounds B and E were tested as potential substrates of recombinant UGT76B1 in vitro. UGT76B1 glucosylated ILA (compound B, 2-hydroxy-3-methylpentanoic acid), whereas it showed no activity toward 2-ethyl-2-hydroxybutyric acid (compound E) (Figures 10A to 10C; see Supplemental Figure 8 online). Thus, ILA turned out to be a substrate of UGT76B1 in vitro, which was in accordance with the observation of plant extracts derived from ugt76b1 knockout and UGT76B1-OE lines. As we had found high levels of SA conjugates in the UGT76B1 overexpression line, we also tested the activity of the recombinant protein toward SA. We could indeed observe the formation of SA Glc conjugate(s), although only a minor peak compared with the substrate SA was detected (Figures 10D to 10F).
Figure 10.
In Vitro Activity Assay of UGT76B1.
Activity of recombinant UGT76B1 was tested with ILA (2-hydroxy-3-methylpentanoic acid, compound B) ([A] to [C]) and SA ([D] to [F]). The reactions were analyzed by mass spectrometry (see Methods). The m/z values of the corresponding substrates and products are indicated. The experiment was independently repeated with similar results.
(A) and (D) Mass spectra of enzyme reactions without substrate.
(B) and (E) Mass spectra of enzyme reactions without enzyme.
(C) and (F) Mass spectra of complete reactions.
The Direct Effect of ILA on Root Growth and Defense Mechanisms
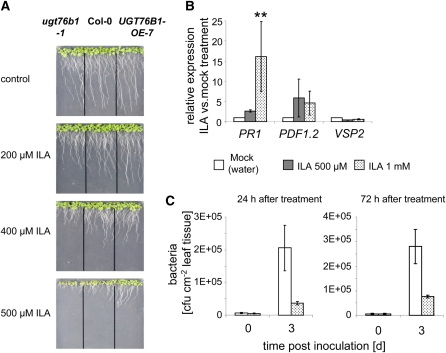
The identification of ILA as a substrate of UGT76B1 raised the question of whether ILA itself was an active compound in planta. Indeed, exogenously applied ILA strongly affects plant root growth and defense marker gene expression. First, ILA inhibits root growth in a concentration-dependent manner (Figure 11A). This repressive effect of ILA on root growth was more pronounced in ugt76b1-1 than in the wild type, but on the other hand was ameliorated by the overexpression of UGT76B1, suggesting that the ILA aglycon was the active agent and glucosylation antagonized its effect. Furthermore, it was crucial to test whether ILA could affect plant defense pathways. Twenty-four hours after spraying an ILA solution onto leaves of 4-week-old plants, PR1 expression was >10-fold induced, indicating a direct and positive influence on the SA pathway. By contrast, the JA marker genes VSP2 and PDF1.2 were not significantly influenced; while VSP2 was only marginally suppressed, PDF1.2 showed a tendency for induction but was highly variable as well (Figure 11B). In addition, ILA could also affect the strong PR1 repression found in UGT76B1-OE-7, which was partially reverted (see Supplemental Figure 9 online). Since ILA induced the defense marker gene PR1, we were interested in determining whether this translated into an enhanced resistance toward P. syringae. Indeed, plants that had been treated with ILA before infection by Ps-avir showed about four- to fivefold less bacterial growth compared with mock treatment (Figure 11C). The increased resistance was persistent for at least 1 to 3 d after ILA spraying. Optimization of the spraying regime and/or the use of surfactants might further enhance the protective effect.
Figure 11.
The Direct Effect of Exogenously Applied ILA.
(A) Root growth inhibition phenotype of ILA inversely correlates with UGT76B1 expression. Photographs were taken 10 d after sowing the seeds on plates containing 0, 200, 400, and 500 μM ILA.
(B) Defense marker gene expression in Col-0 plants after ILA treatment. Transcript levels of PR1, PDF1.2, and VSP2 in leaves of 4-week-old Col-0 plants 24 h after ILA or water treatment were quantified by qRT-PCR. Values are relative to the expression 24 h after mock (water) treatment. Graph represents the mean and sd of three biological replicates. **P value < 0.01.
(C) Bacterial growth in Arabidopsis leaves of wild-type plants sprayed with water (white) or 1 mM ILA (dotted) before infection. Plants were inoculated with 5 × 105 cfu mL−1 of Ps-avir 24 or 72 h after treatment, and bacteria (cfu cm−2) were quantified 0 and 3 d after inoculation. The graphs represent the means and sd of three replicates.
[See online article for color version of this figure.]
DISCUSSION
Plant secondary metabolite glycosyltransferases constitute a large enzyme family. They are presumed to be involved in the biosynthesis, homeostasis, and regulation of the activity of numerous small molecule compounds in plants. However, enzyme-substrate relations and physiological roles of individual isoforms remain mostly obscure. To extend the knowledge of UGTs, we used publicly available databases to identify stress-induced UGT candidates, which might relate them to plant responses to biotic and abiotic stresses. UGT76B1 was the top-ranking isoform among stress-responsive UGTs (Figure 1), and it is present as a single isoform in its subclass (Ross et al., 2001). Analysis of related Brassicaceae genomes revealed a highly conserved, single-copy homolog (M. Das and G. Haberer, personal communication). These features suggested a unique and important function of UGT76B1 in plant stress responses.
Nontargeted Metabolomics Approach Leads to Identification of UGT76B1 Substrate
Despite major advances in plant biology due to genome annotations and omics approaches, a majority of gene products are still orphan enzymes without specific substrates and physiological roles (Fridman and Pichersky, 2005; Saito et al., 2008; Hanson et al., 2010). Although the annotation of an enzyme, for instance as a UGT, most probably denotes its activity as a transferase of an activated sugar onto small-molecule acceptors, this knowledge does not provide a clue toward its native substrate(s) or of its in vivo function. In the case of UGTs, even sequence homology to already known isoforms does not allow to deduce substrate classes (Vogt and Jones, 2000; Bowles et al., 2006). Nevertheless, integration of metabolite profiling with independent evidence, in particular of transcriptional coexpression and comparative genomics, has strongly assisted the elucidation of metabolic pathways and assignment of enzymatic activities (Hirai et al., 2005; Yonekura-Sakakibara et al., 2008; Matsuda et al., 2009; Ohta et al., 2010). In the case of the broadly stress-inducible UGT76B1 gene, coexpression analyses did not indicate an association with particular pathways, which could hint toward a class of potential substrates. Thus, we aimed to use a nontargeted approach employing ultra-high-resolution FT-ICR MS to obtain information on the affected pathway or substrate without any other prior knowledge. Nontargeted FT-ICR MS data are well suited to identify and differentiate metabolic patterns from distinct situations based on multivariate analyses (Ohta et al., 2010). In contrast with this approach, we did a pairwise comparison of m/z values from crude extracts of UGT76B1 overexpression, ugt76b1 loss of function, and wild-type lines. Only two peaks fulfilled the criteria of being both underrepresented in two independent knockout lines (in different accessions as background) and upregulated in two independent overexpression lines (Figure 9A). Thus, this combinatorial approach allowed us to pinpoint informative molecules from the nontargeted metabolome analyses. Since further fragmentation of these m/z peaks indicated that one of them was a hexoside, it was highly suggestive that it indicated the in planta product of UGT76B1. Eventually, enzymatic tests using the recombinant enzyme proved its ability to glucosylate the predicted aglycon in vitro and thereby established ILA as a substrate of UGT76B1.
