This study describes isolation and characterization of loss-of-function mutants of Fused Compound Leaf1 (FCL1) in Medicago truncatula that encodes a class M KNOX transcription factor. Phenotypic and genetic studies demonstrate that FCL1 is required for boundary separation of leaflet primordia and elongation of petioles. Models of genetic interactions of fcl1 with sgl1 and palm1 are discussed.
Abstract
Medicago truncatula is a legume species belonging to the inverted repeat lacking clade (IRLC) with trifoliolate compound leaves. However, the regulatory mechanisms underlying development of trifoliolate leaves in legumes remain largely unknown. Here, we report isolation and characterization of fused compound leaf1 (fcl1) mutants of M. truncatula. Phenotypic analysis suggests that FCL1 plays a positive role in boundary separation and proximal-distal axis development of compound leaves. Map-based cloning indicates that FCL1 encodes a class M KNOX protein that harbors the MEINOX domain but lacks the homeodomain. Yeast two-hybrid assays show that FCL1 interacts with a subset of Arabidopsis thaliana BEL1-like proteins with slightly different substrate specificities from the Arabidopsis homolog KNATM-B. Double mutant analyses with M. truncatula single leaflet1 (sgl1) and palmate-like pentafoliata1 (palm1) leaf mutants show that fcl1 is epistatic to palm1 and sgl1 is epistatic to fcl1 in terms of leaf complexity and that SGL1 and FCL1 act additively and are required for petiole development. Previous studies have shown that the canonical KNOX proteins are not involved in compound leaf development in IRLC legumes. The identification of FCL1 supports the role of a truncated KNOX protein in compound leaf development in M. truncatula.
INTRODUCTION
Leaves are lateral organs initiated from the flanks of the shoot apical meristem (SAM). Leaf development can be divided into three successive and overlapping stages: initiation, in which leaves emerge from the flanks of the SAM; primary morphogenesis, in which the basic leaf form is determined; and secondary morphogenesis or histogenesis, in which leaves differentiate and produce cell types typical of mature leaves (Hagemann and Gleissberg, 1996; Poethig, 1997; Kaplan, 2001; Efroni et al., 2008; Efroni et al., 2010; Shani et al., 2010). Unlike the SAM’s self-renewing nature, the growth of a leaf is determinate and lasts only for a limited duration.
Leaves exhibit a tremendous diversity in shape, size, and arrangement and can be classified as being either simple or compound, according to their complexities. A simple leaf has a single continuous blade, and a compound leaf consists of multiple discontinuous blades, each resembling a simple leaf and known as a leaflet. The margin of a leaf or leaflet can be lobed, serrate, or entire (smooth), further elaborating the complexity of the leaf form.
The class I KNOTTED1-like (KNOXI) homeobox transcription factor genes are required for the establishment and maintenance of the meristematic activity of the SAM (Long et al., 1996). For both simple- and compound-leafed species, leaf initiation requires downregulation of KNOXI genes at incipient leaf primordia (Long et al., 1996; Bharathan et al., 2002). This downregulation is permanent in simple-leafed species such as Arabidopsis thaliana and maize (Zea mays) (Lincoln et al., 1994). In most compound-leafed eudicot species, such as tomato (Solanum lycopersicum) and Cardamine hirsuta (an Arabidopsis relative with compound leaves), KNOXI gene expression is reactivated in developing leaf primordia (Bharathan et al., 2002; Hay and Tsiantis, 2006). Overexpression of KNOXI genes in the tomato-dominant mutants Mouse Ears and Curl (Chen et al., 1997; Parnis et al., 1997) or ectopic expression of KNOXI genes in tomato plants (Hareven et al., 1996; Janssen et al., 1998) results in a dramatic increase in leaf complexity. In C. hirsuta, downregulation of the KNOXI gene SHOOT MERISTEMLESS (STM) results in simplified leaves, whereas ectopic expression of STM results in more complex leaves (Hay and Tsiantis, 2006). These observations indicate that KNOXI genes are required for compound leaf development in these compound-leafed species.
The reactivation of KNOXI genes in leaf primordia in a large number of eudicot species that have compound leaves indicates a requirement for a transient phase of indeterminacy during compound leaf development (Bharathan et al., 2002; Champagne et al., 2007). However, the transient indeterminacy is not sufficient for compound leaf development, since it can also lead to simple leaves as a result of secondary morphogenesis in some species (Bharathan et al., 2002). Depending on the developmental context, ectopic expression of TKNs, the tomato KNOXI genes, has different effects on leaf shape, supporting a role for TKNs in stage-specific suppression of leaf maturation in tomato (Shani et al., 2009). In a large subclade of legumes, the inverted repeat-lacking clade (IRLC), which diverged from the basal Fabaceae ~39 million years ago and includes pea (Pisum sativum) and Medicago truncatula, KNOXI proteins are not detected in compound leaf primordia, suggesting that KNOXI genes are not likely associated with compound leaf development in this group of legumes (Hofer et al., 2001; Champagne et al., 2007).
Although conflicting evidence exists suggesting the expression of KNOXI genes in compound leaf primordia in M. truncatula (Di Giacomo et al., 2008), UNIFOLIATA (UNI) in pea and SINGLE LEAFLET1 (SGL1) in M. truncatula, both orthologous to the floral meristem identity genes FLORICAULA (FLO) and LEAFY (LFY), have been shown to be required for compound leaf development in these legume species (Hofer et al., 1997; Wang et al., 2008). In Lotus japonicus, a legume outside of the IRLC clade, proliferating floral meristem mutants of the FLO/LFY ortholog also exhibit moderately reduced compound leaf phenotypes (Dong et al., 2005). In soybean (Glycine max), downregulation of the two LFY orthologs through RNA interference leads to a moderate reduction in leaflet number (Champagne et al., 2007). In tomato fa mutants, mutations in the tomato LFY ortholog lead to a reduced number of only secondary leaflets (Molinero-Rosales et al., 1999). These results support a role for FLO/LFY orthologs in compound leaf development in species-specific contexts. Interestingly, although KNOXI proteins are not detected in leaf primordia, the developmental program leading to compound leaves in alfalfa (M. sativa), an IRLC legume, is still responsive to ectopic expression of the tomato KNOXI gene Le T6 (Champagne et al., 2007).
Increasing evidence supports that leaf development is responsive to genetic, hormonal, and environmental regulation (Efroni et al., 2010). Recently, it has been shown that local auxin accumulation is required for and precedes the initiation of leaf and leaflet primordia in diverse species (Barkoulas et al., 2008; Koenig et al., 2009). Interruption of auxin accumulation by auxin transport inhibitors or mutations in auxin carrier genes results in compromised initiation and development of leaf/leaflet primordia (DeMason and Chawla, 2004; Wang et al., 2005; Barkoulas et al., 2008; Koenig et al., 2009; Peng and Chen, 2011; Zhou et al., 2011). By contrast, external application of auxin promotes leaflet initiation and blade outgrowth (Koenig et al., 2009). The hormone cytokinin plays an important role in regulating the morphogenetic activity of compound leaves in tomato (Shani et al., 2010). Cytokinin acting downstream from KNOXI can substitute for KNOXI activity at leaf margins and regulates leaf shape in tomato. NAM/CUC genes encoding the NAC domain proteins play a conserved role in leaflet separation and leaf serration in diverse species (Blein et al., 2008; Berger et al., 2009). Downregulation of NAM/CUC genes by virus-induced gene silencing leads to different degrees of leaflet fusion, reduction of leaflet number, and smoothing of leaf margins (Blein et al., 2008). In tomato, loss-of-function mutations in GOBLET, the tomato ortholog of CUC2, lead to cotyledon and leaflet fusions (Blein et al., 2008; Berger et al., 2009). The role of NAM/CUC genes in leaf shape regulation is dependent on its regulation of KNOXI and UNI gene expression (Blein et al., 2008). In the IRLC legume, M. truncatula PALMATE-LIKE PENTAFOLIATA1 (PALM1) has been shown to encode a novel Cys(2)His(2) zinc finger transcription factor and plays a key role in regulating the morphogenetic activity of compound leaves through negative regulation of SGL1 expression (Chen et al., 2010; Ge et al., 2010).
The canonical KNOXI protein contains conserved KNOX1 and KNOX2 domains, collectively called the MEINOX domain at its N terminus and the homeodomain at its C terminus. In Arabidopsis and tomato, class M KNOX proteins that lack the homeodomain have been identified recently (Kimura et al., 2008; Magnani and Hake, 2008). Ectopic expression of the Arabidopsis class M KNOX gene, KNATM-B, results in elongated petioles and narrow lamina (Magnani and Hake, 2008). By contrast, a gain-of-function mutation in the tomato class M KNOX gene, PTS/TKD1, results in a proliferation of compound leaves (Kimura et al., 2008). It has been shown that the class M KNOX genes function to interfere with the canonical KNOXI activity by titrating protein–protein interactions involving KNOXI and its interacting partners, BEL1-like (BELL) proteins, and by regulating nuclear localization of KNOXI-BELL complexes. However, the role of class M KNOXI in leaf development remains to be elucidated, since neither loss-of-function mutants nor knockdown mutants have been isolated in Arabidopsis and tomato (Kimura et al., 2008; Magnani and Hake, 2008). Here, we report the isolation and characterization of loss-of-function fused compound leaf1 (fcl1) mutants from M. truncatula. Our results indicate that the M. truncatula FCL1 gene encodes a class M KNOX protein that has sequence similarity with the Arabidopsis KNATM-B and tomato PTS/TKD1. Our results further indicate that FCL1 plays a key role in boundary separation and proximal-distal axis development of compound leaves in the legume. We show that both FCL1 and SGL1 are required for the development of petioles.
