This work reports the identification of a cytochrome P450 monooxygenase that is responsible for the biosynthesis of glycyrrhizin, a triterpenoid saponin found in licorice. The results reveal a function of CYP72A subfamily proteins as triterpene-oxidizing enzymes and provide proof of concept for engineering the production of high-value triterpenoid products in yeasts.
Abstract
Glycyrrhizin, a triterpenoid saponin derived from the underground parts of Glycyrrhiza plants (licorice), has several pharmacological activities and is also used worldwide as a natural sweetener. The biosynthesis of glycyrrhizin involves the initial cyclization of 2,3-oxidosqualene to the triterpene skeleton β-amyrin, followed by a series of oxidative reactions at positions C-11 and C-30, and glycosyl transfers to the C-3 hydroxyl group. We previously reported the identification of a cytochrome P450 monooxygenase (P450) gene encoding β-amyrin 11-oxidase (CYP88D6) as the initial P450 gene in glycyrrhizin biosynthesis. In this study, a second relevant P450 (CYP72A154) was identified and shown to be responsible for C-30 oxidation in the glycyrrhizin pathway. CYP72A154 expressed in an engineered yeast strain that endogenously produces 11-oxo-β-amyrin (a possible biosynthetic intermediate between β-amyrin and glycyrrhizin) catalyzed three sequential oxidation steps at C-30 of 11-oxo-β-amyrin supplied in situ to produce glycyrrhetinic acid, a glycyrrhizin aglycone. Furthermore, CYP72A63 of Medicago truncatula, which has high sequence similarity to CYP72A154, was able to catalyze C-30 oxidation of β-amyrin. These results reveal a function of CYP72A subfamily proteins as triterpene-oxidizing enzymes and provide a genetic tool for engineering the production of glycyrrhizin.
INTRODUCTION
Triterpenoid saponins consist of a triterpenoid aglycone and one or more sugar moieties and belong to a class of natural plant products that includes various bioactive compounds found in medicinal plants (Waller and Yamasaki, 1996). The roots and stolons of Glycyrrhiza species (Glycyrrhiza uralensis and Glycyrrhiza glabra, in the family Fabaceae; licorice) also constitute one of the most important crude drugs in the world (Gibson, 1978) and contain a large amount (2 to 8% of dry weight) of glycyrrhizin, an oleanane-type triterpenoid saponin. Glycyrrhizin exhibits a wide range of pharmacological activities (Shibata, 2000; Nassiri Asl and Hosseinzadeh, 2008): anti-inflammatory (Finney and Somers, 1958; Kroes et al., 1997), hepatoprotective (Nose et al., 1994; van Rossum et al., 1999; Jeong et al., 2002), antiulcer (He et al., 2001), antiallergy (Park et al., 2004), and antiviral against various DNA and RNA viruses (Fiore et al., 2008), including human immunodeficiency virus (Ito et al., 1987, 1988) and severe acute respiratory syndrome–associated coronavirus (Cinatl et al., 2003). Glycyrrhizin is also 150 times sweeter than Suc (Kitagawa, 1993). Many forms of licorice are commercially available worldwide as medicinal materials and sweetening agents (Hayashi and Sudo, 2009; Kojoma et al., 2010).
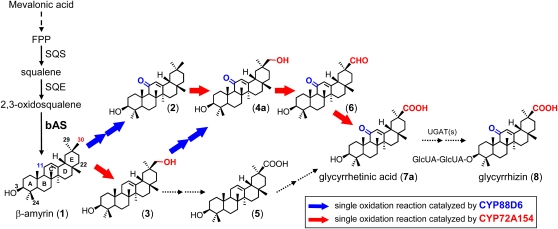
The early stages of triterpenoid saponin biosynthesis involve the dimerization of two farnesyl diphosphate (an intermediate product of the mevalonate pathway) molecules catalyzed by Squalene epoxidase–mediated oxidation then produces 2,3-oxidosqualene, a common substrate of oxidosqualene cyclases (OSCs), as a precursor of both triterpenes and sterols (Abe et al., 1993) (Figure 1). The late stages involve a series of site-specific oxidations of the triterpene skeleton, most probably catalyzed by P450s, followed by glycosylations catalyzed by UDP glycosyltransferases (UGTs). The biosynthesis of glycyrrhizin involves the initial cyclization of 2,3-oxidosqualene by a specific OSC, β-amyrin synthase (bAS), to the triterpene β-amyrin (compound 1 in Figure 1), which is one of the most commonly occurring triterpenes in plants. The subsequent steps involve a series of oxidative reactions at positions C-11 (two-step oxidation) and C-30 (three-step oxidation), followed by glycosyl transfers to the C-3 hydroxyl group (Figure 1). Genes encoding enzymes involved in the early stages of glycyrrhizin biosynthesis, namely, squalene synthase and bAS, have been functionally isolated from G. glabra (Hayashi et al., 1999, 2001). The bAS gene has also been functionally isolated from several other plants, including Arabidopsis thaliana (Shibuya et al., 2009), Avena strigosa (Qi et al., 2004), Lotus japonicus (Sawai et al., 2006), and Medicago truncatula (Suzuki et al., 2002); however, most of the steps in the modification of the β-amyrin skeleton remain uncharacterized at the molecular level. Furthermore, because plant P450s and UGTs are encoded by large multigene families (e.g., a total of 313 putative P450 genes were annotated in the L. japonicus genome; Sato et al., 2008), it is difficult to predict the potential involvement of specific P450s and UGTs in saponin biosynthesis.
Figure 1.
Proposed Pathway for Biosynthesis of Glycyrrhizin.
The structures of possible biosynthetic intermediates between β-amyrin (1) and glycyrrhizin (8) are shown: (2), 11-oxo-β-amyrin; (3), 30-hydroxy-β-amyrin; (4a), 30-hydroxy-11-oxo-β-amyrin; (5), 11-deoxoglycyrrhetinic acid; (6), glycyrrhetaldehyde; and (7a), glycyrrhetinic acid. Solid black arrows indicate a dimerization reaction of two farnesyl diphosphate (FPP) molecules catalyzed by squalene synthase (SQS) originating squalene, oxidation by squalene epoxidase (SQE) to 2,3-oxidosqualene, or cyclization catalyzed by bAS. A dashed arrow between mevalonic acid and farnesyl diphosphate indicates multiple enzyme reactions. The blue arrow indicates a single oxidation reaction catalyzed by the CYP88D6 enzyme (Seki et al., 2008); the red arrow indicates a single oxidation reaction catalyzed by the CYP72A154 enzyme, as described herein; the dotted arrows signify undefined oxidation and glycosylation steps. UGATs, UDP-glucuronosyl transferases.
Recent gene discovery efforts in nonmodel plants using functional genomics-based approaches have successfully identified genes responsible for the production of various plant natural products (reviewed in Yonekura-Sakakibara and Saito, 2009). These include terpenoids with particular importance for human health, such as paclitaxel, a highly effective anticancer drug derived from Taxus species (Jennewein et al., 2001; Schoendorf et al., 2001), and artemisinin, an antimalarial sesquiterpene lactone from Artemisia annua (Ro et al., 2006; Teoh et al., 2006).
As a resource for gene discovery in glycyrrhizin biosynthesis, we generated an EST library from the stolons of G. uralensis; this library comprises 56,857 cDNAs, which have been assembled into 10,474 unique sequences and annotated (Sudo et al., 2009). In our previous work, mining of putative P450 genes from licorice ESTs and subsequent transcript profiling-based selection provided five candidate P450s representing four distinct families (CYP72, CYP83, CYP88, and CYP714); one of these was subsequently identified as a β-amyrin 11-oxidase (CYP88D6), the initial P450 of glycyrrhizin biosynthesis (Seki et al., 2008). CYP88D6 has been shown to catalyze two sequential oxidation steps in the glycyrrhizin pathway: the conversion of β-amyrin (compound 1 in Figure 1) to 11-oxo-β-amyrin (compound 2 in Figure 1) (Seki et al., 2008).