UGT76B1 Affects SA-JA Crosstalk and Is Affected by the SA and JA Pathways
SA and JA defense signaling pathways are known to interact in a mostly antagonistic manner (Kloek et al., 2001; Spoel et al., 2003; Koornneef and Pieterse, 2008). Both UGT76B1 overexpression and loss of function led to a disturbed equilibrium between these two pathways, suggesting a role for UGT76B1 in SA-JA crosstalk. The complete loss of UGT76B1 function led to constitutive enhancement of the SA-dependent defense and repression of the JA pathway, whereas UGT76B1 overexpression led to the opposite effects (Figures 4 and 12). Accordingly, knockout plants were more resistant to infection by a biotrophic pathogen but more susceptible to necrotrophic attack, while the opposite was true for the overexpression line (Figure 3). Knowing that UGT76B1 suppresses the SA-dependent pathway, it seems surprising at first glance that its expression was induced after P. syringae infection within the same time frame as the SA-dependent marker genes PAD4, EDS1, and WRKY70 and prior to SAG13 and PR1 (Figure 7). This finding indicates that UGT76B1 might play a role in suppressing the SA response in unchallenged conditions, while being required to attenuate it after pathogen attack. Controlled suppression of defense responses is important to avoid deleterious consequences and significant costs for the plant (see Introduction).
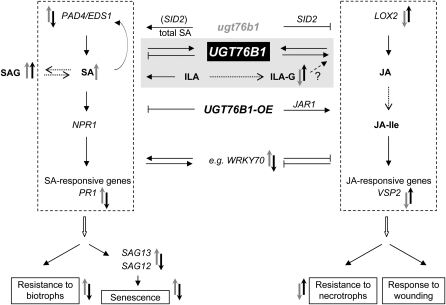
Figure 12.
Model of the Involvement of UGT76B1 as a Novel Mediator in SA- and JA-Dependent Regulation of Defense Responses and Senescence.
The model relates UGT76B1 to SA and JA pathways regulating defense against (hemi-) biotrophic and necrotrophic pathogens. Key steps of both pathways are shown. UGT76B1 induces the JA response and represses the SA-dependent pathway, stimulating defense against necrotrophs and having a negative influence on the resistance to P. syringae and the onset of senescence. The UGT76B1 substrate ILA enhances the SA pathway. The consequences of gain and loss of UGT76B1 function are integrated along with their dependence on SID2 and JAR1. Signaling molecules (SA, SAG, JA, JA-Ile), enzymatic transformations (dashed and open arrows), activation (closed arrows), suppression (line with vertical bar), and important genes are indicated. Positive and negative influences of ugt76b1 (gray) and UGT76B1-OE (black) are shown, respectively.
UGT76B1 was shown to be induced by both SA and methyl jasmonate in microarray expression studies (Zimmermann et al., 2005). Consistent with these findings and its role in promoting the JA pathway, UGT76B1 was induced after wounding (Figure 8J). The constitutive expression of UGT76B1 in hydathodes and young tissues (Figures 8B and 8C) could be involved in the local enhancement of the JA pathway, providing protection against herbivores or necrotrophs at these more vulnerable sites (Hugouvieux et al., 1998; Sprague et al., 2007).
The induction or repression of UGT76B1 in several mutants, in which the SA pathway is affected in unstressed conditions, provided additional evidence that the glucosyltransferase is correlated with this defense pathway (see Supplemental Figure 10 online). Profiling of UGT76B1 expression in Arabidopsis mutants cpr5, mkk1 mkk2, and mpk4 revealed that the gene was highly induced in these plants, which display constitutively enhanced SA-dependent defenses (Bowling et al., 1997; Brodersen et al., 2006; Pitzschke et al., 2009). By contrast, UGT76B1 expression was suppressed in mutants such as eds1, sid2 (eds16), and pad4 (Glazebrook et al., 1996; Feys et al., 2001; Wildermuth et al., 2001), which are impaired in SA-dependent responses (Zimmermann et al., 2005). Many SA-dependent gene regulations are mediated by NPR1 (Vlot et al., 2009). However, the induction of UGT76B1 can at least partially occur in an NPR1-independent manner, since SA could enhance its expression also in the npr1-1 mutant (Blanco et al., 2009). In addition, npr1-1 versus wild type expression analyses indicated an activation of UGT76B1 in the npr1 loss-of-function mutant (see Supplemental Figure 10 online).
There is a considerable overlap between genes involved in defense and senescence signaling. In particular, enhanced SA levels promote senescence-related processes, and some genes involved in the senescence process seem to be directly regulated by SA (Quirino et al., 1999; Morris et al., 2000; Miao and Zentgraf, 2007). Consistently, several of the mutants discussed above that display constitutive expression of SA-related defense responses frequently show an accelerated onset of senescence as well (Yoshida et al., 2002; Barth et al., 2004; Consonni et al., 2006), whereas an SA-deficient NahG line showed delayed developmental senescence (Buchanan-Wollaston et al., 2005). Conversely, plant stress responses are also known to be affected by leaf age (Kus et al., 2002), a phenomenon described as age-related resistance. In the case of ugt76b1, we could exclude age-related resistance (see Supplemental Figure 4 online) and, therefore, further analyzed the link between constitutive induction of SA-dependent defense and the onset of senescence. Both induction of the senescence marker gene SAG13 and early leaf yellowing in ugt76b1-1 were shown to be dependent on SA levels in these plants, since crossing into the sid2 or NahG background reverted these phenotypes (Figure 6; see Supplemental Figure 5 online). On the other hand, PR1 and SAG13 transcript levels in the UGT76B1 overexpression line are comparable to those found in NahG (Figure 6).
Collectively, these data suggest that UGT76B1 is a novel player in the SA-JA crosstalk, acting as a negative modulator and attenuator of the SA response, while it positively affects the JA-dependent pathway (Figure 12). However, the general stress perception and SA/JA signal transduction pathways were preserved and independent of UGT76B1 expression. This was demonstrated by the full inducibility of PR1 after pathogen infection in UGT76B1-OE-7 and by the root growth inhibition upon methyl jasmonate treatment in ugt76b1-1 (Figure 4B; see Supplemental Figure 11 online).
Integration of UGT76B1 in SA-JA Crosstalk
To further examine the integral role of UGT76B1 in SA-JA crosstalk, UGT76B1 knockout and overexpression lines were crossed into mutants compromised in either the SA or JA response. These genetic analyses indicated that both the induction of the SA pathway and the repression of JA responses in ugt76b1 were dependent on SA (Figures 6 and 12). Thus, inhibition of JA-dependent defense in ugt76b1 seemed to be mediated by the known antagonism caused by the enhanced SA level and SA-related signaling in this line. The transcription factor WRKY70 is an important player integrating signals from the antagonistic JA and SA pathways, suggested to be located downstream of NPR1. It acts as a negative regulator of JA-responsive genes and as a positive regulator of SA-induced genes and resistance to P. syringae and might act downstream of the biosynthesis of both hormones (Li et al., 2004; Glazebrook, 2005; Ülker et al., 2007). In this work, enhanced WRKY70 expression in the ugt76B1 knockout was positively correlated with SA signaling but also SA biosynthesis. Therefore, UGT76B1 might overrule the effects of WRKY70 and the latter may only be involved in the SA-mediated suppression of the JA pathway (Figure 12).