RESULTS
Isolation and Characterization of M. truncatula fcl1 Mutants
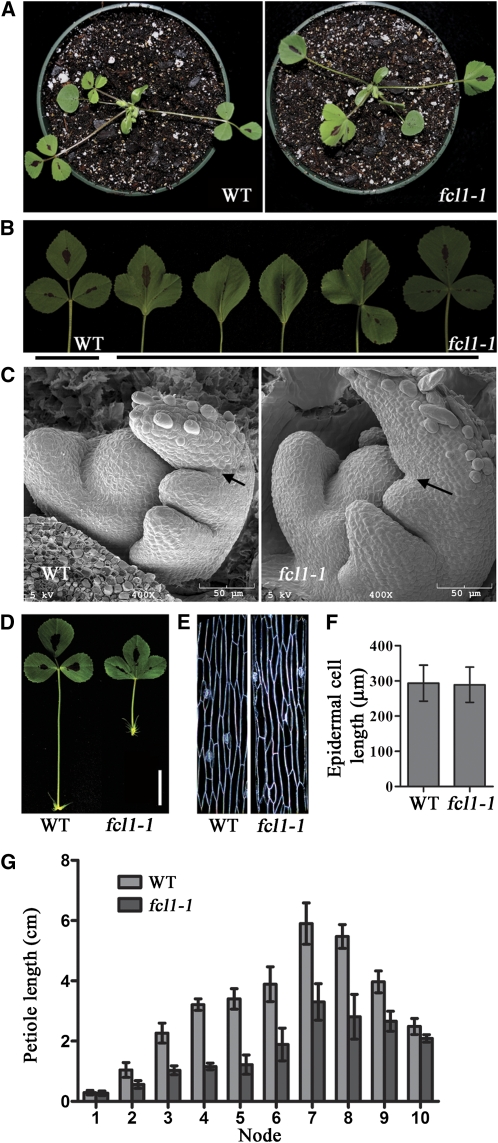
To identify additional regulators of compound leaf development in legumes, we screened and isolated two similar leaf mutants, M368 and M602, from a deletion mutant collection in the model legume M. truncatula (cv Jemalong A17) derived from fast neutron bombardment mutagenesis (Wang et al., 2006). The majority of mature leaves developed in these two mutants were simplified with lobes due to leaflet fusion in contrast with the trifoliolate wild-type leaves (Figures 1A and 1B). A small number of mature leaves in fcl1 mutants were single or had two leaflets fused or three leaflets clustered together without the rachis (Figure 1B). Based on subsequent genetic studies, we named M368 and M602, two recessive mutants, fcl1-1 and fcl1-2, respectively.
Figure 1.
Phenotypic Characterization of M. truncatula fcl1 Mutants.
(A) Three-week-old wild-type (WT) (Jemalong A17; left) and fcl1-1 mutant (right).
(B) Close-up views of mature leaves of 2-month-old wild type and fcl1-1 mutant, showing phenotypic variations within a single mutant plant compared with the trifoliolate wild-type leaf.
(C) Scanning electron microscopy images of wild-type (left) and fcl1-1 (right) compound leaf primordia. Arrows indicate the boundary between terminal and lateral leaflet primordia developed at the P2 stage.
(D) Close-up views of mature leaves of the wild type (left) and the fcl1-1 mutant (right), showing the reduced petiole in the mutant. Bar = 2 cm.
(E) Epidermal peels of petioles of compound leaves on the fourth node of 2-month-old wild type (left) and fcl1-1 mutant (right).
(F) Measurements of petiole epidermal cell lengths of the wild type and the fcl1-1 mutant. Shown are means ± sd; n > 200.
(G) Measurements of petiole lengths of compound leaves at various developmental stages in 2-month-old wild type and fcl1-1 mutant. Shown are means ± sd; n = 10.
To examine the early events in compound leaf development that are altered in fcl1 mutants, we performed scanning electron microscopy analysis. The initiation of common leaf primordia from flanks of the SAM and the initiation of stipule and lateral leaflet primordia from the leaf margin appeared to be normal in the fcl1 mutants compared with wild-type plants (Figure 1C). During a late P2 stage (P for Plastochron) when lateral leaflet primordia separated from the terminal leaflet primordium in wild-type plants (Wang et al., 2008) (Figure 1C), boundaries between lateral and terminal leaflet primordia failed to develop properly, resulting in leaflet fusion observed in fcl1 mutants (Figures 1B and 1C).
In addition to the leaflet fusion phenotype, petiole development was also affected in the fcl1 mutants. In 2-month-old plants, petioles developed on the youngest leaf (the first expanded leaf at the shoot apex) were normal in fcl1 mutants, suggesting that the initial development of petioles was not affected in the mutants. However, petioles developed on the second and older leaves on the stem were significantly shorter in the fcl1 mutants than the wild-type counterparts (Figures 1D and 1G). Since the average length of petiole epidermal cells was indistinguishable between fcl1 mutants and wild-type plants, the reduced petiole elongation was not likely due to reduced cell elongation but instead may be due to compromised cell division activity in fcl1 mutants (Figures 1E and 1F).
Molecular Cloning and Characterization of FCL1
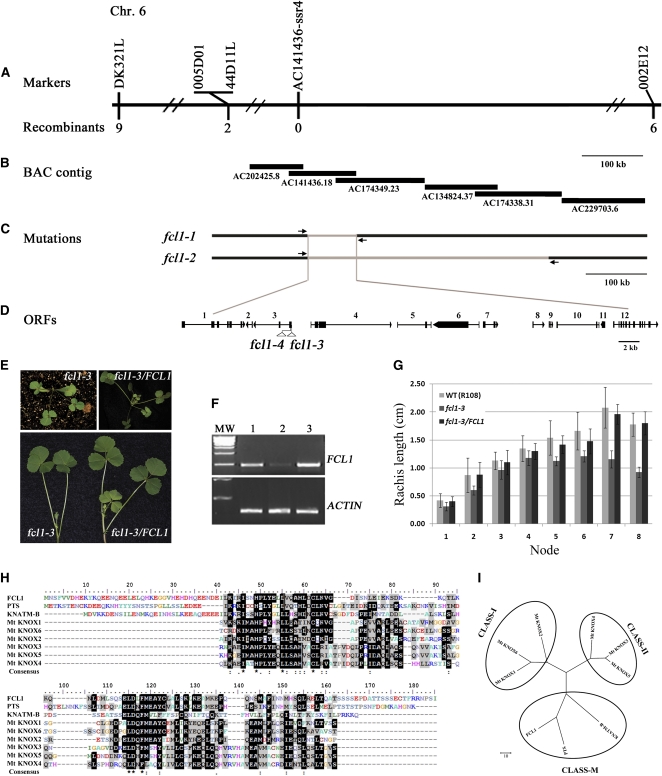
To isolate the FCL1 gene, we took a map-based approach. Genetic mapping results indicate that FCL1 was closely linked to 12 simple sequence repeat (SSR) markers on chromosome 6 (see Supplemental Figure 1 online). To fine-map the FCL1 locus, we generated a large F2 mapping population. Out of 787 F2 segregating plants, 213 plants were mutants and they were used to fine-map the FCL1 locus (see Supplemental Table 1 online). Mapping results showed that the FCL1 locus was flanked by two SSR markers, 005D01 and 002E12, and tightly linked to a newly developed SSR marker, AC141436-ssr4 (Figure 2A; see Supplemental Tables 2, 3, and 4 [for marker information] online). Using chromosomal walking and thermal asymmetric interlaced–PCR, we determined the deletion borders in both fcl1-1 and fcl1-2 mutants (Figures 2B and 2C; see Supplemental Figure 2 online). These results indicate that the region between coordinates 32,712 and 109,431 of the BAC clone AC141436 was deleted in both fcl1-1 and fcl1-2 (Figure 2C; see Supplemental Figure 2 online). Within this region are 12 annotated open reading frames (ORFs) (Figure 2D; see Supplemental Table 5 online). Using tissue-specific RT-PCR, the expression of six ORFs, Medtr6g080670, Medtr6g080690, Medtr6g080700, Medtr6g080720, Medtr6g080770, and Medtr6g080780, in the deletion region could be clearly detected in the vegetative shoot apices of wild-type plants (see Supplemental Figure 2B online).
Figure 2.
Map-Based Cloning of FCL1.
(A) The FCL1 locus was mapped to the region between SSR markers 005D01 and 002E12 on chromosome (Chr.) 6. AC141436-SSR4, a newly developed SSR marker, cosegregated with fcl1. The number of recombinants between markers and the FCL1 locus is shown underneath the markers.
(B) BAC contigs spanning the FCL1 locus.
(C) Deletion borders identified in fcl1-1 and fcl1-2 alleles. Arrows represent primers used to amplify deletion junctions.
(D) Orientation and exon-intron structures of 12 annotated ORFs within the common deletion region in fcl1-1 and fcl1-2 alleles. Numbers above ORFs from 1 to 12 correspond to the following genes: 1, Medtr6g080670; 2, Medtr6g080680; 3, Medtr6g080690; 4, Medtr6g080700; 5, Medtr6g080710; 6, Medtr6g080720; 7, Medtr6g080730; 8, Medtr6g080740; 9, Medtr080750; 10, Medtr6g080760; 11, Medtr6g080770; and 12, Medtr6g080780. Solid boxes denote exons, and horizontal lines denote introns. Open triangles represent Tnt1 insertion sites in ORF3 in fcl1-3 and fcl1-4 alleles.