In this study, we retrieved the previously acquired P450 candidates that did not show β-amyrin–oxidizing activity in in vitro enzyme activity assays and tested their activity against three additional substrates located between β-amyrin (1) and glycyrrhetinic acid (7a) in the proposed pathway (Figure 1). This approach successfully identified a second relevant P450 (CYP72A154), which is responsible for C-30 oxidation in the glycyrrhizin pathway. Our results also showed that CYP72A154 is a multiproduct P450 that may have an important role in generating the structural variety of triterpenoid aglycones in licorice. The results obtained in this study reveal a function of CYP72 family proteins as triterpene-oxidizing enzymes and suggest the potential use of yeast cells in the production of high-value triterpenoid products.
RESULTS
In Vitro CYP72A154 Enzyme Activity Assay
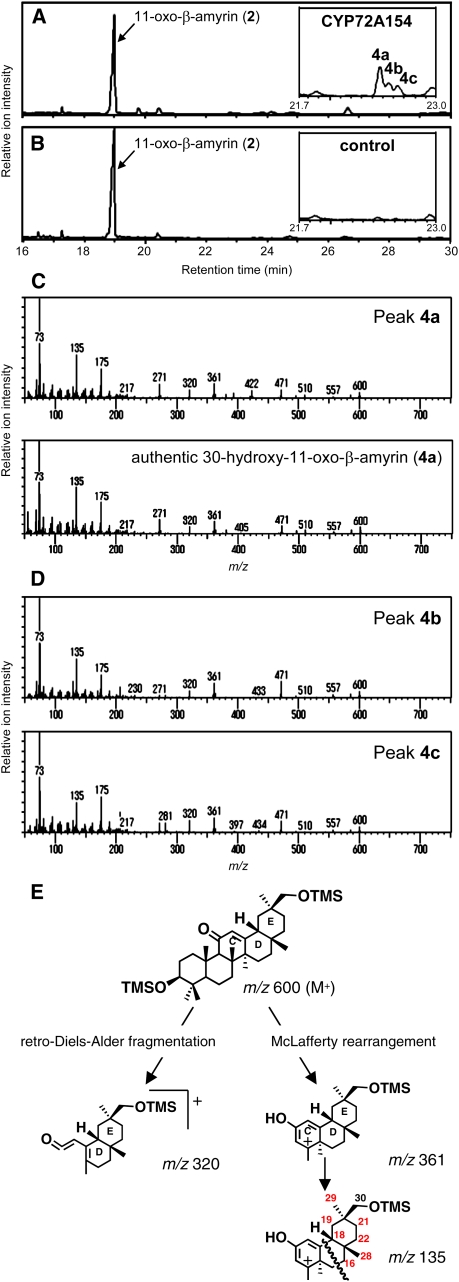
For functional identification, microsomes from Spodoptera frugiperda 9 (Sf9) insect cells expressing each of the candidate P450s were assayed in vitro with the potential substrates (compounds 2, 3, and 4a in Figure 1). The products were analyzed by gas chromatography–mass spectrometry (GC-MS). Of the four candidate P450s, only CYP72A154 (named according to the recommendations of the Cytochrome P450 Nomenclature Committee) showed activity against 11-oxo-β-amyrin (2).
Figure 2A shows the CYP72A154-dependent formation of three GC-MS–detectable compounds (peaks 4a, 4b, and 4c). The mass spectrum of the major product (peak 4a) was an excellent match with that of authentic 30-hydroxy-11-oxo-β-amyrin (4a) (Figure 2C). Both minor products (peaks 4b and 4c; mass spectra shown in Figure 2D) have ion fragmentation patterns similar to that of peak 4a and were identified as isomers of 30-hydroxy-11-oxo-β-amyrin (4a); they are likely to differ in the position of the hydroxyl group introduced by CYP72A154. The mass spectrum of trimethylsilylated 30-hydroxy-11-oxo-β-amyrin (4a) showed fragment ions characteristic of an 11-oxo-olean-12-en derivative at mass-to-charge ratio (m/z) 320, generated by a retro Diels-Alder fragmentation at ring C; at m/z 361, due to a McLafferty rearrangement; and at m/z 135, derived from the fragmentation of m/z 361 (Askam and Bradley, 1971) (Figure 2E). The mass spectra of both isomers (peaks 4b and 4c) also showed abundant fragment ions at m/z 320, 361, and 135, indicating that the introduced hydroxyl group in each isomer is located at one of the available carbon positions (except for C-30; shown in red in Figure 2E) in ring D or E. These peaks were not detected in assays using microsomes from empty vector control Sf9 cells (Figure 2B). These results indicate that CYP72A154 mainly catalyzes the C-30 hydroxylation of 11-oxo-β-amyrin (2) to yield 30-hydroxy-11-oxo-β-amyrin (4a) as the major product, accompanied by its isomers as minor products.
Figure 2.
In Vitro CYP72A154 Enzymatic Activity Assays Containing 11-Oxo-β-Amyrin as Substrate.
(A) and (B) GC-MS analysis (total ion chromatogram [TIC]) of the reaction products resulting from in vitro assays containing 11-oxo-β-amyrin (2) as substrate and microsomal fractions isolated from CYP72A154-expressing Sf9 cells (A) and empty-vector control Sf9 cells (B). Enlargements of the chromatograms corresponding to retention times (Rts) of 21.7 to 23.0 min are shown as insets. GC analyses were performed on an HP-5 column.
(C) The mass spectrum of peak 4a from the GC profile shown in (A) compares well with that of authentic 30-hydroxy-11-oxo-β-amyrin (4a).
(D) Mass spectra of peaks 4b and 4c from the GC profile shown in (A).
(E) Proposed assignment of fragment ions of 30-hydroxy-11-oxo-β-amyrin. The mass spectrum of trimethylsilylated 30-hydroxy-11-oxo-β-amyrin (4a) showed fragment ions characteristic of an 11-oxo-olean-12-en derivative at m/z 320, generated by a retro Diels-Alder fragmentation at ring C; at m/z 361, due to a McLafferty rearrangement; and at m/z 135, derived from the fragmentation of m/z 361.
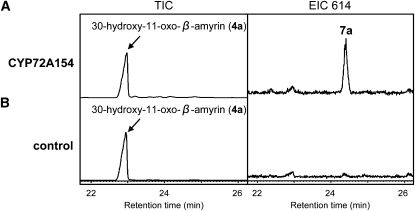
The enzyme activity of CYP72A154 was examined further by testing 30-hydroxy-β-amyrin (3) and 30-hydroxy-11-oxo-β-amyrin (4a) as potential substrates. CYP72A154-containing microsomes exhibited activity against 30-hydroxy-11-oxo-β-amyrin (4a), as indicated in Figure 3. GC-MS analysis showed that the product of the 30-hydroxy-11-oxo-β-amyrin substrate (peak 7a in Figure 3A, EIC 614 panel; mass spectrum shown in Supplemental Figure 1 online) was glycyrrhetinic acid (7a). By contrast, glycyrrhetinic acid was not detected in assays using microsomes from empty vector control Sf9 cells (Figure 3B, EIC 614 panel). These results suggest that CYP72A154 is capable of catalyzing three sequential oxidation steps at C-30 of 11-oxo-β-amyrin (2) to produce glycyrrhetinic acid (7a). No enzymatic activity was found when CYP72A154-containing microsomes were assayed with 30-hydroxy-β-amyrin (3) as the substrate.