Although the pathogen phenotypes and marker gene analyses showed a clearly opposite pattern in ugt76b1 knockout and UGT76B1 overexpression lines, their link to the SA and JA pathways was not simply inverse. In contrast with the SA-mediated ugt76b1 knockout phenotypes, the enhancement of the JA pathway in the UGT76B1 overexpression line was JAR1 dependent and did not rely on the suppression of SID2. Since basal levels of free SA were not significantly repressed in UGT76B1-OE, it is not clear whether the JAR1-dependent enhancement of the JA pathway is related to a suppressive action on the SA pathway (Spoel et al., 2003). Another possibility would be that UGT76B1 directly influences the JA pathway. Such an UGT76B1-dependent stimulation of the JA pathway in the overexpression line could be regulated via enhanced ILA glucoside levels or another UGT76B1-related function, yet both mechanisms would require JAR1 (Figure 12).
The dependence of several aspects of UGT76B1 action on SA and the side activity of the recombinant enzyme toward SA also prompted us to consider UGT76B1 as a SA-conjugating enzyme. Two SA-glucosylating enzymes were described in Arabidopsis. Both UGT74F1 and UGT74F2 were shown to greatly contribute to the formation of SAG and SA Glc ester in vivo, respectively, but the possibility could not be excluded that additional enzymes were involved in SAG biosynthesis (Dean and Delaney, 2008). However, the enhanced levels of free SA and SA glucosides in the ugt76b1 knockout (Figure 5) did not support a role for UGT76B1 as an SA glucosyltransferase, unless the loss of an SA glucosyltransferase would increase free SA and activate other enzymes and therefore lead to even higher glucosylating activity than in the wild type. This scenario, however, is not likely to occur, since neither ugt74f1 nor ugt74f2 single mutants resulted in enhanced glucosylation of exogenously applied SA (Dean and Delaney, 2008). Instead, the enhanced levels of both SA and SAG in ugt76b1 are reminiscent of conditions in other mutants showing constitutive activation of SA-dependent defense, such as cpr20 (Silva et al., 1999), cpr5 (Bowling et al., 1997), and cpr30 (Gou et al., 2009), and of the plant’s normal response to a biotrophic pathogen stimulus.
By contrast, the contribution of the SA-conjugating activity to the UGT76B1 overexpression phenotype cannot be fully excluded. However, the UGT76B1-OE line exhibited the same level of PR1 induction upon P. syringae infection as the wild type (Figure 4B). This is in contrast with the finding for the established SA glucosyltransferase UGT74F2; UGT74F2-OE lines were considerably impaired in their ability to induce PR1 after P. syringae infection, which is in line with UGT74F2’s ability to glucosylate SA and thereby suppress SA-dependent defense (Song et al., 2008).
Our combined data on UGT76B1’s enzymatic activity toward ILA, the correlation of ILA glucoside levels with UGT76B1 expression in vivo, the altered pathogen susceptibility of the ugt76b1 knockout line, as well as the direct effect of exogenous ILA on defense marker gene expression and P. syringae resistance suggest that the endogenous role of UGT76B1 and its relation to the SA and JA pathway is primarily related to its activity on ILA.
Amino Acid–Related ILA and Plant Defense
The direct link between the activity of UGT76B1 toward ILA and SA-dependent defense was established by the finding that exogenously applied ILA enhanced resistance toward PS-avir (Figure 11). PR1 expression was induced to a level similar to the induction found in ugt76b1. Accordingly, the lack of ILA glucosylation in the knockout line could lead to the induction of SA-dependent signaling. On the other hand, UGT76B1 overexpression leads to increased glucosylation of ILA and, therefore, reduced PR1 expression. In line with this, ILA application could partially revert the strong PR1 repression in UGT76B1-OE. These data suggest that the ILA aglycon and not the glucoside is active in enhancing the SA pathway and PR1 expression. ILA was also likely the biologically active compound in another bioassay. Exogenous ILA inhibited root growth, and this effect was more pronounced for ugt76b1 but ameliorated in the UGT76B1-OE line. How ILA is perceived at the molecular level and at which step it activates the SA or other pathways is currently not clear and requires further investigation (see also below). Interestingly, ILA did not significantly reduce the expression of JA marker genes VSP2 and PDF1.2; the latter even showed a tendency for upregulation. Thus, exogenous ILA did not affect the SA and JA pathways in an antagonistic manner, which suggests that it may have biotechnological applications as a plant protective agent. In fact, SA and JA are not exclusively antagonistic, and even synergistic action has been reported for SA and JA dependent on their concentration and timing (Mur et al., 2006). Accordingly, exogenous ILA could interfere with the SA and JA pathways at this level.
Neither ILA itself nor its glucoside had been described before in plants. However, ILA has been characterized in humans as the reduced form of 2-keto-3-methylvaleric acid, a degradation product of the branched-chain amino acid Ile (Mamer and Reimer, 1992; Podebrad et al., 1997). A genetic defect in the further oxidation of this product led to its accumulation along with other degradation products and the amino acids themselves in maple syrup urine disease (Mamer and Reimer, 1992).
A correlation analysis based on microarray data at ATTED-II and VirtualPlant (Obayashi et al., 2009; Katari et al., 2010) provided evidence for a relationship between ILA and amino acid metabolism also in Arabidopsis. UGT76B1 expression correlated with that of LIPOAMIDE DEHYDROGENASE2 (see Supplemental Figure 12 online), a gene encoding a component of the branched-chain keto acid dehydrogenase complex, which catalyzes the oxidative decarboxylation of the α-keto acid derivatives of Val, Leu, or Ile (Binder et al., 2007). The second compound with m/z 279 (C11H20O8) found to be correlated with UGT76B1 expression in our nontargeted metabolomics approach differed from the ILA-glucoside peak (m/z 293, C12H22O8) by one CH2 moiety. This could represent the corresponding glucosylated compound derived from Val metabolism (2-hydroxy-3-methylbutyric acid [valic acid]). Therefore, we evaluated the biological activity of this putative analog of ILA. Indeed, valic acid showed a similar but weaker inhibition of root growth compared with ILA, and recombinant UGT76B1 was able to glucosylate valic acid (see Supplemental Figure 13 online).
Amino acid–derived molecules have also been related to Arabidopsis defense reactions by the involvement of two aminotransferases, ALD1 and AGD2, which supposedly catalyze an amino transfer in opposite directions while acting on an unknown α-keto acid/α-amino acid couple (Song et al., 2004). These authors found that agd2 mutants were more resistant to P. syringae infection, while ald1 showed increased susceptibility. Furthermore, plant hormones are known to be regulated by conjugation with amino acids. In particular, Ile is known to be conjugated to JA to form JA-Ile, the main bioactive form of the hormone (Staswick and Tiryaki, 2004; Fonseca et al., 2009). SA can be also conjugated to amino acids (reviewed in Vlot et al., 2009), and overexpression of GH3.5, an enzyme potentially involved in this conjugation, led to enhanced pathogen resistance and SA accumulation (Park et al., 2007).
Due to the close relationship between ILA and the amino acid Ile, one may also speculate that ILA or its glucoside has an impact on amino acid–related hormone conjugations via a yet unknown mechanism. Thus, future research should focus on the relationship between plant defense pathways, amino acid–derived metabolites, and small-molecule glucosylation.