(E) Genetic complementation of the fcl1-3 mutant. Shown are 3-week-old fcl1-3 mutant (top left), fcl1-3 stably transformed with the full-length wild-type FCL1 genomic sequence (top right), and compound leaves developed from leaf axils in 2-month-old plants (bottom left, fcl1-3; bottom right, fcl1-3 complemented line).
(F) RT-PCR analysis of FCL1 gene expression in the wild type, fcl1-3, and fcl1-3 transformed with the wild-type FCL1 gene. An ACTIN gene was used as an internal control.
(G) Measurements of rachis length of compound leaves of fcl1-3 and fcl1-3 transformed with the wild-type (WT) FCL1 gene.
(H) Amino acid sequence alignments of the full-length FCL1, its close homologs from tomato and Arabidopsis, PTS and KNATM-B, respectively, and the MEINOX domains of six M. truncatula KNOX proteins.
(I) Neighbor-joining, maximum parsimony, and UPGMA phylogenetic tree analysis of FCL1, its close homologs from tomato and Arabidopsis, and the canonical KNOX proteins from M. truncatula.
[See online article for color version of this figure.]
To identify the FCL1 gene from the set of 12 candidate genes deleted in both fcl1-1 and fcl1-2, we first screened a tobacco (Nicotiana tabacum) Tnt1 retrotransposon insertion mutant collection of M. truncatula (cv R108) (Tadege et al., 2008) and isolated one mutant with a weak leaflet fusion phenotype. Leaflets developed in the first three adult leaves were either fused to form simplified leaves or clustered without the rachis structure in the mutant (Figure 2E; see Supplemental Figure 3 online). Although leaflets in older leaves of the mutant were separated from each other, the length of rachis was significantly shorter in later developed leaves in the mutant than in the corresponding wild-type leaves (Figures 2E and 2G). F1 plants derived from genetic crosses between the third mutant and fcl1-1 exhibit the weak leaflet fusion phenotype similar to the third mutant, confirming that it is a new fcl1 allele (see Supplemental Figure 3 online). Accordingly, we named it fcl1-3. Flanking sequence analysis indicated that fcl1-3 carries a tobacco Tnt1 retrotransposon in the 5′ untranslated region (UTR) of the third ORF in the deletion region, corresponding to Medtr6g080690 (see Supplemental Figure 3 online). RT-PCR analysis indicated that the expression of Medtr6g080690 was not abolished but was substantially reduced in fcl1-3 compared with the corresponding wild type (cv R108) (Figure 2F). Thus, the reduced expression of Medtr6g080690 is consistent with the weak leaflet fusion phenotype observed in this mutant.
Using a reverse genetic screen, we uncovered an additional allele, fcl1-4, with the Tnt1 retrotransposon inserted in the first exon of FCL1 (see Supplemental Figure 3 online). Mutant plants exhibit strong compound leaf phenotype similar to the deletion alleles fcl1-1 and fcl1-2 (see Supplemental Figures 3 and 4 online).
To further confirm the identity of FCL1, we performed genetic complementation tests. A 7.8-kb genomic sequence from wild-type plants (cv Jemalong A17), including 2664-bp 5′ upstream sequence, 4242-bp coding region, and 1737-bp 3′ downstream region, was stably introduced into both fcl1-1 and fcl1-3 mutants via Agrobacterium tumefaciens–mediated plant transformation. We show that this wild-type sequence complemented both fcl1-1 and fcl1-3 mutants (Figures 2E to 2G; see Supplemental Figure 5 online). Introducing ORF6, a putative zinc finger transcription factor gene, which is deleted in the fcl1-1 and fcl1-2 mutants, did not rescue the compound leaf phenotype of the fcl1-1 mutant (see Supplemental Figure 6 online). Taken together, these results collectively confirm that the FCL1 gene corresponds to Medtr6g080690.
M. truncatula FCL1 Encodes a Class M KNOX Protein
To identify the full-length transcript, we performed both 5′- and 3′-rapid amplification of cDNA ends (RACE). Sequence analysis indicates that only one full-length cDNA of 794 bp long was amplified, including a 99-bp 5′-UTR, 486-bp coding region, and 209-bp 3′-UTR (see Supplemental Figure 7 online). Comparison of genomic and cDNA sequences revealed that FCL1 is composed of three exons and two introns. FCL1 encodes a polypeptide of 161 amino acids.
BLAST analysis of the National Center for Biotechnology Information (NCBI) protein database (Altschul et al., 1997) revealed that FCL1 encodes a truncated KNOX that contains both KNOX1 and KNOX2 domains (collectively called the MEINOX domain) but lacks the homeodomain (Figure 2H; see Supplemental Figure 7 online). Amino acid sequence comparison indicated that the FCL1 protein shares a high sequence similarity with the Arabidopsis KNATM-B (63% similarities and 43% identities) and tomato PTS/TKD1 (60% similarities and 48% identities), two recently identified class M homeodomainless KNOX proteins in Arabidopsis and tomato, respectively (Kimura et al., 2008; Magnani and Hake, 2008) (Figure 2H). Neighbor-joining, maximum parsimony, and UPGMA phylogenetic tree analysis indicated that FCL1 is grouped with PTS/TKD1 and KNATM-B but separated from the canonical class I and class II KNOX proteins from M. truncatula (Figure 2I; see Supplemental Data Set 1 online). BLAST searches of genome sequences and subsequent sequence annotation further revealed the existence of homologous sequences in closely related species such as L. japonicus and soybean, other eudicot species such as poplar (Populus trichocarpa) and grape (Vitis vinifera), and monocot species such as rice (Oryza sativa) and maize (see Supplemental Figure 8 online). As expected, duplicated genes are present in the soybean and rice genomes due to whole-genome duplications in these species (see Supplemental Figure 8 online).
Expression Pattern of FCL1 and Subcellular Localization of the Encoded Protein
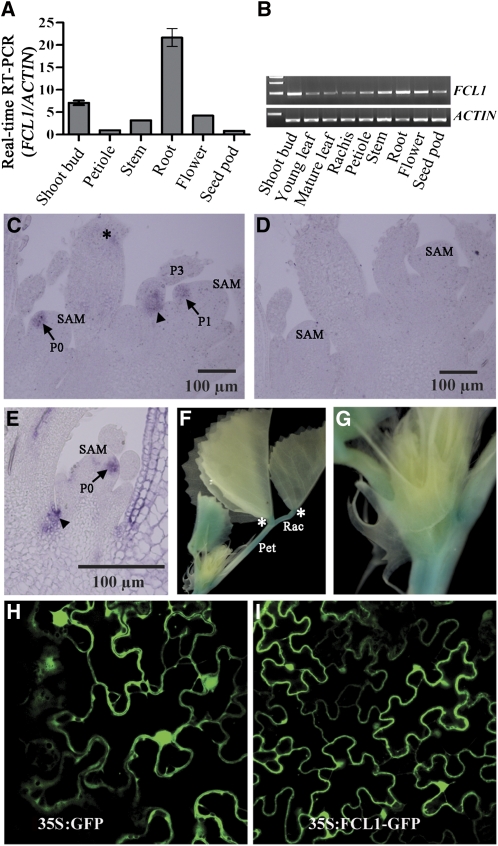
RT-PCR analyses revealed that FCL1 is expressed in vegetative shoot apices, petioles, stems, roots, flowers, and seedpods with the highest level of expression being detected in roots and vegetative shoot buds (Figures 3A and 3B). The tissue-specific expression pattern is consistent with the microarray-based in silico analysis of FCL1 expression (Benedito et al., 2008) (see Supplemental Figure 9 online).
Figure 3.
Tissue-Specific Expression of FCL1 and Subcellular Localization of the Encoded Protein.
(A) and (B) RT-PCR analysis of tissue-specific expression of FCL1.
(C) to (E) RNA in situ hybridization analysis of FCL1 expression in vegetative shoot buds. A sense probe was used as a negative control (D).
(F) and (G) FCL1promoter:GUS reporter gene expression in shoot apices in stable transgenic lines.
(H) Subcellular localization of 35S:GFP in tobacco epidermal cells.
(I) Subcellular localization of 35S:FCL1-GFP in tobacco epidermal cells.
P, plastochron; Pet, petiole; Rac, rachis. Arrowheads in (C) and (E) indicate proximal adaxial domain, and asterisks in (C) and (F) indicate the petiolule.
RNA in situ hybridization was performed to examine tissue- and cell-specific expression patterns of FCL1 in vegetative shoot apices. We show that FCL1 transcripts were detected in leaf primordia at P0 and early P1 stages (Figures 3C and 3E, arrows). At later stages, FCL1 transcripts were detected at boundaries between leaf primordia and the SAM and at the proximal adaxial domain of developing leaf primordia (Figures 3C and 3E, arrowheads), which is the future site of the petiole. In addition, RNA in situ hybridization results show that FCL1 transcripts were not detected in the SAM (Figures 3C and 3E). As a negative control, a sense probe did not give any signal (Figure 3D).
To confirm and further investigate FCL1 expression pattern, we introduced an FCL1promoter:uidA reporter gene construct into wild-type M. truncatula plants (cv R108) via A. tumefaciens–mediated stable plant transformation. We show that the reporter gene expression was detected in the root, cotyledon, petiole, rachis, and young developing leaves in homozygous T3 plants (Figures 3F and 3G). In developing leaves, the reporter gene was weakly expressed in vascular tissues.