Figure 3.
In Vitro CYP72A154 Enzymatic Activity Assays Containing 30-Hydroxy-11-Oxo-β-Amyrin as Substrate.
GC-MS analysis of the reaction products resulting from in vitro assays containing 30-hydroxy-11-oxo-β-amyrin (4a) as substrate and microsomal fractions isolated from CYP72A154-expressing Sf9 cells (A) and empty-vector control Sf9 cells (B). EIC 614, extracted ion chromatograms at m/z 614. Each column of the chromatograms has the same scale. GC analyses were performed on an HP-5 column. The mass spectrum of peak 7a from the GC profile shown in the EIC 614 panel in (A) compared well with that of authentic glycyrrhetinic acid (7a) (see Supplemental Figure 1 online).
In Vivo CYP72A154 Enzyme Activity Assay
To verify the results of our in vitro assays (Figures 2 and 3), we examined the activity of CYP72A154 using an engineered yeast system, which has been proven a successful alternative method for detecting β-amyrin-oxidizing activity of CYP88D6 (Seki et al., 2008). We previously reported the in vivo production of 11-oxo-β-amyrin (2) in an engineered yeast strain cotransformed with bAS, CYP88D6, and Lj CPR1 (CPR, a cytochrome P450 reductase from L. japonicus) as a redox partner for CYP88D6, by redirecting a portion of the native 2,3-oxidosqualene pool from sterol synthesis (Seki et al., 2008). CYP72A154 was introduced into the previously acquired yeast strain endogenously producing 11-oxo-β-amyrin, and the established bAS/CPR/CYP88D6/CYP72A154-transformed yeast strain was cultured in medium containing Gal to induce the expression of CYP88D6, CYP72A154, and CPR under the control of Gal-inducible promoters (GAL10 or GAL1). Following the culture of the transgenic yeast, the ethyl acetate extracts of the yeast cultures were analyzed.
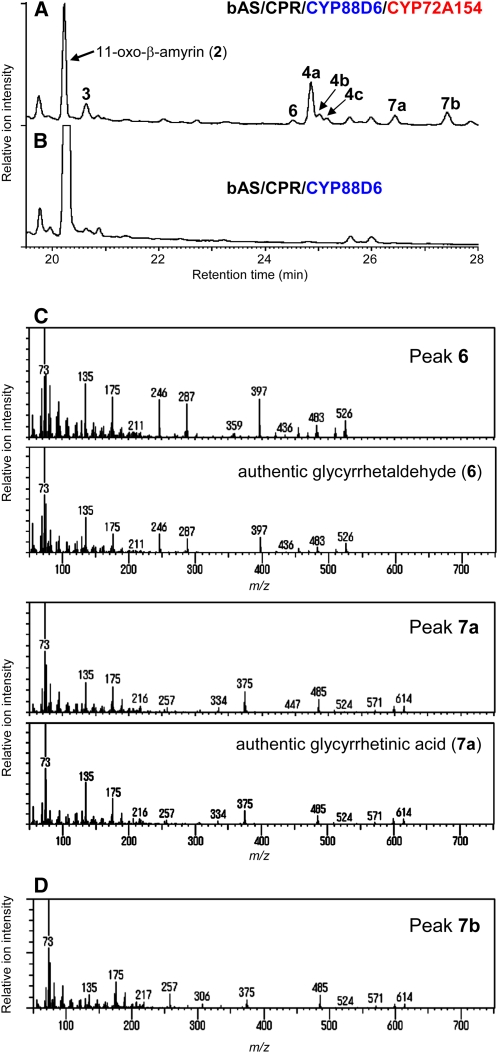
As shown in Figure 4A, the bAS/CPR/CYP88D6/CYP72A154-expressing yeast strain formed seven additional compounds (peaks 3, 6, 4a, 4b, 4c, 7a, and 7b) besides 11-oxo-β-amyrin (2) (the estimated yield of each β-amyrin oxidized product is shown in Supplemental Table 1 online). These compounds were absent from the bAS/CPR/CYP88D6-expressing yeast (Figure 4B). The additional compounds included 30-hydroxy-11-oxo-β-amyrin (4a) (peak 4a; see Supplemental Figure 2B online) and its isomers (peaks 4b and 4c; Supplemental Figure 2B online), as expected from the results of in vitro CYP72A154 enzymatic activity assays using 11-oxo-β-amyrin (2) as the substrate (Figure 2). The yeast-derived 30-hydroxy-11-oxo-β-amyrin (4a) was purified using silica gel column chromatography, and its identity was confirmed using NMR spectroscopy (see Supplemental Figure 2 online).
Figure 4.
In Vivo Production of Glycyrrhetinic Acid in Yeast Coexpressing bAS, CYP88D6, and CYP72A154.
(A) and (B) GC-MS analysis (TIC) of ethyl acetate extracts from yeast cultures. The yeast strains were engineered to express bAS, CPR, CYP88D6, and CYP72A154 (bAS/CPR/CYP88D6/CYP72A154) (A) or bAS, CPR, and CYP88D6 (bAS/CPR/CYP88D6) (B). GC analyses were performed on a DB-1 column.
(C) The mass spectra of peaks 6 and 7a compare well with those of authentic glycyrrhetaldehyde (6) and glycyrrhetinic acid (7a), respectively.
(D) Mass spectrum of peak 7b.
[See online article for color version of this figure.]
The mass spectra of peaks 6 and 7a showed excellent matches with those of authentic glycyrrhetaldehyde (6) and glycyrrhetinic acid (7a), respectively (Figure 4C). The product of peak 7b (mass spectrum shown in Figure 4D), with an ion fragmentation pattern highly similar to that of glycyrrhetinic acid, was identified as an isomer of glycyrrhetinic acid. Although we were unable to determine the precise position of the introduced carboxyl group in peak 7b, GC-MS ionization data indicate that it is most likely located at C-29.
The mass spectrum of peak 3 was an exact match with that of authentic 30-hydroxy-β-amyrin (3) (see Supplemental Figure 2A online). The in vivo production of 30-hydroxy-β-amyrin (3) was also found in bAS/CPR/CYP72A154-transformed yeast (peak 3; see Figure 8B), as confirmed by NMR analysis. These results indicate that CYP72A154 is capable of catalyzing C-30 hydroxylation of β-amyrin (1) supplied in situ to produce 30-hydroxy-β-amyrin (3), even though no enzyme activity was detected when CYP72A154-containing microsomes were assayed in vitro with β-amyrin (1).
Figure 8.
In Vivo Enzyme Assay for CYP72A63.
(A) to (C) GC-MS analysis (TIC) of the ethyl acetate extracts from yeast cultures are shown (chromatograms are scaled to the highest peak). The yeast strains were engineered to coexpress bAS, CPR, and CYP72A63 (bAS/CPR/CYP72A63) (A); bAS, CPR, and CYP72A154 (bAS/CPR/CYP72A154) (B); or bAS and CPR alone (bAS/CPR) (C). The chromatographic peak for β-amyrin (1) is indicated.