METHODS
Plant Materials and Growth Conditions
Two T-DNA insertion lines in two different wild-type backgrounds, ugt76b1-1 (SAIL_1171A11 [Col-0]) and ugt76b1-2 (GT_5_11976 [Ler]), were obtained from the Nottingham Arabidopsis Stock Centre (Scholl et al., 2000). The published position of the T-DNA insertion was confirmed by PCR and DNA sequencing using the primers 5′-TTCATAACCAATCTCGATACAC-3′ and 5′-GTCTGATTATGGGAATGCAGATTA-3′ (76B1_R) for the SAIL (Syngenta Arabidopsis Insertion Library) line and primers 5′-CGTTTTGTATATCCCGTTTCCGT-3′ and 5′-AAGATCCAAGATCAGGGGATAAG-3′ (76B1_F) for the GT-5 line, respectively. A 3:1 segregation of the respective resistance (BASTA for SAIL and kanamycin for GT-5) after backcrossing indicated that the mutation was inherited as a single locus in both cases. RT-PCR analysis using the gene-specific primers 76B1_F and 76B1_R confirmed the lack of UGT76B1 transcripts in both lines (see Supplemental Figure 1 online).
UGT76B1 overexpression lines were produced by Agrobacterium tumefaciens–mediated transformation using plasmids carrying the open reading frame coupled to cauliflower mosaic virus 35S-derived promoters in two different vectors, pB2GW7 and pAlligator2 (Clough and Bent, 1998; Karimi et al., 2002; Bensmihen et al., 2004). The following primers were used for UGT76B1 amplification and cloning using GATEWAY (Invitrogen) recombination: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTACACAATGGAGACTAGAGAAACAAAACCA-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTGATTATGGGAATGCAGATTA-3′. After selection of transformants, segregation analysis was used to identify single insertion lines in the T2 generation.
The point mutation lines sid2-1 (Nawrath and Métraux, 1999) and jar1-1 (Berger, 2002) were both crossed with UGT76B1 loss-of-function and overexpression lines. The NahG transgenic line (Gaffney et al., 1993) was crossed with ugt76b1-1. The homozygosity of the resulting lines was confirmed by its JA-resistant root growth phenotype in the case of jar1. Homozygosity of sid2-1 was confirmed by a cleaved amplified polymorphic sequence marker (Konieczny and Ausubel, 1993). PCR fragments were amplified using primers SID2_f, 5′-TGCTTGGCTAGCACAGTTACA-3′, and SID2_r, 5′-AGCTGATCTGATCCCGACT-3′, and digested with MfeI, which discriminated against a single C-to-T nucleotide substitution in sid2-1.
For infection experiments, qRT-PCR analysis, and plant transformation, plants were grown on soil (Floraton 1 or Floragard B fein; Floragard) under a 12- to 14-h light cycle at 45 μmol m−2 s−1 of light intensity at 18°C in the dark and 20°C in the light. For metabolic analysis of root material, plants were grown hydroponically at 120 μmol m−2 s−1 of light intensity. Seeds were surface sterilized and grown on plates with half-strength Murashige and Skoog medium (M5519; Sigma-Aldrich), 1% Suc, and 0.25% Gelrite. Seedlings were transplanted after 7 d in a floating hydroponic system (Battke et al., 2003) and grown for two more weeks. Each Vitro Vent box contained 300 mL liquid medium and 250 mL polypropylene granulate as the floating material.
Chemicals
Compounds A, E, F, and valic acid [(S)-(+)-2-hydroxy-3-methylbutyric acid] were obtained from Sigma-Aldrich and compound B [(2S, 3S)-2-hydroxy-3-methylpentanoic acid] from Interchim. Compounds C and D were not commercially available. Therefore, they were synthesized according to previously described protocols (Yabuuchi and Kusumi, 1999; Caille et al., 2009).
Dark-Induced Senescence
Excised leaves from 5-week-old plants grown under short-day photoperiodic conditions were kept for 5 d in the dark at room temperature (Oh et al., 1996).
Data Mining of Public Expression Data
A complete collection of 122 UGT genes from Arabidopsis thaliana was extracted via the CAZY database (www.cazy.org). No Arabidopsis Genome Initiative locus was associated with the two pseudogenes listed (UGT85A6P and UGT90A3P), and UGT89B1 (At1g73880) was not represented on the ATH1 array. Among the residual 119 probe sets, 105 targeted individual members specifically, whereas seven did not discriminate between two highly homologous isoforms each. Normalized microarray data for all 119 probe sets comprising abiotic and biotic (without elicitors) stressors applied to Col wild-type seedlings were downloaded from the BAR database.
For a number of treatments, two or more time points had been deposited. In cases where both up- and downregulations were recorded, the difference between the total number of significant inductions (>1.5-fold) and repressions (<0.67) was calculated. A specific gene was assigned as “induced” when inductions were present in at least two time points and the number of inductions exceeded repressions in at least two consecutive time points. In cases where the number of inductions equaled repressions, genes were nevertheless assigned as induced if clear induction kinetics had been observed. In cases with only two experimental time points, induction in one instance was sufficient as long as no repression had been observed in the second time point. For final classification, the maximal induction among different time points was selected for each treatment. The total number of significant stress inductions was separately indicated for abiotic and biotic stress cues, and a mutual rank MR [MR = (rank abiotic × rank biotic)0.5] (Obayashi et al., 2009)] for both biotic and abiotic stress inductions was calculated for each UGT isoform to sort genes from highest to lowest combined stress inducibility.
Real-Time qRT-PCR
Rosette leaves of the indicated age were collected. Total RNA was isolated using the RNeasy plant mini kit (Qiagen). RNA integrity and amount were analyzed by gel electrophoresis and spectrophotometry. One microgram of total RNA was reverse transcribed using a SuperScript II RT-PCR Kit (Invitrogen) according to the manufacturer’s instructions.
Gene-specific primer pairs were designed using the Primer Express 3.0 software. Primer pairs are listed in Supplemental Table 1 online. All primer pairs were evaluated for amplification specificity and an efficiency of over 80% using a serial cDNA dilution. Real-time quantification was performed using a 7500 real-time PCR system (Applied Biosystems). Individual PCR reaction mixtures contained 4 μL of diluted cDNA, 10 μL of Sybr Green Mastermix (Thermo Scientific), and 250 μM of each primer in a final volume of 20 μL. In all experiments, three biological replicates of each sample and two technical (PCR) replicates were performed. The amount of target gene was normalized over the abundance of the constitutive UBQ5 and S16 genes. The stability of the reference genes was tested, and normalization was performed using GeNorm (Vandesompele et al., 2002). For qRT-PCR of infected material, plants were infected as described below. Three biological replicates were analyzed, each consisting of six individually infected leaves. Plant material was harvested before infection and mock treatments (time point 0) and at the indicated time points after treatment. Each experiment was repeated with similar results.
For marker gene analysis on uninfected material and senescent leaves, methods for paired or grouped data were applied, namely the paired t test and repeated-measurement analysis of variance (ANOVA; linear mixed-effect models), in order to control for interplate variation (each replicate was measured on a different qPCR plate). Two-way ANOVA was used to join results from two independent analyses. First, a model with interaction was fitted. If the interaction effect was significant, one-way ANOVAs were performed for the single experiments; otherwise a two-way ANOVA without an interaction effect was fitted. All analyses (P value, arithmetic mean) were performed on log10-transformed data as recommended in the literature (Rieu and Powers, 2009). For all calculations, R software with the nlme package was used (Pinheiro et al., 2009; R Development Core Team, 2009).