The FCL1 protein does not contain any known subcellular localization signals. To determine FCL1 subcellular localization, we transiently expressed FCL1 fused to the green fluorescent protein (FCL1-GFP) under control of the constitutive cauliflower mosaic virus 35S promoter in tobacco leaves. The FCL1-GFP fusion protein was detected in both cytoplasm and nucleus in tobacco epidermal cells similarly to the localization pattern of free GFP (35S:GFP) (Figures 3H and 3I). The subcellular localization pattern suggests that FCL1 may move freely between nucleus and cytoplasm likely due to its small size.
Expression of Class I KNOX Genes Was Not Altered in Loss-of-Function fcl1 Mutants
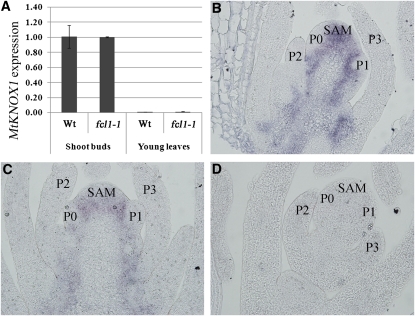
Previous studies have shown that class I KNOX proteins are not detectable in compound leaf primordia in IRLC legumes (Champagne et al., 2007). To test whether loss-of-function mutations in FCL1 affect spatial-temporal expression of class I KNOX genes in M. truncatula, we examined expression of three class I KNOX genes, KNOX1, KNOX2, and KNOX6, in the fcl1-1 mutant. Real-time RT-PCR analysis indicated that KNOX1, KNOX2, and KNOX6 were similarly expressed in the vegetative shoot buds in the wild type and fcl1-1 mutant (Figure 4A; see Supplemental Figures 10A and 10B online). Also, their expression was similarly diminished in young leaves in the wild type and fcl1-1 mutant (Figure 4A; see Supplemental Figures 10A and 10B online). RNA in situ hybridization analysis indicated that KNOX1 transcripts were present in the SAM but excluded from incipient and subsequent leaf primordia in both the wild type and fcl1-1 mutant (Figures 4B and 4C), consistent with the quantitative RT-PCR results and with previous studies. As a negative control, a sense probe did not detect any signals (Figure 4D). These results indicate that FCL1 does not appear to regulate temporal-spatial expression of class I KNOX genes in M. truncatula.
Figure 4.
KNOX1 Expression in the fcl1-1 Mutant.
(A) Real-time RT-PCR analysis of KNOX1 expression in vegetative shoot buds and young leaves in the wild type (Wt) and fcl1-1 mutant. An M. truncatula ACTIN gene was used as an internal control.
(B) and (C) RNA in situ hybridization analysis of KNOX1 expression in shoot apices of the wild type (B) and fcl1-1 mutant (C).
(D) A negative control for RNA in situ hybridization using a sense probe and an adjacent section of shoot apices of the fcl1-1 mutant. P, plastochron.
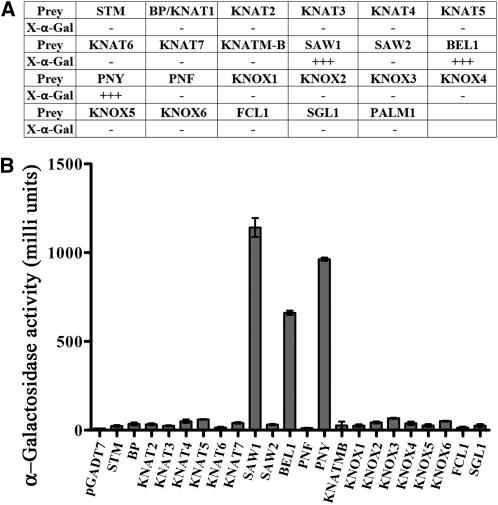
FCL–BELL and FCL–KNOX Protein Interactions
Arabidopsis KNATM-B contains a full MEINOX domain and has been shown to interact with the homeodomain BELL proteins, including BEL1, SAW1, SAW2, and PNY, but not the PNY paralog PNF (Magnani and Hake, 2008). The M. truncatula FCL1 is predicted to contain an intact MEINOX domain (Figure 2H; see Supplemental Figure 7 online). To investigate whether FCL1 interacts similarly as KNATM-B, we performed yeast two-hybrid assays using FCL1 as the bait and Arabidopsis BELL proteins as preys. To our surprise, unlike KNATM-B, which activates reporter gene expression by itself in the yeast two-hybrid assays (Magnani and Hake, 2008), FCL1 did not activate reporter gene expression by itself (Figures 5A and 5B). Furthermore, we observed that FCL1 strongly interacted with a subset of the BELL proteins, including PNY, SAW1, and BEL1, but not with SAW2 and PNF (Figures 5A and 5B). These interactions were further confirmed by quantification of the reporter gene expression in yeast (Figure 5B). Thus, FCL1 interacts with BELL proteins with slightly different specificities compared with the Arabidopsis KNATM-B. Interestingly, both FCL1 and KNATM-B did not interact with PNF, the PNY paralog (Magnani and Hake, 2008) (Figures 5A and 5B).
Figure 5.
Yeast Two-Hybrid Assays for Protein–Protein Interactions.
(A) X-α-gal activity staining for protein–protein interactions between FCL1 and the following proteins: STM, BP/KNAT1,KNAT2, KNAT3, KNAT4, KNAT5, KNAT6, KNAT7, KNATM-B, SAW1, SAW2, BEL1, PNF, PNY, KNOX1, KNOX2, KNOX3, KNOX4, KNOX5, KNOX6, FCL1, SGL1, and PALM1.
(B) Quantification of protein–protein interactions. Shown are means ± sd; n = 3.
KNATM-B has been shown to interact strongly with BP/KNAT1 and weakly with KNAT3 and KNAT4 in the yeast two-hybrid assays (Magnani and Hake, 2008). By contrast, FCL1 did not interact with BP/KNAT1 and other KNOX proteins from Arabidopsis (Figure 5). We also tested FCL1 interactions with KNOX proteins from M. truncatula. We show that FCL1 did not interact with the M. truncatula KNOX proteins in the yeast two-hybrid assays. Although we observed weak X-gal staining from interactions between FCL1 and M. truncatula KNOX3 and KNOX4 in a prolonged incubation, quantification results suggest that the reporter gene expression was only slightly above the background level (Figure 5B). In addition, FCL1 did not interact with itself, KNATM-B, SGL1, or PALM1 (Figure 5).
Genetic Interactions between fcl1 and sgl1
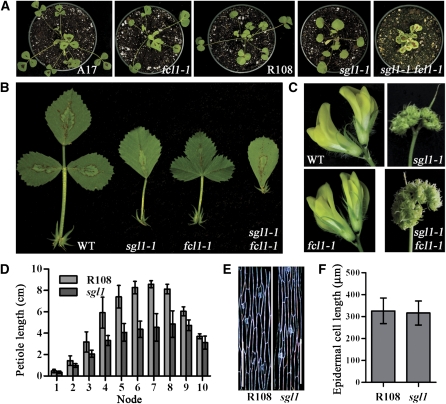
Our genetic analysis suggests that FCL1 is required for the development of boundaries between terminal and lateral leaflet primordia at the P2 stage and development of petioles at a later stage during compound leaf development in M. truncatula (Figure 1). Previously, we have shown that SGL1, the M. truncatula FLO/LFY/UNI ortholog, is required for the initiation of lateral leaflet primordia and development of petioles (Wang et al., 2008). The involvement of SGL1 in petiole development appears to be similar to that of FCL1. To examine genetic interactions between fcl1 and sgl1, we made crosses between sgl1-1 and fcl1-1 and generated sgl1 fcl1 double mutants (see Supplemental Table 6 online). Mature leaves of sgl1 fcl1 double mutants were simple, resembling those of sgl1 mutants (Figures 6A and 6B). Flowers developed in the double mutants also resembled those of sgl1 mutants (Figure 6C). These results indicate that sgl1 is genetically epistatic to fcl1 in both leaf patterning and flower development. By contrast, both sgl1 and fcl1 had about a 50% reduction in petiole length compared with their wild-type counterparts, and sgl1 fcl1 double mutants completely lacked petioles (Figures 6A, 6B, and 6D), suggesting that SGL1 and FCL1 act additively and both are required for petiole development. Since there were no significant differences in epidermal cell length of petioles between the wild type and sgl1 mutants (Figures 6E and 6F), the reduced petiole development in sgl1 mutants is likely due to reduced cell division as in fcl1 mutants (Figures 1D to 1G). RT-PCR analysis of sgl1 and fcl1 mutants suggests that SGL1 and FCL1 do not appear to regulate each other’s transcript levels (Figures 7B and 7D). In addition, SGL1 and FCL1 did not interact with each other in the yeast two-hybrid assays (Figure 5).
Figure 6.
Genetic Interactions between fcl1 and sgl1.
(A) Three-week-old seedlings (from left to right: Jemalong A17, fcl1-1, R108, sgl1-1, and sgl1 fcl1 plants).
(B) Comparison of compound leaf morphology (from left to right: Jemalong A17, sgl1-1, fcl1-1, and sgl1-1 fcl1-1). WT, wild type.
(C) Comparison of flower morphology (clockwise from top left: Jemalong A17, sgl1-1, sgl1-1 fcl1-1, and fcl1-1).
(D) Measurements of petiole lengths of the wild type (R108) and sgl1-1. Shown are means ± sd; n = 10.