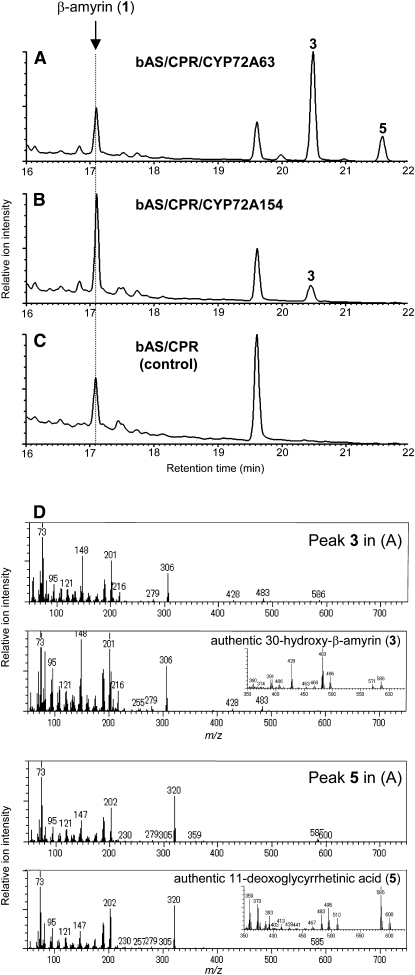
(D) The mass spectra of peaks 3 and 5 shown in (A) compare well with those of authentic 30-hydroxy-β-amyrin (3) and 11-deoxoglycyrrhetinic acid (5), respectively. The identity of yeast-derived 30-hydroxy-β-amyrin (peak 3) in (B) was confirmed by NMR analysis. 30-Hydroxy-β-amyrin (3) derived from bAS/CPR/CYP72A154-transformed yeast: 1H NMR (500 MHz, CDCl3): δ 0.792 (3H, s), 0.833 (3H, s), 0.899 (3H, s), 0.937 (3H, s), 0.962 (3H, s), 1.000 (3H, s), 1.151 (3H, s), 3.225 (1H, dd, J = 11.5, 4.6 Hz), 3.484 (1H, d, J = 10.9 Hz), 3.560 (1H, d, J = 10.9 Hz), 5.191 (1H, t, J = 3.4 Hz); 13C NMR (125 MHz, CDCl3): δ 15.49, 15.57, 16.79, 18.36, 23.51, 25.98, 26.07, 27.21, 27.47, 28.09, 28.21, 29.62, 32.43, 32.63, 35.52, 36.49, 36.94, 38.57, 38.77, 39.78, 41.72, 41.93, 46.73, 47.61, 55.16, 66.75, 79.01, 122.29, 144.56.
Two additional CYP72A subfamily members identified from licorice ESTs, CYP72A153 (60.0% identity with CYP72A154) and CYP72A155 (50.9% identity) (for phylogenetic relationships, see Figure 7), were also tested for potential β-amyrin (1) and 11-oxo-β-amyrin (2) oxidizing activity in vitro and in vivo (in bAS/CPR/CYP88D6-expressing yeast background). However, neither of these P450s showed detectable activity against these substrates.
Figure 7.
Phylogenetic Relationships among the CYP72A Proteins in Dicots.
A phylogenetic tree was generated based on probable entire amino acid sequences of CYP72A proteins from Arabidopsis (At), C. roseus (Cr), G. max (Gm), G. uralensis (Gu), Solanum lycopersicum (Le), M. truncatula (Mt), and Solanum tuberosum (St). The CYP734A1 of Arabidopsis was included as the outgroup. The numbers above the branches are the bootstrap values for 1000 replicates. The scale bar shows the amino acid substitution ratio. The closed arrowheads indicate the P450s functionally defined in this study. The open arrowheads indicate the G. uralensis and M. truncatula P450s analyzed, but no obvious activity was detected in our enzyme assays. The Cr-CYP72A1 (49.2% identity with CYP72A154) from C. roseus, the only CYP72A subfamily P450 whose biochemical function has been elucidated thus far, is indicated by an asterisk.
CYP72A154 Catalyzes Multiple Oxidations at C-30
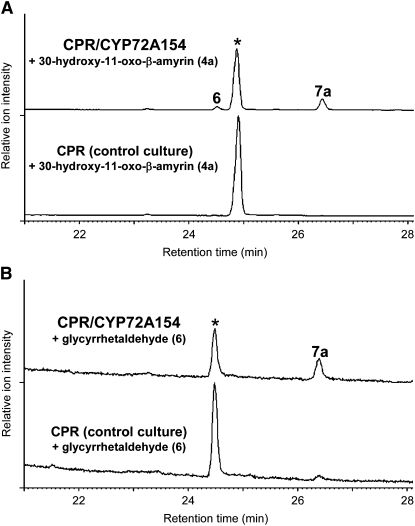
To verify whether CYP72A154 catalyzes all three oxidation steps at C-30 of 11-oxo-β-amyrin (2) to produce glycyrrhetinic acid (7a), we conducted feeding experiments with the CPR/CYP72A154-transformed yeast strain, using 30-hydroxy-11-oxo-β-amyrin (4a) or glycyrrhetaldehyde (6) as a substrate. The CPR/CYP72A154-transformed yeast fed with 30-hydroxy-11-oxo-β-amyrin (4a) converted it to a mixture of glycyrrhetinic acid (7a) as the major product (peak 7a; mass spectrum shown in Supplemental Figure 3B online) and glycyrrhetaldehyde (6) as a minor product (peak 6; mass spectrum shown in Supplemental Figure 3A online) (Figure 5A). The identities of the yeast-derived glycyrrhetinic acid and glycyrrhetaldehyde were confirmed by NMR analysis (see Supplemental Figure 3 online). These compounds were not observed when 30-hydroxy-11-oxo-β-amyrin (4a) was fed to a control strain expressing CPR alone. When glycyrrhetaldehyde (6) was fed as a substrate (Figure 5B), substantial conversion to glycyrrhetinic acid was detected with CPR/CYP72A154-transformed yeast, whereas only trace amounts of glycyrrhetinic acid were reproducibly found with the control yeast strain, presumably owing to some weak endogenous activity or an oxygen-dependent nonenzymatic reaction during incubation. Collectively, these results demonstrate that CYP72A154 performed multiple oxidations at C-30 of 11-oxo-β-amyrin (2) to form glycyrrhetinic acid (7a), although we cannot completely rule out the possibility that other enzymes are involved in the second and third oxidations in planta.
Figure 5.
In Vivo Formation of Glycyrrhetinic Acid in CYP72A154-Expressing Yeast Fed with Precursors of Glycyrrhetinic Acid.
GC-MS analysis (TIC) of ethyl acetate extracts of CPR/CYP72A154-transformed yeast cultures (CPR/CYP72A154) and CPR-transformed control cultures (CPR) fed with 30-hydroxy-11-oxo-β-amyrin (4a) (A) or glycyrrhetaldehyde (6) (B). Substrate additions to the media are marked with asterisks in the GC profiles. The mass spectra of peaks 6 and 7a compare well with those of authentic glycyrrhetaldehyde (6) and glycyrrhetinic acid (7a), respectively (see Supplemental Figure 3 online).
Expression of CYP72A154
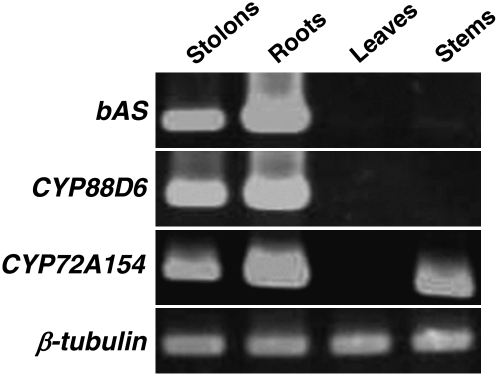
The organ specificity of CYP72A154 expression was analyzed with RT-PCR (Figure 6). Transcripts of CYP72A154 were detected in roots, stolons, and stems, whereas no transcripts were observed in leaves. It is well known that glycyrrhizin accumulates predominantly in the underground parts of Glycyrrhiza plants. The expression of CYP72A154 in the roots and stolons at levels comparable to those of bAS and CYP88D6, combined with data from our functional analyses, suggests a role for CYP72A154 in glycyrrhizin biosynthesis, although we currently do not have a good explanation for the possible function of CYP72A154 expressed in stem tissues.