Pathogen Infections
Bacterial strains used in this study include Pseudomonas syringae pv tomato DC3000 (Ps-vir) and P. syringae pv tomato DC3000 (avrRpm1) (Ps-avir). Bacteria were grown overnight at 28°C in King's B medium with appropriate antibiotics (100 μg/mL rifampicin for Ps-avir and Ps-vir and 50 μg/mL kanamycin for Ps-avir) and diluted to 5 × 105 cfu mL−1 with 10 mM MgCl2 for plant inoculation. Whole leaves of 5- to 6-week-old plants were infiltrated using a 1-mL syringe without a needle. Control plants were infiltrated with 10 mM MgCl2. Leaf discs from control-treated and infected plants were harvested from inoculated leaves at 0, 1, and 3 d after infiltration. Bacterial growth was assessed as described previously (Katagiri et al., 2002). For each time point, three samples were made by pooling six leaf discs from three different treated plants.
Alternaria brassicicola strain CBS 125088 (Pogány et al., 2009) spores were used for fungal infections. Conidial suspensions (1.5 × 106 conidia mL−1 of distilled water) for the inoculation were prepared from 10- to 15-d-old A. brassicicola cultures grown on malt extract agar medium (15 g of malt extract and 7.5 g of agar per L; Merck) at 22°C with a 12-h-light/12-h-dark cycle. A 5-μL droplet of the spore suspension was transferred onto the adaxial surface of leaves (sixth to eleventh leaf). Plants were kept in the container covered with plexiglass before and after inoculation to maintain a high ambient humidity. Distilled water was used for mock treatment. Lesions on the inoculated leaves were recorded 10 to 13 d after inoculation.
Histochemical Localization of Gene Expression
A fragment upstream of the UGT76B1 start codon was amplified from genomic DNA (accession Col-0) by PCR using primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCGGTTAAACATAAACCATGT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTCTCCATTTTTGTTGTGAAT-3′ and introduced into vector pBGWFS7 (Karimi et al., 2002). The resulting UGT76B1pro:GUS-GFP fusion construct was transformed into Col-0 Arabidopsis plants (Clough and Bent, 1998). After selection of transformants, segregation analysis was used to identify lines with single insertions in the T2 generation. Thus, two independent lines, UGT76B1pro:GUS-GFP-2 and UGT76B1pro:GUS-GFP-12, were selected for further analysis.
Histochemical analysis of the GUS reporter gene was performed at different developmental stages according to Lagarde et al. (1996) using 1 mM each of potassium ferro- and ferricyanide. Ethanol (70%) was used for destaining chlorophyll. To gain more detailed information about UGT76B1 expression in roots, the same UGT76B1pro:GUS-GFP lines were analyzed with a confocal laser scanning microscope (LSM 510 Axiovert 100M; Carl Zeiss). For cell wall staining, 1-week-old seedlings grown on vertical agar plates were immersed in 50 μg mL−1 propidium iodide for 30 min, washed twice with double-distilled water, and then observed. Staining was performed on two independent single insertion lines, showing consistent results.
SA Determination
Metabolites of pooled 5-week-old rosette leaves from four to six individual plants (snap-frozen in liquid nitrogen and stored at −80°C) were extracted with a 1+2 mixture of methanol and 2% (v/v) formic acid. The extract was split into three aliquots for separate determination of free SA, SA glucosides, and SA esters. For determination of the SA conjugates, the extract was digested overnight with β-glucosidase (Roth) or with esterase (Sigma-Aldrich). SA from undigested and digested samples was extracted under acidic conditions using reversed-phase sorbent cartridges (Oasis HLB 1 cc; Waters), recovered under basic conditions, and subsequently analyzed via HPLC. Quantification was based on SA fluorescence (excitation 305 nm/emission 400 nm) with o-anisic acid added as an internal standard during metabolite extraction and authentic SA standards. Thus, the content in free SA, in Glc-conjugated SA, and in esterified SA could be determined.
Nontargeted Metabolome Analysis
Three biological replicates and two technical replicates each were used for each genotype. Frozen root tissue was individually disrupted using a dismembrator. Metabolite extraction was performed as described previously (Weckwerth et al., 2004) with slight modifications. Loganin (44 μg mL−1) and nitrophenol (3 μg mL−1) were added to the extraction buffer (methanol/chloroform/water 2.5:1:1 [v/v/v]) as internal standards. After extraction, the aqueous phase was divided in several 200-μL aliquots and dried completely using a Speedvac. For FT-ICR MS analysis, one dried aliquot from each sample was redissolved in 70% methanol and diluted 1:25 in 70% methanol containing 35 pmol mL−1 dialanin.
Ultra-high-resolution mass spectra were acquired on a Bruker APEX Qe Fourier transform ion cyclotron resonance mass spectrometer (Bruker) equipped with a 12-T superconducting magnet and an APOLLO II electrospray ionization source. Measurements were performed in the negative ionization mode. Measurements in the positive ionization mode were also performed. However, in this case, there were no significant changes (at P value < 0.01), which were consistent in two independent knockout or overexpression lines, respectively. Samples were introduced into the electrospray source at a flow rate of 120 μL/h with a nebulizer gas pressure of 20 p.s.i. and a drying gas pressure of 15 p.s.i. (at 200°C). Spectra were externally calibrated based on Arg cluster ions (10 ppm). The spectra were acquired with a time domain of 1 megaword over a mass range between 146 and 2000 atomic mass units. Three hundred scans were accumulated for each spectrum. Internal mass calibration was performed using the internal standards (loganin and dialanin; Sigma-Aldrich) in addition to endogenous plant metabolites with calibration accuracy smaller than 0.01 ppm. Internal standards were also used to detect variation in the extraction procedure, matrix effects, and variation in the ionization efficiency in the electrospray source.
Mass lists were calibrated using the data analysis program (Bruker) and exported to ascii files. Mass list matrices for statistical analysis were produced using a custom-made program (M. Frommberger, Helmholtz Zentrum München). Masses, which were detected in only two or less out of six measurements in both genotypes, were deleted. Pearson correlation analysis (excluding missing values) was used to check extract reproducibility (correlation r2 > 0.9). The sum of total peak intensities was monitored to detect variation in the ionization efficiency (in addition to internal standards). Nondetectable peaks were replaced by 200,000 counts, which were considered as the detection limit, to enable calculation of mean values and ratios. A two-sample Wilcoxon rank-sum test was performed for each mass separately to detect significant peak intensity differences between wild-type and mutant plants (detailed m/z lists are available in Supplemental Data Sets 1 and 2 online). The significance level was set to 1%. Measurements were repeated twice to filter for reproducible metabolites, and only those peaks were selected that correlated with UGT76B1 expression in both knockout and overexpression lines. Statistical analysis was performed in R (R Development Core Team, 2009).
Fragmentation Studies Using FT-ICR MS
For MS/MS fragmentation studies, the plant extract from UGT76B1-OE-7 was partially cleaned and concentrated using a Strata NH2 column (3 mL; Phenomenex). The targeted ions were trapped in a first hexapole for 200 ms prior to their mass selection inside a quadrupole mass filter. Once isolated, the targeted ions were accelerated and allowed to collide with argon atoms inside a second hexapole, which serves as a collision cell. The second hexapole had a relatively high pressure of 5 × 10−3 mbar. As a result of the collisions between the accelerated isolated ions and argon atoms in the second hexapole, product ions were produced and were forwarded to the ICR cell via a couple of accelerating and decelerating lenses. The ion accumulation time inside the collision cell was 500 ms.