(E) Epidermal peels of petioles in the wild type (R108; left) and sgl1-1 mutant (right).
(F) Measurements of epidermal cell lengths of petioles in the wild type (R108) and sgl1-1 mutant. Shown are means ± sd; n > 200.
[See online article for color version of this figure.]
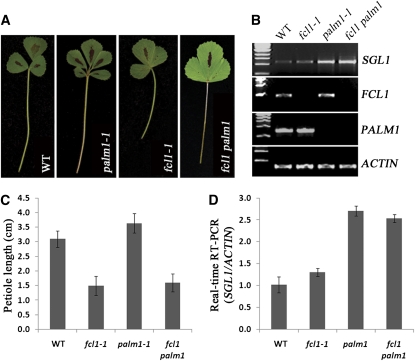
Figure 7.
Genetic Interactions between fcl1 and palm1.
(A) Compound leaf morphology (from left to right: Jemalong A17, palm1-1, fcl1-1, and fcl1-1 palm1-1). WT, wild type.
(B) RT-PCR analysis of SGL1, FCL1, PALM1, and ACTIN gene expression in vegetative shoot buds of the wild type (Jemalong A17), fcl1-1, palm1-1, and fcl1-1 palm1-1 mutants.
(C) Measurements of petiole length of the wild type (Jemalong A17), fcl1-1, palm1-1, and fcl1-1 palm1-1 mutants.
(D) Real-time RT-PCR analysis of SGL1 expression in the wild type (Jemalong A17), fcl1-1, palm1-1, and fcl1-1 palm1-1 mutants. Shown are means ± sd; n = 3.
[See online article for color version of this figure.]
Genetic Interactions between fcl1 and palm1
Another key regulator of compound leaf development in M. truncatula is the recently identified PALM1 gene, which encodes a Cys(2)His(2) zinc finger transcription factor (Chen et al., 2010; Ge et al., 2010). Mature leaves of loss-of-function palm1 mutants are pentafoliolate with longer petioles, in contrast with trifoliolate wild-type leaves (Chen et al., 2010) (Figure 7A). To investigate genetic interactions between fcl1 and palm1, we generated fcl1 palm1 double mutants (see Supplemental Table 6 online). The fcl1 palm1 double mutants exhibited fused compound leaves and short petioles, resembling those of fcl1 mutants (Figures 7A and 7C). These results indicate that fcl1 is genetically epistatic to palm1 in both leaf complexity and petiole development. Previous studies have shown that the increase in leaflet number and petiole elongation is attributed to a significantly increased level of SGL1 expression in palm1 mutants (Chen et al., 2010) (Figures 7B and 7D). We show that the increase in SGL1 expression was not affected in the fcl1 palm1 double mutants (Figures 7B, 7D, and 8). Interestingly, despite an increase in SGL1 expression, fcl1 palm1 double mutants did not produce extra leaflets (Figures 7A and 8). In loss-of-function fcl1 mutants, the PALM1 transcript level was not altered as indicated by RT-PCR data (Figures 7B and 7D). Although these results suggest no direct transcriptional regulation of PALM1 by FCL1, it is still possible that subtle or tissue-specific differences in PALM1 expression might occur but were not detected in fcl1 mutants due to limitations of RT-PCR. Our data also show that FCL1 and PALM1 did not interact in yeast two-hybrid assays (Figure 5). Taken together, these results suggest that, although FCL1 is not required for the upregulation of SGL1 expression in the loss-of-function palm1 mutants, it is required for the development of extra leaflet primordia in the absence of the PALM1 gene (Figure 8).
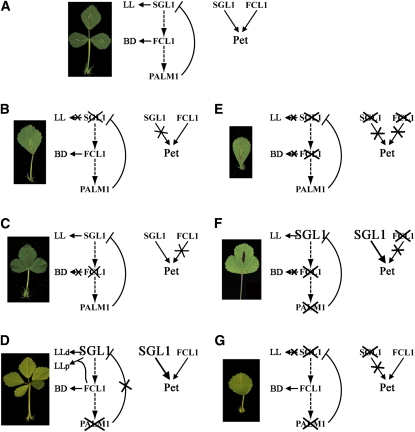
Figure 8.
A Genetic Model of Compound Leaf Development in M. truncatula.
(A) Wild-type M. truncatula plants exhibit trifoliolate compound leaves. Double mutant analyses indicate that sgl1 is epistatic to fcl11, and fcl1 is epistatic to palm1 in terms of leaf complexity (dashed arrows; also see below). In terms of leaf proximal-distal axis development, SGL1 and FCL1 are additive (arrows). It has been shown that PALM1 represses SGL1 transcription by directly binding to its promoter sequence (Chen et al., 2010). Together, SGL1, FCL1, and PALM1 define the trifoliolate morphology of wild-type leaves. BD, boundary between terminal and lateral leaflet primordia; LL, lateral leaflet primordia; Pet, petiole.
(B) Loss-of-function mutations in SGL1 result in plants with simple leaves and reduced petioles, consistent with its role in the initiation of lateral leaflet primordia at an early stage and the leaf proximal-distal axis development at a late stage (Wang et al., 2008).
(C) Loss-of-function mutations in FCL1 result in fusion of leaflets and reduced petioles, consistent with its role in boundary separation and the proximal-distal axis development (this study).
(D) Loss-of-function mutations in PALM1 results in proliferation of compound leaves and slightly elongated petioles. In palm1 mutants, the SGL1 transcript level is significantly increased, consistent with its negative role in transcriptional regulation of SGL1 expression in wild-type plants (Chen et al., 2010).
(E) The sgl1 fcl1 double mutants exhibit simple leaves, indicating that sgl1 is epistatic to fcl1 in terms of leaf complexities. sgl1 and fcl1 single mutants have ~50% reduction in petiole length, and sgl1 fcl1 double mutants completely lack petioles, indicating an additive interaction between SGL1 and FCL1 in leaf proximal-distal axis development.
(F) The fcl1 palm1 double mutants exhibit leaflet fusion similar to the fcl1 mutants, indicating that fcl1 is epistatic to palm1 (this study). Interestingly, in fcl1 palm1 double mutants, the SGL1 transcript level is still significantly increased but no extra leaflets are developed, suggesting that FCL1 is not required for the upregulation of SGL1 but required for the development of extra leaflets in palm1 single mutants (this study).
(G) The sgl1 palm1 double mutants exhibit simple leaves with reduced petioles similarly as the sgl1 single mutants, indicating that sgl1 is epistatic to palm1 (Chen et al., 2010).
[See online article for color version of this figure.]
DISCUSSION
Phylogenetic analysis of leaf evolution suggests that compound leaves arose numerous times from the ancestral form, during the evolution of angiosperms (Bharathan et al., 2002; Champagne and Sinha, 2004). Consistent with this, recent molecular genetic studies demonstrate that KNOX1- and UNI/SGL1-dependent processes are required for compound leaf development in most compound-leafed species, including tomato and legume species belonging to the IRLC. In some legume species with compound leaves, KNOXI transcripts and encoded proteins are excluded from the incipient and developing leaf primordia, supporting that they are not associated with compound leaf development in these legume species. Interestingly, developmental programs are still responsive to ectopic expression of KNOXI genes in legumes, consistent with the notion that UNI/SGL1 transcription factors are recruited to function in place of KNOXI proteins in compound leaf development in IRLC legumes. However, molecular mechanisms of compound leaf development in legumes remain to be elucidated.
Here, we report isolation of two deletion mutants in M. truncatula, in which FCL1 is completely deleted, and two insertion mutants, in which FCL1 expression is partially or completely compromised. Loss-of-function fcl1 mutants exhibit various compound leaf defects, ranging from leaflet fusion to simple leaves and leaflet clustering. In addition to leaflet patterning defects, petiole elongation is also affected in fcl1 mutants. In the partial loss-of-function fcl1 mutant (fcl1-3), compound leaf development is only mildly affected. In the first three adult leaves developed on the main stem and from axils of primary leaves, leaflets lack rachis and are clustered. Although leaflets of later developed leaves are separated from each other, the length of both rachis and petiole is significantly reduced in the weak fcl1-3 mutant compared with the wild type. The effect of FCL1 on compound leaf development suggests that FCL1 plays two distinct roles in compound leaf development (i.e., boundary separation of leaflet primordia at the P2 stage and the proximal-distal axis development at later stages [after P4]). Because the petiole epidermal cell length is not affected, the reduced petiole development in fcl1 mutants is likely due to a compromised cell division activity but not cell elongation. We hypothesize that FCL1 is required to maintain windows of organogenetic activities at the leaf margin at an early stage for boundary separation and at a later stage for petiole development. Mutations in FCL1 shorten the windows of organogenetic activities and promote leaf maturation, leading to clustering and fusion of leaflets, simple leaves, and reduced petiole development. This hypothesis is supported by genetic analyses of two other compound leaf mutants. Loss-of-function sgl1 mutants exhibit simple leaves and reduced petioles, and loss-of-function palm1 mutants exhibit an increased level of SGL1 gene expression and proliferation of leaflets and increased petioles (Wang et al., 2008; Chen et al., 2010) (Figure 8). During early stages of leaf development, SGL1 is epistatic to FCL1, highlighting a requirement for SGL1 in the development of trifoliolate compound leaves (Figure 8). During later stages of leaf development, SGL1 and FCL1 are involved in separate processes and both are required for petiole development (Figure 8). Previous studies suggest that the proliferation of compound leaves in palm1 mutants is attributed to an increase in SGL1 expression (Chen et al., 2010). However, our double mutant analyses indicate that an increase in SGL1 gene expression alone is not sufficient for the proliferation of compound leaves in palm1 mutants. Instead, both SGL1 and FCL1 are required for the development of pentafoliolate leaves in the absence of PALM1. The results are consistent with a role of FCL1 in maintaining windows of organogenetic activities at the leaf margin required for compound leaf development in M. truncatula.