Figure 6.
Expression Pattern of CYP72A154.
RT-PCR analysis of bAS, CYP88D6, and CYP72A154 mRNA levels in stolons, roots, leaves, and stems, with β-tubulin as an internal control.
Isolation and Functional Analysis of M. truncatula CYP72As
Both licorice and M. truncatula belong to the family Fabaceae. More than 30 different triterpenoid saponins, which are mainly derived from five different triterpenoid aglycones based on the β-amyrin skeleton, have been detected from roots and/or aerial parts of the model legume M. truncatula (Huhman and Sumner, 2002). To further suggest the possible involvement of CYP72As in triterpenoid saponin biosynthesis, additional CYP72As identified from M. truncatula were tested for potential β-amyrin–oxidizing activity by in vivo assay in a bAS/CPR-expressing yeast background.
Recently, 151 putative P450 genes, including nine CYP72As, were assigned in M. truncatula (Li et al., 2007). Of the nine CYP72As, full-length cDNAs of six were obtained by RT-PCR. In addition, we successfully isolated a full-length cDNA for CYP72A63, although CYP72A63 has been recognized as a pseudogene. The obtained seven full-length P450s included three members (CYP72A62v2, CYP72A63, and CYP72A65v2) with >75% amino acid sequence identity with CYP72A154 (for phylogenetic relationships, see Figure 7).
To test for potential β-amyrin–oxidizing activity in vivo, each CYP72A was coexpressed with bAS and CPR in yeast. Of the seven M. truncatula CYP72As tested, CYP72A63 (75.5% identity with CYP72A154) showed activity against β-amyrin (Figure 8A). As shown in Figure 8A, the bAS/CPR/CYP72A63-expressing yeast strain formed two additional compounds (peaks 3 and 5) besides β-amyrin (1). The mass spectra of peaks 3 and 5 compare well with those of authentic 30-hydroxy-β-amyrin (3) and 11-deoxoglycyrrhetinic acid (5), respectively (Figure 8D). By contrast, the expression of bAS and CPR alone did not yield detectable levels of 30-hydroxy-β-amyrin (3) or 11-deoxoglycyrrhetinic acid (5) (Figure 8C). These analyses show that CYP72A63 catalyzed C-30 hydroxylation of β-amyrin. Moreover, in contrast with CYP72A154, which only catalyzed monohydroxylation of β-amyrin (1) to 30-hydroxy-β-amyrin (3) (Figure 8B), CYP72A63 was also able to catalyze further oxidation steps to produce 11-deoxoglycyrrhetinic acid (5).
DISCUSSION
The data presented here clearly show that CYP72A154 is responsible for C-30 oxidation in the glycyrrhizin pathway. However, the relatively complex reaction product profile of CYP72A154 (Figures 2A and 4A) suggests that it has a role in generating structural variations in triterpene aglycones derived from β-amyrin. Both the in vitro and in vivo analyses showed that CYP72A154 is able to catalyze oxidation in at least three different carbon positions (including C-30) in ring D or E (Figure 2E). Consistent with this complex reaction product profile, it has been reported that licorice root contains a variety of oleanane-type triterpenoid saponins, called licorice saponins (Kitagawa, 1993), in addition to glycyrrhizin as the major saponin. The elucidated structures of 11 licorice saponins (Kitagawa, 1993) clearly indicate that in G. uralensis, the oxidation of the β-amyrin skeleton can occur at each of the positions C-11, C-22, C-24, C-29, and C-30 (Figure 1). We previously identified CYP88D6 and CYP93E3 from G. uralensis as β-amyrin 11-oxidase and β-amyrin 24-hydoxylase, respectively (Seki et al., 2008). Hence, it is reasonable to speculate that CYP72A154 is responsible for the oxidation at positions C-22 and C-29 as well as C-30. It is generally well known that glycyrrhizin is the major saponin contained in licorice (2 to 8% of the dry weight). Surprisingly, the bAS/CPR/CYP88D6/CYP72A154-expressing yeast produced comparable amounts of glycyrrhetinic acid (7a) (C-30 carboxylated form; Figure 4A, peak 7a) and its isomer, which differs in the position of an introduced carboxyl group (presumably a C-29 carboxylated form; Figure 4A, peak 7b). It is possible that the C-30 carboxylated form is the preferred substrate for the yet unidentified UGT in planta.
Here, we revealed a function of a CYP72A subfamily protein as a triterpene-oxidizing enzyme. Our observation indicates that in G. uralensis, and most probably in other legumes, the P450 enzymes involved in triterpenoid saponin biosynthesis are recruited from at least three distinct CYP families representing both major P450 clades, A-type and non-A type (Durst and Nelson, 1995): CYP72 (CYP72 clan) and CYP88 (CYP85 clan) of the non-A type P450s and CYP93 (CYP71 clan) of the A-type P450s. Unlike the CYP93E and CYP88D subfamilies, which are likely to be restricted to the Fabaceae (based on extensive database searches), the CYP72A subfamily P450s are distributed widely among plants. There are at least 221 CYP72As, which have been designated as CYP72A1 to CYP72A221 (http://drnelson.uthsc.edu/biblioD.html), including nine and 17 CYP72As found in the genomes of Arabidopsis and rice (Oryza sativa), respectively. The functional identification of CYP72A154 hints at the involvement of other CYP72A subfamily P450s in the biosynthesis of triterpenoid saponins with possibly unique reaction specificity. However, the CYP72A1 (49.2% amino acid sequence identity with CYP72A154) from Catharanthus roseus, the only CYP72A subfamily P450 whose biochemical function has been elucidated thus far, is a secologanin synthase, a ring-opening enzyme in the biosynthesis of the seco-iridoid unit of terpene indole alkaloids (Irmler et al., 2000).
More than 30 different triterpenoid saponins have been detected from roots and/or aerial parts of the model legume M. truncatula, and these are mainly derived from five different triterpenoid aglycones based on the β-amyrin skeleton: soyasapogenol B and E, medicagenic acid, hederagenin, and bayogenin (Huhman and Sumner, 2002). Recently, based on comprehensive gene expression clustering analysis, Naoumkina et al. (2010) identified six M. truncatula P450s, with expression patterns very similar to that of bAS, as strong candidates for involvement in triterpenoid saponin biosynthesis. Four of these were CYP72A subfamily P450s: two CYP72A61s, CYP72A67, and CYP72A68 (for expression profiles, see Supplemental Figure 4 online). Operon-like gene clusters in Arabidopsis and oat (Avena strigosa) are required for triterpene biosynthesis (Qi et al., 2004; Field and Osbourn, 2008). Of these P450s, the two CYP72A61s were tightly linked to the glucosyltransferase UGT73K1 on chromosome 4 and had expression profiles very similar to that of bAS, although bAS is not tightly linked to these genes (Naoumkina et al., 2010). UGT73K1 is known to glycosylate soyasapogenol B and E as well as hederagenin (Achnine et al., 2005).
Although we failed to detect β-amyrin–oxidizing activity of CYP72A61v2, CYP72A67v2, and CYP72A68v2 in our enzyme activity assays, we did detect β-amyrin 30-oxidase activity of CYP72A63 (Figure 8A). To our knowledge, no C-30 oxidated triterpene aglycones have been identified from M. truncatula; thus, CYP72A63 may be responsible for the biosynthesis of minor saponins and may be restricted to specific tissues of a plant. Consistent with this speculation, the expression of CYP72A63 appeared to be rather low in all tissues analyzed and was restricted to specific tissues: flowers and NaCl-treated roots (for expression profile, see Supplemental Figure 4 online). Overall, these observations, combined with our functional analysis data, strongly suggest the involvement of CYP72A subfamily P450s in triterpenoid saponin biosynthesis in legumes. Molecular phylogenetic analysis showed that CYP72As identified from legumes (licorice, M. truncatula, and soybean [Glycine max]) are grouped separately from the CYP72As of other dicots, and these could be further divided into three subgroups, Groups I, II, and III (Figure 7). As both CYP72A154 (licorice) and CYP72A63 (M. truncatula) are included in Group II, further analyses of Group II CYP72As are a high priority.