For targeted ions with m/z < 200 amu, no quadrupole MS/MS fragmentation was done. Instead, the ions were forwarded as normal to the ICR cell and then they were isolated inside the cell by applying a frequency sweep to eject all ions but those that should be selected for further fragmentation events. Once isolated inside the ICR cell, the targeted ions could be excited in the radial plane, which was perpendicular to the magnetic field lines, by applying an on-resonance radial single-shot excitation pulse with a duration of 400 μs and a power of 4.5 Vp-p. A pulsed valve opened at the same time for 5 ms to inject argon atoms inside the ICR cell for collision-induced dissociation experiments. The produced fragment ions were then allowed to thermalize inside the cell before accelerating them in the radial plane for detection.
Recombinant UGT76B1 and Glucosyltransferase Assay
A glutathione S-transferase–UGT761 expression plasmid was constructed using pDEST15 (Invitrogen). The UGT76B1 open reading frame was amplified with the same primers as used for the construction of the overexpression lines. The recombinant protein was affinity purified using glutathione-coupled sepharose beads, according to the manufacturer’s instructions (GE Healthcare), concentrated by membrane filtration (Amicon Ultra-4; Millipore) and supplemented with 20% glycerol for storage at −20°C (Messner et al., 2003).
To analyze the UGT enzyme activity assay, mixtures contained 0.1 M Tris–HCl, pH 7.5, 5 mM UDP-Glc, 0.5 mM aglycone, and ~1 μg fusion protein in a final volume of 50 μL. After incubation for 1 h at 30°C, the reaction was stopped by the addition of 200 μL methanol and cleared by centrifugation. Reactions were diluted 1:50 in 70% methanol (except for valic acid; see Supplemental Figure 13 online) and analyzed on an API4000 mass spectrometer using direct injection into the electrospray source at a flow rate of 30 μL. A total of 150 scans were accumulated for each measurement in the dual ion monitoring mode, which was adjusted to monitor ions at the nominal m/z ratios of the corresponding expected substrate and product peaks with a mass range of ±5 D.
ILA Treatment
For ILA treatment, 4-week-old plants were sprayed with 0.5 or 1 mM ILA (diluted in water) or only water for mock treatments. Plants were covered with a plexiglass lid until the surface of the leaves became dry. The fifth to eighth true leaves of each plant were harvested 24 h after treatment. Four leaves from three independent plants were pooled for each replicate and analyzed by real-time PCR.
Accession Numbers
Sequence data related to the genes described and analyzed in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: UGT76B1 (At3g11340), PR1 (At2g14610), PDF1.2 (At5g44420), VSP2 (At5g24770), SAG13 (At2g29350), SAG12 (At5g45890), LOX2 (At3g45140), WRKY70 (At3g56400), EDS1 (At3g48090), PAD4 (At3g52430), JAR1 (At2g46370), SID2 (At1g74710), UBQ5 (At3g62250), and S16 (At5g18380/At2g09990).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Molecular Characterization of ugt76b1 Knockout and UGT76B1 Overexpression Lines.
Supplemental Figure 2. Growth Phenotype of ugt76b1 Knockout Mutants and the UGT76B1-OE-7 Overexpression Line.
Supplemental Figure 3. Early Senescence and Pathogen Resistance of ugt76b1-2 Compared with Ler (Wild-Type Background).
Supplemental Figure 4. Relative Quantification of PR1 Expression at an Early Time Point.
Supplemental Figure 5. The Impact of UGT76B1 Expression on the Onset of Senescence Is Dependent on SID2 but Independent on JAR1.
Supplemental Figure 6. Detection of m/z 293.124 and 279.108 in Leaves of Col-0 and UGT76B1-OE-7 Plants.
Supplemental Figure 7. Fragmentation Patterns of the Unknown Aglycon (Derived from m/z 293) from the Plant Extract and of Putative C6H12O3 Isomers.
Supplemental Figure 8. In Vitro Activity Assay of UGT76B1 toward 2-Ethyl-2-Hydroxybutyric Acid.
Supplemental Figure 9. Isoleucic Acid Treatment in ugt76b1-1 and UGT76B1-OE-7.
Supplemental Figure 10. UGT76B1 Expression in Several Mutant Backgrounds.
Supplemental Figure 11. ugt76b1-1 Knockout and UGT76B1-OE-7 Plants Showed Inhibition of Root Growth on MeJA-Containing Medium Similar to Wild-Type Plants.
Supplemental Figure 12. Gene Network Suggesting a Relationship between UGT76B1 and Branched-Chain Amino Acid Degradation Pathways.
Supplemental Figure 13. Valic Acid, a Putative Substrate of UGT76B1 in Addition to ILA.
Supplemental Table 1. Primer Pairs Used for qRT-PCR Analysis.
Supplemental Data Set 1. Nontargeted Metabolome Analyses of ugt76b1 Knockout Lines versus Their Respective Wild Type.
Supplemental Data Set 2. Nontargeted Metabolome Analyses of UGT76B1 Overexpression Lines versus Col-0.
Acknowledgments
We thank A. Corina Vlot, Jörg Durner, and Werner Heller (Helmholtz Zentrum München) for valuable advice and discussion. Paul Knochel and Vladimir Malakhov (Ludwig-Maximilians Universität München) synthesized and generously provided 2-hydroxy-2,3-dimethyl-butanoic acid (compound C). Cornelia Prehn (Helmholtz Zentrum München) provided access to an API4000 Q-Trap mass spectrometer. Lucia Gößl (Helmholtz Zentrum München) helped with the SA measurements. V.v.S.P. was supported by a Kekulé Fellowship of the Verband der Chemischen Industrie and W.Z. by a fellowship of the China Scholarship Council. Finally, we thank the three reviewers for their helpful comments, which improved this manuscript.
AUTHOR CONTRIBUTIONS
V.v.S.P. and A.R.S. designed the research. W.Z. contributed to the design of research (Alternaria infection, ILA action, and double mutant analyses). V.v.S.P., W.Z., B.G., and B.K. performed the research. B.K. and P.S.-K. contributed analytical tools (FT-ICR MS). V.v.S.P., W.Z., A.R.S., and T.F.-K. analyzed the data. V.v.S.P. and A.R.S. wrote the article with assistance from W.Z.