FCL1 encodes a truncated KNOX that contains both KNOX1 and KNOX2 domains (collectively called the MEINOX domain) but lacks the homeodomain. Phylogenetic analysis indicates that FCL1 is a member of the class M KNOX proteins, including two recently identified members from tomato and Arabidopsis, PTS/TKD1 and KNATM-B, respectively. The function of PTS/TKD1 and KNATM-B needs to be further elucidated because no loss-of-function mutants of these two genes have been isolated so far. Nonetheless, ectopic expression of KNATM-B results in elongated petioles and narrower, shorter, and serrated leaves in Arabidopsis (Magnani and Hake, 2008). In tomato plants, the semidominant Petroselinum (Pts) mutant, caused by upregulation of PTS/TKD1 due to mutations in its promoter region, exhibits a proliferation of compound leaves, affecting both primary and secondary leaflets (Kimura et al., 2008). The gain-of-function phenotypes in Arabidopsis with simple leaves and tomato with compound leaves are consistent with expected results from compound leaf defects of the loss-of-function M. truncatula fcl1 mutants. Taken together, these observations support a general role of the class M KNOX proteins in the development of both simple and compound leaves in diverse species.
The fcl1-1 and fcl1-2 mutant alleles derived from fast neutron bombardment mutagenesis carry a common deletion of ~77 kb on chromosome 6. Commonly deleted in fcl1-1 and fcl1-2 alleles are 12 putative genes, six of which are expressed in vegetative shoot buds and possibly play a role in shoot development. In this study, we showed that FCL1 plays important roles in compound leaf development. Interestingly, one of the deleted genes, Medtr6g080720, is also expressed in the shoot apices and encodes a putative zinc finger domain transcription factor that belongs to the highly conserved human NF-X1 family of transcription factors. Although our data indicate that Medtr6g080720 is not involved in compound leaf development, previous studies have shown that the two Arabidopsis homologs, NFXL1 and NFXL2, play antagonistic roles in abiotic and biotic stress responses in plants (Lisso et al., 2006; Asano et al., 2008). It would be interesting to test whether Medtr6g080720 plays a similar role as its Arabidopsis homolog does.
How class M KNOX proteins regulate leaf development remains to be elucidated. It has been shown that KNATM-B and PTS/TKD1 form heterodimers with BELL proteins (Kimura et al., 2008; Magnani and Hake, 2008). The Arabidopsis BELL proteins SAW1 and SAW2 interact with the canonical KNOXI proteins and act redundantly to repress KNOX expression in leaves (Kumar et al., 2007). Through interactions with the BELL protein BIPINNATA (BIP), PTS/TKD1 antagonizes BIP and KNOX1 interactions and interferes with BELL and KNOX regulatory networks (Kimura et al., 2008). In a large number of compound-leafed legume plants that belong to the IRLC, KNOXI proteins are permanently downregulated in leaf primordia and are likely not associated with compound leaf development in these species (Champagne et al., 2007). The involvement of a truncated KNOX protein (FCL1) in compound leaf development in M. truncatula, an IRLC legume, is surprising and poses an interesting question of the role of FCL1 in compound leaf development. Although further investigation is required to understand the role of FCL1 in compound leaf development, we have shown that FCL1 selectively interacts with a subset of BELL proteins, including SAW1, BEL1, and PNY, but not with SAW2 and PNF in yeast two-hybrid assays. In Arabidopsis, SAW1 and BEL1 transcripts can be detected in developing leaves (Kumar et al., 2007). It is possible that M. truncatula SAW1 and BEL1 homologs are also expressed in leaves and play a role in compound leaf development through interactions with FCL1. The selectivity of FCL1 interactions with BELL proteins is slightly different from KNATM-B since the latter strongly interacts with SAW2 (Magnani and Hake, 2008). In addition, unlike KNATM-B, FCL1 does not interact with the KNOXI protein, BP/KNAT1. RNA in situ hybridization results indicate that FCL1 is expressed in the incipient leaf primordia (P0) but not in the SAM. Thus, FCL1 expression appears not to overlap with that of KNOXI genes in M. truncatula. These results, together with others, collectively suggest that FCL1 may regulate compound leaf developmental processes through specific protein–protein interactions with BEL1-like homeodomain proteins. Alternately, FCL1 may be involved in regulating NAM/CUC transcription factor–dependent and/or auxin transport-dependent regulatory networks known to be required for boundary separation and various developmental processes in diverse species. Future investigation will be aimed at elucidating the regulatory roles of the class M KNOX protein in leaf development in legume.
METHODS
Plant Materials
Seeds of Medicago truncatula cv Jemalong A17 were exposed to fast neutron radiation at the dosage level of 40 gray and germinated in a greenhouse with a controlled environment. Approximately 30,000 M2 plants derived from 5000 M1 lines were screened, resulting in the isolation of fcl1-1 (M368) and fcl1-2 (M602). The fcl1-3 and fcl1-4 alleles were isolated from a collection of tobacco (Nicotiana tabacum) Tnt1 retrotransposon insertion mutants as previously described (Wang et al., 2008; Chen et al., 2010; Cheng et al., 2011). fcl1-1 was backcrossed to the parental line (cv Jemalong A17) for two generations. BC2 plants were used for phenotypic characterization. fcl1-2 was backcrossed to the Jemalong A17 line for four generations. BC4 plants were used.
Scanning Electron Microscopy
Shoot apices of 2- to 4-week-old seedlings were subjected to vacuum infiltration in a fixative solution (3% glutaraldehyde in 25 mM phosphate buffer, pH 7.0) for 1 h and then incubated at 4°C overnight. Plant tissues were further fixed with 1.0% osmium tetroxide in the same phosphate buffer overnight and dehydrated in a graded ethanol series. Scanning electron microscopy was performed as described previously (Wang et al., 2008).
Genetic Mapping
F2 mapping populations were generated from crosses between fcl1-2 and a polymorphic ecotype, M. truncatula cv Jemalong A20. Initially, an F2 population of 254 individuals (175 wild-type-like and 79 mutants; see Supplemental Table 1 online) was used in a bulk segregant analysis to construct a linkage map of the fcl1 locus. A total of 267 SSR markers distributed across the eight chromosomes of M. truncatula (http://www.medicagohapmap.org/?genome) were used to construct the linkage map. Subsequently, an F2 population of 533 individuals (399 wild-type-like and 134 mutants; Supplemental Table 1 online) was generated. A total of 213 fcl1 mutants were used to fine map the FCL1 locus. Total genomic DNA was isolated from leaf tissues of individual plants grown in a greenhouse using a protocol previously described (Saghai-Maroof et al., 1984). PCR amplification and product separation were performed as previously described (Yu et al., 2006).
For genetic mapping, we made four DNA bulks, two from 15 wild-type plants each and two from 15 fcl1-1 mutant plants each by pooling equal amounts of DNA samples prepared from individual F2 plants. DNA samples from the two wild-type parental lines M. truncatula cv Jemalong A17 and Jemalong A20 were also included in the analysis to validate the markers.
Recombination frequency was calculated using the maximum likelihood estimator, P = (h + 2b)/2n, where P is the recombination frequency, n is homozygous recessive individuals in F2, h is the number of recombinant homozygotes, and b is the number of recombinant heterozygotes (Allard, 1956; Hinze et al., 1991). The Kosambi mapping function was used to estimate genetic distances between markers and the fcl1-2 locus as previously described (Koornneef et al., 1983). We constructed the linkage map using JoinMap (v3.0) (van Ooijen and Voorrips, 2001). For chromosomal walking, we designed PCR primers based on BAC sequences in the mapped region using Primer3 (Rozen and Skaletsky, 2000).
Bulked segregant analysis revealed that 12 of the 267 SSR markers located on chromosome 6 are cosegregated with the fcl1-2 locus. To construct a linkage map, we determined recombination frequencies and map positions for the 12 SSR markers using 254 individuals from an F2 mapping population. Based on this analysis, the fcl1-2 locus was mapped to the vicinity of four SSR markers, Mitc250, 005D01, 002E12, and 002F12, on chromosome 6 (see Supplemental Figure 1A online).
To fine-map the fcl1-2 locus, we examined a total of 213 fcl1-2 mutant plants identified from two F2 mapping populations of 787 individuals using the following SSR markers located in the region covered by three BAC clones (AC174349, AC141436, and AC202465) on chromosome 6: DK321L, 44D11L, 005D01, 002E12, 002F12, 18A5R, 19O4L, and AC141436-ssr4 (see Supplemental Tables 2 and 3 and Supplemental Figures 1B and 2 online). AC141436-SSR4 was designed based on the BAC clone AC141436 (see Supplemental Table 4 online).
Twenty mutant plants were identified carrying recombination between the parental Jemalong A20 allele and the fcl1-2 locus (see Supplemental Table 3 online). Two plants carried both parental alleles of the SSR markers 44D11L and 005D01, and six plants carried both parental alleles of the SSR marker 002E12 (see Supplemental Table 3 online; highlighted in red). These results indicate that two recombination events occurred between 44D11L and fcl1-2, and 005D01 and fcl1-2, and six recombination events occurred between 002E12 and fcl1-2. In addition, these eight individuals only carried the mutant allele of the AC141436-ssr4 marker. These fine-mapping results indicate that the fcl1-2 locus was likely located within the BAC clone AC141436 and tightly linked to the SSR marker AC141436-ssr4 (see Supplemental Figure 1B online).