In this study, we identified CYP72A154 as a second important P450 in the glycyrrhizin biosynthetic pathway. Moreover, we engineered the coexpression of bAS, CYP88D6, CYP72A154, and CPR in a wild-type yeast background to produce glycyrrhetinic acid through the redirection of a portion of the native 2,3-oxidosqualene pool from sterol synthesis. Despite low production (~15 μg/L culture during a 48-h culture after Gal induction), these results illustrate the potential for using yeast and a synthetic biology approach for the production of glycyrrhetinic acid. Further engineering efforts to enhance the availability of 2,3-oxidosqualene by overexpressing a truncated form of 3-hydroxy-3-methylglutaryl CoA reductase and downregulating ERG7 (lanosterol synthase) may improve yields. Taking this approach, Kirby et al. (2008) achieved β-amyrin levels of 6 mg/L culture. Furthermore, as CYP72A154 did not fully consume its preferred substrate, 11-oxo-β-amyrin, there is potential for yield improvements through protein engineering of CYP72A154 to enhance its catalytic activity and/or modulate its product specificity. Comparative studies of structure-function relationships among the CYP72A subfamily P450s are needed to obtain structural insight into the reaction specificity of CYP72A154.
METHODS
NMR Analysis
The NMR spectra were recorded on a Bruker DSX-300 or JEOL ECA-500 spectrometer in deuterated chloroform (CDCl3). Analytical thin layer chromatography was performed on Merck silica gel 60F-254 plates (0.25 mm, precoated). β-Amyrin (1) was synthesized by a previously described method (Ohyama et al., 2007). Glycyrrhetinic acid (7a) was purchased from Sigma-Aldrich. Other standard samples were synthesized from either β-amyrin or glycyrrhetinic acid as described previously (Seki et al., 2008) or as in Supplemental Figure 5 online (glycyrrhetaldehyde). 1H- and 13C-NMR chemical shifts are reported in δ values based on the internal tetramethylsilane (δH = 0) and reference solvent signals (CDCl3 δC = 77.0).
In Vitro Enzyme Assays
Thermococcus kodakaraensis-plus DNA polymerase (TOYOBO) was used for PCR, according to the manufacturer’s instructions. The open reading frame of each licorice (Glycyrrhiza uralensis) CYP72A gene was amplified by RT-PCR using primers 1 and 2 for CYP72A154, primers 3 and 4 for CYP72A153, and primers 5 and 6 for CYP72A155 (for primer sequences, see Supplemental Table 2 online). The resulting PCR products were cloned via pENTR/D-TOPO (Invitrogen) into a pDEST 8 vector to generate insect cell baculovirus expression clones. The resulting constructs were used to generate the recombinant bacmid DNAs by transformation of Escherichia coli strain DH10Bac (Invitrogen). The preparation of recombinant bacmid DNA and transfection of Sf9 cells were performed according to the manufacturer’s instructions (Invitrogen). The expression of plant P450s in Sf9 cells and in vitro enzyme assays were performed as previously described (Ohnishi et al., 2006; Seki et al., 2008).
GC-MS Analysis
GC-MS was conducted using a JMS-AM SUN200 mass spectrometer (JEOL) connected to a gas chromatograph (6890A; Agilent Technologies), with an HP-5 (30 m × 0.32 mm, 0.25-μm film thickness; JandW Scientific) or DB-1 (30 m × 0.25 mm, 0.25-μm film thickness; JandW Scientific) capillary column. The injection temperature was 250°C, and the column temperature program was as follows: 80°C for 1 min, followed by a rise to 300°C at a rate of 20°C/min, and a hold at 300°C for 20 or 28 min. The carrier gas was He; the flow rate was 1.2 or 1.0 mL/min; and the interface temperature was 300°C, with a splitless injection. All samples were trimethylsilylated with N-methyl-N-trimethylsilyltrifluoroacetamide (Sigma-Aldrich) at 80°C for 30 min.
Constructs for Yeast in Vivo Assays
The plasmids pYES3-ADH-OSC1, for constitutive expression of bAS (Lj OSC1; Sawai et al., 2006) under the control of the ADH1 promoter, and pELC, for Gal-inducible expression of Lj CPR1 alone, were constructed as previously described (Seki et al., 2008). For Gal-inducible dual expression of Lj CPR1 and CYP88D6, the plasmid pELC88BN was constructed as follows. The entire coding region of CYP88D6 was PCR amplified from the CYP88D6 entry clone (Seki et al., 2008) using primers 7 and 8, digested with BamHI-XbaI, and cloned into BamHI-NheI–digested (XbaI and NheI are compatible) pESC-LEU (Stratagene). The fragment containing the CYP88D6 coding sequence was then excised from the resulting plasmid as a NotI-PacI fragment and cloned into NotI-PacI–digested pELC to produce pELC88BN. To create the CYP72A154 yeast expression vector pDEST52-CYP72A154 for the Gal-inducible expression, CYP72A154 cDNA was transferred via pENTR/D-TOPO into pYES-DEST52 (Invitrogen) using Gateway LR Clonase Enzyme Mix (Invitrogen). The same cloning strategy was used for CYP72A153 and CYP72A155 to create pDEST52-CYP72A153 and pDEST52-CYP72A155, respectively.
Isolation of CYP72A Subfamily P450s from M. truncatula
The entire coding sequences of M. truncatula CYP72As were identified in either the GenBank database or Dana-Farber Cancer Institute Medicago Gene Index (Release 8.0; http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=medicago). The cDNA fragments containing the open reading frame of each gene were amplified from whole 4-week-old Medicago truncatula (R108-1) plants by RT-PCR with primers 9 and 10 for CYP72A59v2, 11 and 12 for CYP72A61v2, 13 and 14 for CYP72A63 and CYP72A62v2, 15 and 16 for CYP72A65v2, 17 and 18 for CYP72A67v2, and 19 and 20 for CYP72A68v2. The amplified fragment was cloned into pENTR/D-TOPO (Invitrogen), sequenced, and transferred into the Gateway-adopted version of pELC vector (Seki et al., 2008) to generate a construct for Gal-inducible dual expression of Lj CPR1 and each P450 in yeast.
Yeast in Vivo Assays in an Engineered Yeast Strain That Endogenously Produces 11-Oxo-β-Amyrin
Saccharomyces cerevisiae BJ2168 (MATa, prc1-407, prb1-1122, pep4-3, leu2, trp1, ura3-52, gal2; Nippon Gene) harboring pYES3-ADH-OSC1 was transformed with pELC or pELC88BN. The resulting yeast strains were further transformed with pDEST52-CYP72A153, pDEST52-CYP72A154, or pDEST52-CYP72A155 or with empty pYES2 vector (Invitrogen) as a control. Recombinant yeast cells were cultured as described previously (Seki et al., 2008). Yeast cultures were extracted with ethyl acetate, and portions of the extracts were analyzed by GC-MS after trimethylsilylation.
Yeast Feeding Assay
Yeast strains pretransformed with pELC were further transformed with pDEST52-CYP72A154 or empty pYES2 vector (Invitrogen) as a control. Recombinant yeast cells were cultured in synthetic complete medium containing 2% Glc without Leu and uracil (SC−L−U) for 2 d at 28°C. The cells were collected and resuspended in SC−L−U medium containing hemin, 2% Gal instead of Glc, and 2 μM of either 30-hydroxy-11-oxo-β-amyrin (4a) or glycyrrhetaldehyde (6) as substrate. After 2 d of culture at 28°C, the yeast cultures were extracted with ethyl acetate, and the extracts were analyzed by GC-MS after trimethylsilylation.