References
- Balagué C., Lin B., Alcon C., Flottes G., Malmström S., Köhler C., Neuhaus G., Pelletier G., Gaymard F., Roby D. (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandu M.L., Grubbs T., Kater M., Desaire H. (2006). Collision induced dissociation of alpha hydroxy acids: Evidence of an ion-neutral complex intermediate. Int. J. Mass Spectrom. 251: 40–46 [Google Scholar]
- Barth C., Moeder W., Klessig D.F., Conklin P.L. (2004). The timing of senescence and response to pathogens is altered in the ascorbate-deficient Arabidopsis mutant vitamin c-1. Plant Physiol. 134: 1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battke F., Schramel P., Ernst D. (2003). A novel method for in vitro culture of plants: Cultivation of barley in a floating hydroponic system. Plant Mol. Biol. Rep. 21: 405–409 [Google Scholar]
- Bensmihen S., To A., Lambert G., Kroj T., Giraudat J., Parcy F. (2004). Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561: 127–131 [DOI] [PubMed] [Google Scholar]
- Berger S. (2002). Jasmonate-related mutants of Arabidopsis as tools for studying stress signaling. Planta 214: 497–504 [DOI] [PubMed] [Google Scholar]
- Binder S., Knill T., Schuster J. (2007). Branched-chain amino acid metabolism in higher plants. Physiol. Plant. 129: 68–78 [Google Scholar]
- Blanco F., Salinas P., Cecchini N.M., Jordana X., Van Hummelen P., Alvarez M.E., Holuigue L. (2009). Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 70: 79–102 [DOI] [PubMed] [Google Scholar]
- Bolton M.D. (2009). Primary metabolism and plant defense—Fuel for the fire. Mol. Plant Microbe Interact. 22: 487–497 [DOI] [PubMed] [Google Scholar]
- Bowles D., Isayenkova J., Lim E.K., Poppenberger B. (2005). Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 8: 254–263 [DOI] [PubMed] [Google Scholar]
- Bowles D., Lim E.K., Poppenberger B., Vaistij F.E. (2006). Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 57: 567–597 [DOI] [PubMed] [Google Scholar]
- Bowling S.A., Clarke J.D., Liu Y., Klessig D.F., Dong X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling S.A., Guo A., Cao H., Gordon A.S., Klessig D.F., Dong X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier-Hicks M., Edwards R. (2005). Functional importance of the family 1 glucosyltransferase UGT72B1 in the metabolism of xenobiotics in Arabidopsis thaliana. Plant J. 42: 556–566 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Bjørn Nielsen H., Zhu S., Newman M.A., Shokat K.M., Rietz S., Parker J., Mundy J. (2006). Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47: 532–546 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.F., Wu S.H., Swidzinski J., Ishizaki K., Leaver C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Caille S., Cui S., Hwang T.L., Wang X., Faul M.M. (2009). Two asymmetric syntheses of AMG 221, an inhibitor of 11beta-hydroxysteroid dehydrogenase type 1. J. Org. Chem. 74: 3833–3842 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Consonni C., Humphry M.E., Hartmann H.A., Livaja M., Durner J., Westphal L., Vogel J., Lipka V., Kemmerling B., Schulze-Lefert P., Somerville S.C., Panstruga R. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38: 716–720 [DOI] [PubMed] [Google Scholar]
- Dean J.V., Delaney S.P. (2008). Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant. 132: 417–425 [DOI] [PubMed] [Google Scholar]
- Devadas S.K., Enyedi A., Raina R. (2002). The Arabidopsis hrl1 mutation reveals novel overlapping roles for salicylic acid, jasmonic acid and ethylene signalling in cell death and defence against pathogens. Plant J. 30: 467–480 [DOI] [PubMed] [Google Scholar]
- Feys B.J., Moisan L.J., Newman M.A., Parker J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-Iso-jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Fridman E., Pichersky E. (2005). Metabolomics, genomics, proteomics, and the identification of enzymes and their substrates and products. Curr. Opin. Plant Biol. 8: 242–248 [DOI] [PubMed] [Google Scholar]
- Gachon C.M.M., Langlois-Meurinne M., Saindrenan P. (2005). Plant secondary metabolism glycosyltransferases: The emerging functional analysis. Trends Plant Sci. 10: 542–549 [DOI] [PubMed] [Google Scholar]
- Gaffney T., Friedrich L., Vernooij B., Negrotto D., Nye G., Uknes S., Ward E., Kessmann H., Ryals J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Genger R.K., Jurkowski G.I., McDowell J.M., Lu H., Jung H.W., Greenberg J.T., Bent A.F. (2008). Signaling pathways that regulate the enhanced disease resistance of Arabidopsis “defense, no death” mutants. Mol. Plant Microbe Interact. 21: 1285–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Rogers E.E., Ausubel F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M., Su N., Zheng J., Huai J., Wu G., Zhao J., He J., Tang D., Yang S., Wang G. (2009). An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 60: 757–770 [DOI] [PubMed] [Google Scholar]
- Greenberg J.T., Guo A., Klessig D.F., Ausubel F.M. (1994). Programmed cell death in plants: A pathogen-triggered response activated coordinately with multiple defense functions. Cell 77: 551–563 [DOI] [PubMed] [Google Scholar]
- Grubb C.D., Zipp B.J., Ludwig-Müller J., Masuno M.N., Molinski T.F., Abel S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40: 893–908 [DOI] [PubMed] [Google Scholar]
- Hanson A.D., Pribat A., Waller J.C., de Crecy-Lagard V. (2010). 'Unknown' proteins and 'orphan' enzymes: The missing half of the engineering parts list--and how to find it. Biochem. J. 425: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel A.J., Clarke J.D., Antonovics J., Dong X.N. (2004). Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 168: 2197–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M., Baldwin I.T. (2002). Fitness costs of induced resistance: Emerging experimental support for a slippery concept. Trends Plant Sci. 7: 61–67 [DOI] [PubMed] [Google Scholar]
- Hirai M.Y., et al. (2005). Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 280: 25590–25595 [DOI] [PubMed] [Google Scholar]
- Hugouvieux V., Barber C.E., Daniels M.J. (1998). Entry of Xanthomonas campestris pv. campestris into hydathodes of Arabidopsis thaliana leaves: A system for studying early infection events in bacterial pathogenesis. Mol. Plant Microbe Interact. 11: 537–543 [DOI] [PubMed] [Google Scholar]
- Jackson R.G., Kowalczyk M., Li Y., Higgins G., Ross J., Sandberg G., Bowles D.J. (2002). Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: Phenotypic characterisation of transgenic lines. Plant J. 32: 573–583 [DOI] [PubMed] [Google Scholar]
- Jackson R.G., Lim E.K., Li Y., Kowalczyk M., Sandberg G., Hoggett J., Ashford D.A., Bowles D.J. (2001). Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 276: 4350–4356 [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jones P., Messner B., Nakajima J., Schäffner A.R., Saito K. (2003). UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 278: 43910–43918 [DOI] [PubMed] [Google Scholar]
- Jones P., Vogt T. (2001). Glycosyltransferases in secondary plant metabolism: Tranquilizers and stimulant controllers. Planta 213: 164–174 [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N., Somssich I.E., Roby D., Kroj T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Katagiri F., Thilmony R., He S. (2002). The Arabidopsis thaliana–Pseudomonas syringae interaction. The Arabidopsis Book 1: e0039, doi/10.1199/tab.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari M.S., Nowicki S.D., Aceituno F.F., Nero D., Kelfer J., Thompson L.P., Cabello J.M., Davidson R.S., Goldberg A.P., Shasha D.E., Coruzzi G.M., Gutiérrez R.A. (2010). VirtualPlant: A software platform to support systems biology research. Plant Physiol. 152: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2008). Jasmonate signaling: Toward an integrated view. Plant Physiol. 146: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloek A.P., Verbsky M.L., Sharma S.B., Schoelz J.E., Vogel J., Klessig D.F., Kunkel B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26: 509–522 [DOI] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Koornneef A., Leon-Reyes A., Ritsema T., Verhage A., Den Otter F.C., Van Loon L.C., Pieterse C.M. (2008). Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147: 1358–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A., Pieterse C.M. (2008). Cross talk in defense signaling. Plant Physiol. 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kus J.V., Zaton K., Sarkar R., Cameron R.K. (2002). Age-related resistance in Arabidopsis is a developmentally regulated defense response to Pseudomonas syringae. Plant Cell 14: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde D., Basset M., Lepetit M., Conejero G., Gaymard F., Astruc S., Grignon C. (1996). Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9: 195–203 [DOI] [PubMed] [Google Scholar]