To delineate deletion borders, we initiated chromosomal walking. PCR primers were designed to scan the region covered by BAC clones AC174349, AC141436, and AC202465 of chromosome 6 (see Supplemental Table 4 online). Regular PCR and thermal asymmetric interlaced–PCR amplifications and subsequent sequencing of deletion junctions indicate that the deletion in the fcl1-1 allele (M368) occurred within an ~80-kb interval between the coordinates 29,233 and 109,431 of the BAC clone AC141436, and the deletion in the fcl1-2 allele (M602) occurred within a large genomic interval starting from the coordinate 32,712 of the BAC clone AC141436 and ending in the BAC clone AC174338 (see Supplemental Figure 2 online). Thus, a genomic interval of ~76 kb between 32,712 and 109,431 of the BAC clone AC141436 was deleted in both the fcl1-1 and fcl1-2 alleles.
5′- and 3′-RACE, RT-PCR, and Real-Time RT-PCR
Total RNA was prepared using the RNeasy plant mini kit (Qiagen). Residual genomic DNA was removed using a DNA-free kit (Ambion). The 5′- or 3′-RACE was performed using an RLM-RACE kit (Invitrogen) according to the manufacturer’s instructions. cDNA synthesis for RT-PCR and real-time RT-PCR was performed using SuperScript III reverse transcriptase (Invitrogen) starting with 2 μg of total RNA in a 20-μL reaction mix with oligo(dT)15 primer (Promega). Real-time RT-PCR was performed as previously described (Laxmi et al., 2008). Real-time RT-PCR was performed using a 7900HT Fast real-time PCR system (Applied Biosystems). SDS2.2.1 software (Applied Biosystems) was used to analyze the melt curve to confirm the single amplification. PCR efficiency was estimated using the LinRegPCR program (Ramakers et al., 2003), and the transcript level was determined in reference to the expression of the M. truncatula ACTIN gene (Medtr3g095530).
Genetic Complementation of M. truncatula fcl1 Mutants
The BAC clone AC141436 was digested with HindIII to generate a 7.8-kb genomic DNA fragment, which was subsequently ligated to HindIII-digested pCAMBIA3300 vector (CAMBIA). The final construct designated as pFCL1:FCL1 was sequenced to confirm no sequence errors. The construct was introduced into the Agrobacterium tumefaciens EHA105 strain using electroporation. The fcl1-1 and fcl1-3 alleles were transformed following the protocol previously reported (Cosson et al., 2006).
Epidermal Cell Length Measurement
Epidermal peels were made from the petiole of the fifth compound leaf of 2-month-old R108, sgl1-1, Jemalong A17, and fcl1-1 plants. More than 200 epidermal cells from multiple independent plants of each genotype were measured using Image J. Student’s t tests were used for statistical analysis.
RNA in Situ Hybridization
RNA in situ hybridization was performed as previously described (Coen et al., 1990) with minor modifications. Briefly, FCL1 and KNOX1 transcripts were transcribed from cDNA clones corresponding to the nucleotide sequences from 60 to 459 and from 60 to 498 downstream of the translation initiation codon ATG of FCL1 and KNOX1, respectively, and labeled with digoxigenin. Ten-micrometer sections from shoot apices of 2-week-old wild-type Jemalong A17 and fcl1-1 mutant were hybridized with the digoxigenin-labeled sense or antisense probes. Primer sequences used are listed in Supplemental Table 4 online.
Histochemical Analysis of the GUS Reporter Gene Expression
GUS activity staining was performed as previously described (Jefferson et al., 1987). Briefly, shoot buds were incubated in a GUS staining solution (100 mM sodium phosphate, pH 7.0, 1 mM EDTA, 0.05% Triton X-100, 1 mM potassium ferricyanide and potassium ferrocyanide, and 1 mg/mL X-glucuronide) at 37°C overnight. Samples were then cleared in 70% (v/v) ethanol. Images taken using a digital camera mounted on a dissection microscope (SMZ1500; Nikon) were assembled using Adobe Photoshop.
Subcellular Localization of FCL1
To generate the FCL1-GFP fusion construct, the coding region of the full-length FCL1 cDNA without the stop codon was amplified using primers FCL1-F and FCL1-R and cloned into pENTR/D vector using TOPO cloning technology (Invitrogen). Subsequently, the FCL1 ORF was cloned into pMDC83 vector using LR clonase II (Invitrogen). The final construct was introduced into the A. tumefaciens GV3101 strain and then used to infiltrate tobacco leaves. FCL1-GFP signal was examined using a confocal laser scanning microscope (TCS SP2 AOBS; Leica). Primer sequences are listed in Supplemental Table 4 online.
Phylogenetic Analysis
Phylogenetic trees were constructed using neighbor-joining, maximum parsimony, and UPGMA algorithms implemented in the MEGA software suite (Tamura et al., 2007) (http://www.megasoftware.net/) with 1000 bootstrap replications. Multiple sequences were aligned using ClustalX (Thompson et al., 1994).
Yeast Two-Hybrid Assay
The Matchmaker Gold System (Clontech) was used in yeast two-hybrid assays. The coding sequence of FCL1 was PCR amplified from cDNA templates, cloned into the pGBKT7 vector, and used as the bait. Coding sequences of STM, BP, KNAT2, KNAT3, KNAT4, KNAT5, KNAT6, KNAT7, KNATM-B, SAW1, SAW2, BEL1, PNF, PNY, KNOX1, KNOX2, KNOX3, KNOX4, KNOX5, KNOX6, FCL1, SGL1, and PALM1 were PCR amplified from cDNA templates, cloned into the pGADT7 vector, and used as prey. Positive clones grown on double selection media (SD-Leu-Trp) were tested on three different media (quadra dropout [QD], QD + aureobasidin A [AbA], QD + AbA + X-α-gal) for protein–protein interactions and subsequent quantification of the X-α-galactosidase activity following the manufacturer’s instructions (Clontech). All primers used for PCR amplification are listed in Supplemental Table 4 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or NCBI GenBank under the following accession numbers: FCL1(HQ695002), Gm FCL1 (HQ695005), Gm FCL2 (HQ695006), Lj FCL1 (HQ695007), Os FCL1 (HQ695003), Os FCL2 (HQ695004), Zm FCL1 (HQ695008), Pt FCL1 (JN794532), Vv FCL1 (JN794533), Mt KNOX1 (EF128056.1), Mt KNOX2 (EF128057.1), Mt KNOX3 (EF128058.1), Mt KNOX4 (EF128059.1), Mt KNOX5 (EF128060.1), Mt KNOX6 (EF128061.1), SGL1/UNI (AY928184.1), PALM1 (HM038482.1), PTS/TKD1 (EU352653.1), KNATM-B (NM_001160868.1), PNY (AT5G02030), PNF (AT2G27990), SAW1 (AT4G36870), SAW2 (AT2G23760), BEL1 (AT5G41410), STM (AT1G62360), BP/KNAT1 (AT4G08150), KNAT2 (AT1G70510), KNAT6 (AT1G23380), KNAT3 (AT5G25220), KNAT4 (AT5G11060), KNAT5 (AT4G32040), and KNAT7 (AT1G62990).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Genetic Map Position of the fcl1 Locus on Chromosome 6
Supplemental Figure 2. Confirmation of Deletion Junctions and Expression Analysis of Genes Deleted in fcl1-1 and fcl1-2 Alleles.
Supplemental Figure 3. Allelic Series of fcl1 Mutants.
Supplemental Figure 4. Compound Leaf Phenotypes of the fcl1-4 Mutant.
Supplemental Figure 5. Genetic Complementation of fcl1-1.
Supplemental Figure 6. Zinc Finger Domain Transcription Factor Medtr6g080720 Did Not Rescue fcl1-1 Compound Leaf Defects.
Supplemental Figure 7. Full-Length FCL1 cDNA and the Deduced Amino Acid Sequences.
Supplement Figure 8. Amino Acid Sequence Alignments of FCL1 and Its Close Homologs.
Supplemental Figure 9. Microarray-Based in Silico Analysis of FCL1 Tissue-Specific Expression.
Supplemental Figure 10. Quantitative RT-PCR Analysis of KNOX2 and KNOX6 Expression in the Wild Type and the fcl1-1 Mutant.
Supplemental Table 1. Segregation Ratio of fcl1-2 Mutants in F2 Mapping Populations.
Supplemental Table 2. Recombination Frequency and Genetic Distance between the fcl1-2 Locus and Molecular Markers on Chromosome 6.
Supplemental Table 3. Genotypes of 20 fcl1-2 Mutant Plants That Carry Recombination between fcl1 and Closely Linked Markers.
Supplemental Table 4. Primer Sequences Used in This Study.
Supplemental Table 5. Arabidopsis thaliana Homologs of Deleted Genes in fcl1 Mutants.
Supplemental Table 6. Double Mutant Analyses of F2 Plants.
Supplemental Data Set 1. Amino Acid Sequence Alignment of FCL1 with PTS, KNATM-B, and the MEINOX Domain of Six Canonical KNOX Proteins from Medicago truncatula.