Yeast in Vivo Assays in an Engineered Yeast Strain That Endogenously Produces β-Amyrin
A yeast strain pretransformed with pYES3-ADH-OSC1 was further transformed with pELC-CYP72A63 (dual expression of Lj CPR1 and CYP72A63) or pELC-CYP72A154 (dual expression of Lj CPR1 and CYP72A154) or with pELC (expression of Lj CPR1 alone) as a control. Recombinant yeast cells were cultured in synthetic complete medium containing 2% Glc without Trp and Leu (SC−W−L) for 2 d at 28°C. The cells were collected, resuspended in SC−W−L medium containing 13 μg/mL hemin and 2% Gal instead of Glc, and cultured at 28°C for 2 d. The yeast cells were harvested and extracted with ethyl acetate, and portions of the extracts were analyzed by GC-MS after trimethylsilylation.
RT-PCR Analysis
Preparation of total RNAs and subsequent first-strand cDNA synthesis were performed as described previously (Seki et al., 2008). PCR was performed using primers 21 and 22 for bAS, primers 23 and 24 for CYP88D6, primers 25 and 4 for CYP72A154, primers 26 and 27 for β-tubulin. The exponential ranges of the reactions were determined by removing 5 μL of PCR product every three cycles for 24 to 36 cycles to allow quantitative comparisons. We selected conditions of 33 cycles for bAS, CYP88D6, and CYP72A154 and 30 cycles for β-tubulin, so that the amplified products were clearly visible on an agarose gel, yet amplification was still in the exponential range and had not reached a plateau.
Phylogenetic Analysis
Probable entire amino acid sequences of CYP72As were taken from the Cyt P450 website (http://drnelson.uthsc.edu/biblioD.html#89A) and soybean (Glycine max) P450s website (http://drnelson.uthsc.edu/soybean.html) provided by David R. Nelson, from the CYPedia-Cytochrome P450 Expression Database using Arabidopsis thaliana (http://www-ibmp.u-strasbg.fr/~CYPedia/), or from GenBank database. The alignment was performed using ClustalW (Thompson et al., 1994) with a gap open cost of 10 and gap extension cost of 0.2 (see Supplemental Data Set 1 online). The phylogenetic tree was constructed in PHYLIP using the neighbor-joining method with bootstrap analysis of 1000 replicates and was displayed using NJPlot software (version 2.3). Arabidopsis CYP734A1, a member of CYP734 family that belongs to the CYP72 clan, was used as the outgroup.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: AB558144 (CYP72A59v2), AB558145 (CYP72A61v2), AB558146 (CYP72A63), AB558147 (CYP72A62v2), AB558148 (CYP72A65v2), AB558149 (CYP72A67v2), AB558150 (CYP72A68v2), AB558152 (CYP72A153), AB558153 (CYP72A154), and AB558154 (CYP72A155).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mass Spectrum of the Product Resulting from in Vitro CYP72A154 Enzymatic Activity Assays Containing 30-Hydroxy-11-Oxo-β-Amyrin as Substrate.
Supplemental Figure 2. Mass Spectra of the Products from Yeast Coexpressing β-Amyrin Synthase, CYP88D6, and CYP72A154.
Supplemental Figure 3. Mass Spectra of Glycyrrhetaldehyde and Glycyrrhetinic Acid Formed by CPR/CYP72A154-Transformed Yeast Fed with 30-Hydroxy-11-Oxo-β-Amyrin.
Supplemental Figure 4. Expression Profiles of M. truncatula bAS and Candidate P450s (CYP72As) Potentially Involved in the Triterpenoid Saponin Biosynthesis.
Supplemental Figure 5. Synthesis of Glycyrrhetaldehyde (6) from Glycyrrhetinic Acid (7a).
Supplemental Table 1. Estimated Yield of Each β-Amyrin–Oxidized Product Formed by bAS/CPR/CYP88D6/CYP72A154-Transformed Yeast.
Supplemental Table 2. Oligonucleotides Used for PCR.
Supplemental Data Set 1. Multiple Alignments Used to Construct the Phylogenetic Tree in Figure 7.
Acknowledgments
We thank Jun Ogihara and Masayuki Shimamura (Nihon University) and Tomoko Nishizawa (Kihara Institute for Biological Research) for their technical assistance. This work was supported in part by a grant from the Bio-oriented Technology Research Advancement Institution; a grant from the New Energy and Industrial Technology Development Organization of Japan; grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and research funding from the Kato Memorial Bioscience Foundation.
AUTHOR CONTRIBUTIONS
H. Seki, S.S., K.O., M.M., T.O., H. Sudo, and E.O.F. designed and performed research and analyzed data. T. Akashi and T. Aoki contributed new reagents/analytic tools. H. Seki, S.S., and K.O. wrote the article. K.S. and T.M. designed and supervised research.
References
- Abe I., Rohmer M., Prestwich G.D. (1993). Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem. Rev. 93: 2189–2206 [Google Scholar]
- Achnine L., Huhman D.V., Farag M.A., Sumner L.W., Blount J.W., Dixon R.A. (2005). Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 41: 875–887 [DOI] [PubMed] [Google Scholar]
- Askam V., Bradley D.M. (1971). Transformations in ring A of methyl glycyrrhetinate. J. Chem. Soc. 1895–1901 [Google Scholar]
- Asl M.N., Hosseinzadeh H. (2008). Review of pharmacological effects of Glycyrrhiza sp. and its bioactive compounds. Phytother. Res. 22: 709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. (2003). Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 361: 2045–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durst F., Nelson D.R. (1995). Diversity and evolution of plant P450 and P450-reductases. Drug Metabol. Drug Interact. 12: 189–206 [DOI] [PubMed] [Google Scholar]
- Field B., Osbourn A.E. (2008). Metabolic diversification—Independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Finney R.S.H., Somers G.F. (1958). The antiinflammatory activity of glycyrrhetinic acid and derivatives. J. Pharm. Pharmacol. 10: 613–620 [DOI] [PubMed] [Google Scholar]
- Fiore C., Eisenhut M., Krausse R., Ragazzi E., Pellati D., Armanini D., Bielenberg J. (2008). Antiviral effects of Glycyrrhiza species. Phytother. Res. 22: 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M.R. (1978). Glycyrrhiza in old and new perspectives. Lloydia 41: 348–354 [PubMed] [Google Scholar]
- Hayashi H., Hirota A., Hiraoka N., Ikeshiro Y. (1999). Molecular cloning and characterization of two cDNAs for Glycyrrhiza glabra squalene synthase. Biol. Pharm. Bull. 22: 947–950 [DOI] [PubMed] [Google Scholar]
- Hayashi H., Huang P., Kirakosyan A., Inoue K., Hiraoka N., Ikeshiro Y., Kushiro T., Shibuya M., Ebizuka Y. (2001). Cloning and characterization of a cDNA encoding β-amyrin synthase involved in glycyrrhizin and soyasaponin biosyntheses in licorice. Biol. Pharm. Bull. 24: 912–916 [DOI] [PubMed] [Google Scholar]
- Hayashi H., Sudo H. (2009). Economic importance of licorice. Plant Biotechnol. 26: 101–104 [Google Scholar]
- He J.X., Akao T., Nishino T., Tani T. (2001). The influence of commonly prescribed synthetic drugs for peptic ulcer on the pharmacokinetic fate of glycyrrhizin from Shaoyao-Gancao-tang. Biol. Pharm. Bull. 24: 1395–1399 [DOI] [PubMed] [Google Scholar]