- Langlois-Meurinne M., Gachon C.M.M., Saindrenan P. (2005). Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol. 139: 1890–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brader G., Palva E.T. (2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang Y., Clarke J.D., Li Y., Dong X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98: 329–339 [DOI] [PubMed] [Google Scholar]
- Li Y., Baldauf S., Lim E.K., Bowles D.J. (2001). Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 276: 4338–4343 [DOI] [PubMed] [Google Scholar]
- Lim E.K., Doucet C.J., Hou B., Jackson R.G., Abrams S.R., Bowles D.J. (2005). Resolution of (+)-abscisic acid using an Arabidopsis glycosyltransferase. Tetrahedron Asymmetry 16: 143–147 [Google Scholar]
- Lim E.K., Doucet C.J., Li Y., Elias L., Worrall D., Spencer S.P., Ross J., Bowles D.J. (2002). The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J. Biol. Chem. 277: 586–592 [DOI] [PubMed] [Google Scholar]
- Lorrain S., Vailleau F., Balagué C., Roby D. (2003). Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8: 263–271 [DOI] [PubMed] [Google Scholar]
- Mackenzie P.I., et al. (1997). The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7: 255–269 [DOI] [PubMed] [Google Scholar]
- Mamer O.A., Reimer M.L. (1992). On the mechanisms of the formation of L-alloisoleucine and the 2-hydroxy-3-methylvaleric acid stereoisomers from L-isoleucine in maple syrup urine disease patients and in normal humans. J. Biol. Chem. 267: 22141–22147 [PubMed] [Google Scholar]
- Matsuda F., Yonekura-Sakakibara K., Niida R., Kuromori T., Shinozaki K., Saito K. (2009). MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 57: 555–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner B., Thulke O., Schäffner A.R. (2003). Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta 217: 138–146 [DOI] [PubMed] [Google Scholar]
- Miao Y., Zentgraf U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K., MacKerness S.A.H., Page T., John C.F., Murphy A.M., Carr J.P., Buchanan-Wollaston V. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Mur L.A.J., Kenton P., Atzorn R., Miersch O., Wasternack C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Métraux J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37(Database issue): D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.A., Lee S.Y., Chung I.K., Lee C.H., Nam H.G. (1996). A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 30: 739–754 [DOI] [PubMed] [Google Scholar]
- Ohta D., Kanaya S., Suzuki H. (2010). Application of Fourier-transform ion cyclotron resonance mass spectrometry to metabolic profiling and metabolite identification. Curr. Opin. Biotechnol. 21: 35–44 [DOI] [PubMed] [Google Scholar]
- Park J.E., Park J.Y., Kim Y.S., Staswick P.E., Jeon J., Yun J., Kim S.Y., Kim J., Lee Y.H., Park C.M. (2007). GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 282: 10036–10046 [DOI] [PubMed] [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Pieterse C.M., Leon-Reyes A., Van der Ent S., Van Wees S.C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Pinheiro J., Bates D., DebRoy S., Sarkar D., and the R Development Core Team (2009). nlme: Linear and Nonlinear Mixed Effects Models. R package version 31–94 [Google Scholar]
- Pitzschke A., Djamei A., Bitton F., Hirt H. (2009). A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol. Plant 2: 120–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podebrad F., Heil M., Leib S., Geier B., Beck T., Mosandl A., Sewell A.C., Bohles H. (1997). Analytical approach in diagnosis of inherited metabolic diseases: Maple syrup urine disease (MSUD) - Simultaneous analysis of metabolites in urine by enantioselective multidimensional capillary gas chromatography mass spectrometry (enantio-MDGC-MS). J. High Resolut. Chromatogr. 20: 355–362 [Google Scholar]
- Pogány M., von Rad U., Grün S., Dongó A., Pintye A., Simoneau P., Bahnweg G., Kiss L., Barna B., Durner J. (2009). Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol. 151: 1459–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B., Fujioka S., Soeno K., George G.L., Vaistij F.E., Hiranuma S., Seto H., Takatsuto S., Adam G., Yoshida S., Bowles D. (2005). The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA 102: 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino B.F., Normanly J., Amasino R.M. (1999). Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol. Biol. 40: 267–278 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2009). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Rieu I., Powers S.J. (2009). Real-time quantitative RT-PCR: Design, calculations, and statistics. Plant Cell 21: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Li Y., Lim E., Bowles D.J. (2001). Higher plant glycosyltransferases. Genome Biol. 2: REVIEWS3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C., Aviv D.H., Holt B.F., III, Dangl J.L., Parker J.E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13: 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Hirai M.Y., Yonekura-Sakakibara K. (2008). Decoding genes with coexpression networks and metabolomics - ‘majority report by precogs’. Trends Plant Sci. 13: 36–43 [DOI] [PubMed] [Google Scholar]
- Sasaki Y., et al. (2001). Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: Self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 8: 153–161 [DOI] [PubMed] [Google Scholar]
- Scholl R.L., May S.T., Ware D.H. (2000). Seed and molecular resources for Arabidopsis. Plant Physiol. 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J., Kachroo P., Klessig D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11: 191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva H., Yoshioka K., Dooner H.K., Klessig D.F. (1999). Characterization of a new Arabidopsis mutant exhibiting enhanced disease resistance. Mol. Plant Microbe Interact. 12: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Song J.T., Koo Y.J., Seo H.S., Kim M.C., Choi Y.D., Kim J.H. (2008). Overexpression of AtSGT1, an Arabidopsis salicylic acid glucosyltransferase, leads to increased susceptibility to Pseudomonas syringae. Phytochemistry 69: 1128–1134 [DOI] [PubMed] [Google Scholar]
- Song J.T., Lu H., Greenberg J.T. (2004). Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, aberrant growth and death2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Johnson J.S., Dong X. (2007). Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 104: 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague S.J., Watt M., Kirkegaard J.A., Howlett B.J. (2007). Pathways of infection of Brassica napus roots by Leptosphaeria maculans. New Phytol. 176: 211–222 [DOI] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The Botany Array Resource: e-Northerns, expression angling, and promoter analyses. Plant J. 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Uknes S., Mauch-Mani B., Moyer M., Potter S., Williams S., Dincher S., Chandler D., Slusarenko A., Ward E., Ryals J. (1992). Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B., Shahid Mukhtar M., Somssich I.E. (2007). The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226: 125–137 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.A., Klessig D.F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Vogt T., Jones P. (2000). Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 5: 380–386 [DOI] [PubMed] [Google Scholar]
- Weaver L.M., Gan S., Quirino B., Amasino R.M. (1998). A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37: 455–469 [DOI] [PubMed] [Google Scholar]
- Weckwerth W., Wenzel K., Fiehn O. (2004). Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics 4: 78–83 [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Yabuuchi T., Kusumi T. (1999). A convenient one-pot synthesis of quaternary alpha-methoxy- and alpha-hydroxycarboxylic acids. Chem. Pharm. Bull. (Tokyo) 47: 684–686 [Google Scholar]
- Yonekura-Sakakibara K., Tohge T., Matsuda F., Nakabayashi R., Takayama H., Niida R., Watanabe-Takahashi A., Inoue E., Saito K. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Ito M., Nishida I., Watanabe A. (2002). Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Plant J. 29: 427–437 [DOI] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Tsui F., Klessig D.F., Glazebrook J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hennig L., Gruissem W. (2005). Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 10: 407–409 [DOI] [PubMed] [Google Scholar]