Given the importance of auxin, it has been surprisingly difficult to elucidate its biosynthetic pathways in plants. Several Trp-dependent routes have been proposed, in which CYTOCHROME P450 79B2/B3 (CYP79B2/B3), YUCCA (YUC), and TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1/TRYPTOPHAN AMINOTRANSFERASE RELATED (TAA1/TAR) genes have been thought to act in separate pathways (reviewed in Zhao, 2010). Recent data have suggested that YUC and TAA1/TAR genes may in fact be part of a single pathway, and new work from Stepanova et al. (pages 3961–3973) provides solid evidence for this using genetic, pharmacological, and biochemical approaches in Arabidopsis thaliana.
Stepanova et al. tested whether YUC1 functions in a pathway involving CYP79B2/B3 or TAA1/TAR proteins. Overexpressing YUC1 in a cyp79b2/b3 mutant led to high-auxin phenotypes, demonstrating that CYP79B2/B3 function is not required for YUC1-mediated auxin biosynthesis. Furthermore, YUC1 overexpression led to additive phenotypes in the superroot2 mutant, in which auxin biosynthesis via the CYP79B2/B3 pathway is increased. Thus, YUC1 and CYP79B2/B3 appear not to operate in the same auxin biosynthetic pathway.
Arabidopsis encodes 11 YUC and three TAA1/TAR genes, and Stepanova et al. generated higher-order mutants, finding that reduced YUC function in backgrounds with already reduced TAA1/TAR function did not lead to additive phenotypes. Additionally, treatment with a newly described inhibitor of TAA1/TAR activity (He et al., pages 3944–3960) reversed the effects of YUC1 overexpression, supporting the idea that YUCs and TAA1/TARs function in the same pathway. The authors also found that cells in which TAA1 is normally expressed contain all of the necessary machinery for YUC1-associated auxin biosynthesis and that YUC1 overexpression induced high-auxin phenotypes in a TAA1/TAR-dependent manner, adding further credence to the single YUC-TAA1/TAR pathway hypothesis.
Direct quantification of IAA and IAA metabolites gave evidence that overexpression of YUC1 led to increased auxin biosynthetic activity in the wild-type background. This effect was absent in a TAA1/TAR-impaired background, again consistent with YUCs functioning in the TAA1/TAR pathway. The authors went on to show that pyruvate decarboxylases, which are key IAA biosynthetic enzymes in auxin-producing bacteria, are unlikely to play a role in auxin biosynthesis in Arabidopsis.
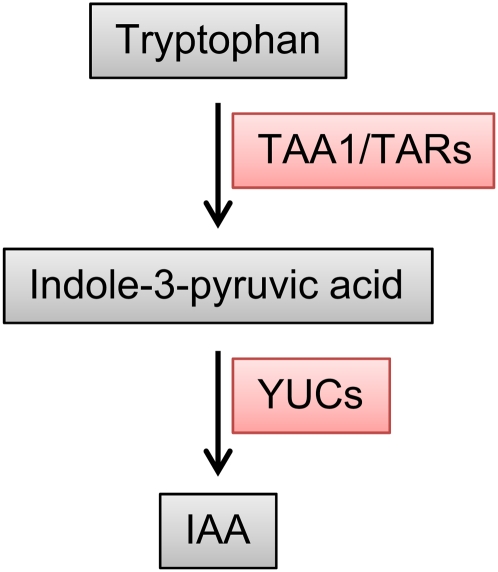
This work from Stepanova et al. nicely complements two newly published reports that use different mutant combinations and different approaches to similarly demonstrate that YUCs function in the same pathway as TAA1/TARs, likely by converting indole-3-pyruvic acid produced by TAA1/TARs into IAA (see figure; Mashiguchi et al., 2011; Won et al., 2011). Together, these studies represent an important step forward in understanding Trp-dependent auxin biosynthesis in plants.
A single pathway model for auxin (indole-3-acetic acid) biosynthesis via YUC and TAA1/TAR activities. (Adapted from Stepanova et al. [2011] and Won et al. [2011].)
References
- He W., et al. (2011). A small-molecule screen identifies l-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K., et al. (October 24, 2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA http://dx.doi.org/10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Robles L.M., Novak O., He W., Guo H., Ljung K., Alonso J.M. (2011). The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y. (October 24, 2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA http://dx.doi.org/10.1073/pnas.1108436108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]