- Huhman D.V., Sumner L.W. (2002). Metabolic profiling of saponins in Medicago sativa and Medicago truncatula using HPLC coupled to an electrospray ion-trap mass spectrometer. Phytochemistry 59: 347–360 [DOI] [PubMed] [Google Scholar]
- Irmler S., Schröder G., St-Pierre B., Crouch N.P., Hotze M., Schmidt J., Strack D., Matern U., Schröder J. (2000). Indole alkaloid biosynthesis in Catharanthus roseus: New enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J. 24: 797–804 [DOI] [PubMed] [Google Scholar]
- Ito M., Nakashima H., Baba M., Pauwels R., De Clercq E., Shigeta S., Yamamoto N. (1987). Inhibitory effect of glycyrrhizin on the in vitro infectivity and cytopathic activity of the human immunodeficiency virus [HIV (HTLV-III/LAV)]. Antiviral Res. 7: 127–137 [DOI] [PubMed] [Google Scholar]
- Ito M., Sato A., Hirabayashi K., Tanabe F., Shigeta S., Baba M., De Clercq E., Nakashima H., Yamamoto N. (1988). Mechanism of inhibitory effect of glycyrrhizin on replication of human immunodeficiency virus (HIV). Antiviral Res. 10: 289–298 [DOI] [PubMed] [Google Scholar]
- Jennewein S., Rithner C.D., Williams R.M., Croteau R.B. (2001). Taxol biosynthesis: Taxane 13 α-hydroxylase is a cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. USA 98: 13595–13600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.G., You H.J., Park S.J., Moon A.R., Chung Y.C., Kang S.K., Chun H.K. (2002). Hepatoprotective effects of 18beta-glycyrrhetinic acid on carbon tetrachloride-induced liver injury: Inhibition of cytochrome P450 2E1 expression. Pharmacol. Res. 46: 221–227 [DOI] [PubMed] [Google Scholar]
- Kirby J., Romanini D.W., Paradise E.M., Keasling J.D. (2008). Engineering triterpene production in Saccharomyces cerevisiae-β-amyrin synthase from Artemisia annua. FEBS J. 275: 1852–1859 [DOI] [PubMed] [Google Scholar]
- Kitagawa I. (1993). Licorice root. A natural sweetener and an important ingredient in Chinese medicine. Pure Appl. Chem. 74: 1189–1198 [Google Scholar]
- Kojoma M., Ohyama K., Seki H., Hiraoka Y., Asazu S.N., Sawa S., Sekizaki H., Yoshida S., Muranaka T. (2010). In vitro proliferation and triterpenoid characteristics of licorice (Glycyrrhiza uralensis Fischer, Leguminosae) stolons. Plant Biotechnol. 27: 59–66 [Google Scholar]
- Kroes B.H., Beukelman C.J., van den Berg A.J., Wolbink G.J., van Dijk H., Labadie R.P. (1997). Inhibition of human complement by β-glycyrrhetinic acid. Immunology 90: 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Cheng H., Gai J., Yu D. (2007). Genome-wide identification and characterization of putative cytochrome P450 genes in the model legume Medicago truncatula. Planta 226: 109–123 [DOI] [PubMed] [Google Scholar]
- Naoumkina M.A., Modolo L.V., Huhman D.V., Urbanczyk-Wochniak E., Tang Y., Sumner L.W., Dixon R.A. (2010). Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula. Plant Cell 22: 850–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose M., Ito M., Kamimura K., Shimizu M., Ogihara Y. (1994). A comparison of the antihepatotoxic activity between glycyrrhizin and glycyrrhetinic acid. Planta Med. 60: 136–139 [DOI] [PubMed] [Google Scholar]
- Ohnishi T., Nomura T., Watanabe B., Ohta D., Yokota T., Miyagawa H., Sakata K., Mizutani M. (2006). Tomato cytochrome P450 CYP734A7 functions in brassinosteroid catabolism. Phytochemistry 67: 1895–1906 [DOI] [PubMed] [Google Scholar]
- Ohyama K., Suzuki M., Masuda K., Yoshida K., Muranaka T. (2007). Chemical phenotypes of the hmg1 and hmg2 mutants of Arabidopsis demonstrate the in planta role of HMG-CoA reductase in triterpene biosynthesis. Chem. Pharm. Bull. 55: 1518–1521 [DOI] [PubMed] [Google Scholar]
- Park H.Y., Park S.H., Yoon H.K., Han M.J., Kim D.H. (2004). Anti-allergic activity of 18beta-glycyrrhetinic acid-3-O-beta-D-glucuronide. Arch. Pharm. Res. 27: 57–60 [DOI] [PubMed] [Google Scholar]
- Qi X., Bakht S., Leggett M., Maxwell C., Melton R., Osbourn A. (2004). A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA 101: 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro D.K., et al. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440: 940–943 [DOI] [PubMed] [Google Scholar]
- Sato S., et al. (2008). Genome structure of the legume, Lotus japonicus. DNA Res. 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai S., Shindo T., Sato S., Kaneko T., Tabata S., Ayabe S., Aoki T. (2006). Functional and structural analysis of genes encoding oxidosqualene cyclases of Lotus japonicus. Plant Sci. 170: 247–257 [Google Scholar]
- Schoendorf A., Rithner C.D., Williams R.M., Croteau R.B. (2001). Molecular cloning of a cytochrome P450 taxane 10 β-hydroxylase cDNA from Taxus and functional expression in yeast. Proc. Natl. Acad. Sci. USA 98: 1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H., Ohyama K., Sawai S., Mizutani M., Ohnishi T., Sudo H., Akashi T., Aoki T., Saito K., Muranaka T. (2008). Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc. Natl. Acad. Sci. USA 105: 14204–14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S. (2000). A drug over the millennia: Pharmacognosy, chemistry, and pharmacology of licorice. Yakugaku Zasshi 120: 849–862 [DOI] [PubMed] [Google Scholar]
- Shibuya M., Katsube Y., Otsuka M., Zhang H., Tansakul P., Xiang T., Ebizuka Y. (2009). Identification of a product specific β-amyrin synthase from Arabidopsis thaliana. Plant Physiol. Biochem. 47: 26–30 [DOI] [PubMed] [Google Scholar]
- Sudo H., et al. (2009). Expressed sequence tags from rhizomes of Glycyrrhiza uralensis. Plant Biotechnol. 26: 105–107 [Google Scholar]
- Suzuki H., Achnine L., Xu R., Matsuda S.P., Dixon R.A. (2002). A genomics approach to the early stages of triterpene saponin biosynthesis in Medicago truncatula. Plant J. 32: 1033–1048 [DOI] [PubMed] [Google Scholar]
- Teoh K.H., Polichuk D.R., Reed D.W., Nowak G., Covello P.S. (2006). Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 580: 1411–1416 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum T.G.J., Vulto A.G., Hop W.C., Brouwer J.T., Niesters H.G., Schalm S.W. (1999). Intravenous glycyrrhizin for the treatment of chronic hepatitis C: A double-blind, randomized, placebo-controlled phase I/II trial. J. Gastroenterol. Hepatol. 14: 1093–1099 [DOI] [PubMed] [Google Scholar]
- Waller G.R., Yamasaki K. (1996). Saponins Used in Traditional and Modern Medicine. Advances in Experimental Medicine and Biology, Vol. 404 (New York: Plenum Press; ). [Google Scholar]
- Yonekura-Sakakibara K., Saito K. (2009). Functional genomics for plant natural product biosynthesis. Nat. Prod. Rep. 26: 1466–1487 [DOI] [PubMed] [Google Scholar]