Acknowledgments
We thank members of the Chen laboratory for helpful discussions and comments on the manuscript. We also thank Junying Ma and Yuhong Tang, Jianghua Chen, Xiaofei Cheng and Jiangqi Wen, and Shuirong Zhang for assistance with RNA in situ hybridization, scanning electron microscopy, reverse genetic screens, and plant care, respectively, as well as Douglas Cook and Kirankumar Mysore for providing BAC clones and Tnt1 lines, respectively. Financial support for work done in the Chen laboratory was provided in part by the Samuel Roberts Noble Foundation and the National Science Foundation (DBI 0703285).
AUTHOR CONTRIBUTIONS
J.P., J.Y., H.W., and R.C. designed the research. J.P., J.Y., H.W., Y.G., and G.L. performed the experiments. J.P., J.Y., H.W., G.B., and R.C. analyzed the data. J.P. and R.C. wrote the article.
References
- Allard R.W. (1956). Formulas and tables to facilitate the calculation of recombination values in heredity. Hilgardia 24: 235–278 [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T., Masuda D., Yasuda M., Nakashita H., Kudo T., Kimura M., Yamaguchi K., Nishiuchi T. (2008). AtNFXL1, an Arabidopsis homologue of the human transcription factor NF-X1, functions as a negative regulator of the trichothecene phytotoxin-induced defense response. Plant J. 53: 450–464 [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Hay A., Kougioumoutzi E., Tsiantis M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40: 1136–1141 [DOI] [PubMed] [Google Scholar]
- Benedito V.A., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Berger Y., Harpaz-Saad S., Brand A., Melnik H., Sirding N., Alvarez J.P., Zinder M., Samach A., Eshed Y., Ori N. (2009). The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136: 823–832 [DOI] [PubMed] [Google Scholar]
- Bharathan G., Goliber T.E., Moore C., Kessler S., Pham T., Sinha N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860 [DOI] [PubMed] [Google Scholar]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., Johansen I.E., Tsiantis M., Laufs P. (2008). A conserved molecular framework for compound leaf development. Science 322: 1835–1839 [DOI] [PubMed] [Google Scholar]
- Champagne C., Sinha N. (2004). Compound leaves: Equal to the sum of their parts? Development 131: 4401–4412 [DOI] [PubMed] [Google Scholar]
- Champagne C.E., Goliber T.E., Wojciechowski M.F., Mei R.W., Townsley B.T., Wang K., Paz M.M., Geeta R., Sinha N.R. (2007). Compound leaf development and evolution in the legumes. Plant Cell 19: 3369–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., et al. (2010). Control of dissected leaf morphology by a Cys(2)His(2) zinc finger transcription factor in the model legume Medicago truncatula. Proc. Natl. Acad. Sci. USA 107: 10754–10759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.J., Janssen B.J., Williams A., Sinha N. (1997). A gene fusion at a homeobox locus: Alterations in leaf shape and implications for morphological evolution. Plant Cell 9: 1289–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Wen J., Tadege M., Ratet P., Mysore K.S. (2011). Reverse genetics in Medicago truncatula using Tnt1 insertion mutants. Methods Mol. Biol. 678: 179–190 [DOI] [PubMed] [Google Scholar]
- Coen E.S., Romero J.M., Doyle S., Elliott R., Murphy G., Carpenter R. (1990). floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Cosson V., Durand P., d’Erfurth I., Kondorosi A., Ratet P. (2006). Medicago truncatula transformation using leaf explants. Methods Mol. Biol. 343: 115–127 [DOI] [PubMed] [Google Scholar]
- DeMason D.A., Chawla R. (2004). Roles for auxin during morphogenesis of the compound leaves of pea ( Pisum sativum). Planta 218: 435–448 [DOI] [PubMed] [Google Scholar]
- Di Giacomo E., Sestili F., Iannelli M.A., Testone G., Mariotti D., Frugis G. (2008). Characterization of KNOX genes in Medicago truncatula. Plant Mol. Biol. 67: 135–150 [DOI] [PubMed] [Google Scholar]
- Dong Z.C., Zhao Z., Liu C.W., Luo J.H., Yang J., Huang W.H., Hu X.H., Wang T.L., Luo D. (2005). Floral patterning in Lotus japonicus. Plant Physiol. 137: 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Eshed Y., Lifschitz E. (2010). Morphogenesis of simple and compound leaves: A critical review. Plant Cell 22: 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Blum E., Goldshmidt A., Eshed Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Chen J., Chen R. (2010). Palmate-like Pentafoliata1 encodes a novel Cys(2)His(2) zinc finger transcription factor essential for compound leaf morphogenesis in Medicago truncatula. Plant Signal. Behav. 5: 1134–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann W., Gleissberg S. (1996). Organogenetic capacity of leaves: The significance of marginal blastozones in angiosperms. Plant Syst. Evol. 199: 121–152 [Google Scholar]
- Hareven D., Gutfinger T., Parnis A., Eshed Y., Lifschitz E. (1996). The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 84: 735–744 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38: 942–947 [DOI] [PubMed] [Google Scholar]
- Hinze K., Thompson R.D., Ritter E., Salamini F., Schulze-Lefert P. (1991). Restriction fragment length polymorphism-mediated targeting of the ml-o resistance locus in barley (Hordeum vulgare). Proc. Natl. Acad. Sci. USA 88: 3691–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J., Gourlay C., Michael A., Ellis T.H. (2001). Expression of a class 1 knotted1-like homeobox gene is down-regulated in pea compound leaf primordia. Plant Mol. Biol. 45: 387–398 [DOI] [PubMed] [Google Scholar]
- Hofer J., Turner L., Hellens R., Ambrose M., Matthews P., Michael A., Ellis N. (1997). UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7: 581–587 [DOI] [PubMed] [Google Scholar]
- Janssen B.J., Lund L., Sinha N. (1998). Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 117: 771–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.R. (2001). Fundamental concepts of leaf morphology and morphogenesis: A contribution to the interpretation of molecular genetic mutants. Int. J. Plant Sci. 162: 465–474 [Google Scholar]
- Kimura S., Koenig D., Kang J., Yoong F.Y., Sinha N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18: 672–677 [DOI] [PubMed] [Google Scholar]
- Koenig D., Bayer E., Kang J., Kuhlemeier C., Sinha N. (2009). Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Koornneef M., van Eden J., Hanhart C.J., Stam P., Braaksma F.J., Feenstra W.J. (1983). Linkage map of Arabidopsis thaliana. Heredity 74: 265–272 [Google Scholar]
- Kumar R., Kushalappa K., Godt D., Pidkowich M.S., Pastorelli S., Hepworth S.R., Haughn G.W. (2007). The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell 19: 2719–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmi A., Pan J., Morsy M., Chen R. (2008). Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS One 3: e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C., Long J., Yamaguchi J., Serikawa K., Hake S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisso J., Altmann T., Müssig C. (2006). The AtNFXL1 gene encodes a NF-X1 type zinc finger protein required for growth under salt stress. FEBS Lett. 580: 4851–4856 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Magnani E., Hake S. (2008). KNOX lost the OX: The Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20: 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero-Rosales N., Jamilena M., Zurita S., Gómez P., Capel J., Lozano R. (1999). FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J. 20: 685–693 [DOI] [PubMed] [Google Scholar]
- Parnis A., Cohen O., Gutfinger T., Hareven D., Zamir D., Lifschitz E. (1997). The dominant developmental mutants of tomato, Mouse-ear and Curl, are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene. Plant Cell 9: 2143–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Chen R. (2011). Auxin efflux transporter MtPIN10 regulates compound leaf and flower development in Medicago truncatula. Plant Signal. Behav. 6: 1537–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. (1997). Leaf morphogenesis in flowering plants. Plant Cell 9: 1077–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J.M., Deprez R.H., Moorman A.F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof M.A., Soliman K.M., Jorgensen R.A., Allard R.W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81: 8014–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Ben-Gera H., Shleizer-Burko S., Burko Y., Weiss D., Ori N. (2010). Cytokinin regulates compound leaf development in tomato. Plant Cell 22: 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Burko Y., Ben-Yaakov L., Berger Y., Amsellem Z., Goldshmidt A., Sharon E., Ori N. (2009). Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21: 3078–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., Cayrel A., Endre G., Zhao P.X., Chabaud M., Ratet P., Mysore K.S. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen J., Voorrips R. (2001). JoinMap3.0 Software for Calculation of Genetic Linkage Maps. (Wageningen, The Netherlands: Plant Research International; ). [Google Scholar]
- Wang H., Chen J., Wen J., Tadege M., Li G., Liu Y., Mysore K.S., Ratet P., Chen R. (2008). Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 146: 1759–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Jones B., Li Z., Frasse P., Delalande C., Regad F., Chaabouni S., Latché A., Pech J.C., Bouzayen M. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li G., Chen R. (2006). Fast neutron bombardment (FNB) induced deletion mutagenesis for forward and reverse genetic studies in plants. Floriculture, Ornamental and Plant Biotechnology: Advances and Topical Issues, 1st ed, Teixeira da Silva J., (Isleworth, UK: Global Science Books; ), pp. 629–639 [Google Scholar]
- Yu J.B., Bai G.H., Cai S.B., Ban T. (2006). Marker-assisted characterization of Asian wheat lines for resistance to Fusarium head blight. Theor. Appl. Genet. 113: 308–320 [DOI] [PubMed] [Google Scholar]
- Zhou C., Han L., Hou C., Metelli A., Qi L., Tadege M., Mysore K.S., Wang Z.Y. (2011). Developmental analysis of a Medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development. Plant Cell 23: 2106–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]