This work identifies an enzyme, named Like Sex Four2 (LSF2), that hydrolyzes the phosphate group from the C3-position of glucosyl residues of starch. The lsf2 mutation in Arabidopsis thaliana leads to modified starch with elevated levels of C3-bound phosphate.
Abstract
Starch contains phosphate covalently bound to the C6-position (70 to 80% of total bound phosphate) and the C3-position (20 to 30%) of the glucosyl residues of the amylopectin fraction. In plants, the transient phosphorylation of starch renders the granule surface more accessible to glucan hydrolyzing enzymes and is required for proper starch degradation. Phosphate also confers desired properties to starch-derived pastes for industrial applications. In Arabidopsis thaliana, the removal of phosphate by the glucan phosphatase Starch Excess4 (SEX4) is essential for starch breakdown. We identified a homolog of SEX4, LSF2 (Like Sex Four2), as a novel enzyme involved in starch metabolism in Arabidopsis chloroplasts. Unlike SEX4, LSF2 does not have a carbohydrate binding module. Nevertheless, it binds to starch and specifically hydrolyzes phosphate from the C3-position. As a consequence, lsf2 mutant starch has elevated levels of C3-bound phosphate. SEX4 can release phosphate from both the C6- and the C3-positions, resulting in partial functional overlap with LSF2. However, compared with sex4 single mutants, the lsf2 sex4 double mutants have a more severe starch-excess phenotype, impaired growth, and a further change in the proportion of C3- and C6-bound phosphate. These findings significantly advance our understanding of the metabolism of phosphate in starch and provide innovative options for tailoring novel starches with improved functionality for industry.
INTRODUCTION
Starch is the major storage carbohydrate in higher plants and a key resource for humankind both as the main component of our staple crops and as a renewable industrial material. Amylopectin (a polymer comprised of α-1,4-linked glucan chains, branched via α-1,6-bonds) is the major constituent of starch, accounting for 70% or more of the granule. Additionally, starch is composed of amylose, which is essentially a linear 1,4-linked Glc polymer interspersed between the amylopectin molecules. By virtue of its branched architecture, chains of amylopectin form double helices that align into a layered, semicrystalline matrix (Zeeman et al., 2010). Many industrial applications require chemical modification of native starches (e.g., oxidation and esterification) to stabilize the constituent glucan polymers during processing or to introduce required functional groups (BeMiller, 1997). Starch phosphorylation is the only known modification of starch to occur in vivo. The extent of phosphorylation varies from a relatively high level in potato (Solanum tuberosum) tuber starch (0.5% of glucosyl units) to almost undetectable amounts in the cereal starches (Blennow et al., 2000). The amount of covalently bound phosphate influences the molecular structure, crystallinity, and physico-chemical properties of starch. For example, high-phosphate starches have a very high swelling power when heated in water, forming transparent, viscous, and freeze-thaw stable pastes, which are desired in many industrial applications (Santelia and Zeeman, 2011). Knowledge of the molecular details of starch metabolism and in vivo modification provides options for tailoring novel starches with improved functionality.
The reversible phosphorylation of starch is an integral part of its normal metabolism (Zeeman et al., 2010). In leaves, starch is produced during the day from photoassimilated carbon and accumulates inside chloroplasts. The surface of the starch granule is resistant to degradation by most enzymes, such as exoamylases (β-amylases) and debranching enzymes. It is proposed that starch phosphorylation increases the hydration status of the granule-stroma interface, disrupting its crystallinity and thereby facilitating the actions of the glucan-hydrolyzing enzymes (Edner et al., 2007). In wild-type starches, phosphate esters are exclusively found on the amylopectin portion of the granule, mostly at the C6-position (70 to 80%) and, to a lesser extent, at the C3-position (20 to 30%) of glucosyl units (Blennow et al., 1998; Ritte et al., 2006). Starch Excess1/glucan water dikinase (SEX1/GWD) phosphorylates the C6-position of glucosyl residues (Ritte et al., 2002, 2006), while phosphoglucan water dikinase (PWD) phosphorylates the C3 position. PWD acts on glucan chains prephosphorylated by GWD (Baunsgaard et al., 2005; Kötting et al., 2005; Ritte et al., 2006). Starch from Arabidopsis thaliana sex1 (gwd) null mutants is essentially phosphate free, whereas pwd starch is only phosphorylated at C6-positions (Ritte et al., 2006). Both mutant plants display impaired starch degradation, leading to a starch-excess (sex) phenotype, which is severe in sex1 and more moderate in pwd (Yu et al., 2001; Kötting et al., 2005). Similarly, in potato tubers, inhibition of GWD leads to starch with low phosphate and the valuable repression of starch degradation during cold storage (Lorberth et al., 1998).
Removal of the phosphate groups, at both the C3- and C6-positions, by the phosphoglucan phosphatase SEX4 is also required for proper starch metabolism (Kötting et al., 2009). Although phosphate groups promote the solubilization of the starch granule surface, they can also obstruct glucan hydrolytic enzymes, as demonstrated for β-amylase, which removes maltosyl units sequentially from the nonreducing end of an α-1,4-linked glucan chain. β-Amylase is required for starch degradation but cannot degrade past a phosphate group (Takeda and Hizukuri, 1981; Fulton et al., 2008). This suggests an interdependence between reversible starch phosphorylation and glucan hydrolysis (Edner et al., 2007; Kötting et al., 2009; Hejazi et al., 2010). SEX4 possesses a carbohydrate binding module (CBM) and a phosphatase domain of the dual-specificity (DSP) class. Both domains are required for activity toward soluble and insoluble phosphoglucan substrates (Niittylä et al., 2006; Gentry et al., 2007; Hejazi et al., 2010). sex4 mutants have impaired starch degradation, causing the sex phenotype to develop over repeated diurnal cycles (Niittylä et al., 2006; Kötting et al., 2009). The decreased glucan phosphatase activity results in the accumulation of phosphoglucans, mostly in the form of soluble phospho-oligosaccharides released from the starch granule surface by α-amylase 3 (AMY3) and the debranching enzyme isoamylase 3 (ISA3). These phospho-oligosaccharides may be normal intermediates of starch breakdown but are below the limit of detection in the wild type (Kötting et al., 2009).
Bioinformatics analyses revealed that the Arabidopsis genome encodes two further proteins with a SEX4-like phosphatase domain: Like Sex Four1 (LSF1) and LSF2. As with SEX4, LSF1 possesses a CBM and is targeted to the chloroplast (Comparot-Moss et al., 2010). Mutation of this gene also results in elevated amounts of leaf starch, but there is no direct evidence that LSF1 acts as a glucan phosphatase. There is no measurable reduction in glucan-dephosphorylating activity in lsf1 mutants and no phospho-oligosaccharides accumulate (Comparot-Moss et al., 2010). Thus, LSF1 might have a regulatory function during starch degradation. Here, we report the functional characterization of the third Arabidopsis homolog LSF2. We show that LSF2 is a chloroplastic phosphoglucan phosphatase specific for C3-glucosyl residues of starch and that mutation of LSF2 leads to starch with elevated levels of C3-bound phosphate. We investigate the function of LSF2 in transitory starch metabolism and discuss the potential biotechnological value of LSF2 for the creation of novel starches.
RESULTS
LSF2 Is a Chloroplastic Protein Homolog of SEX4
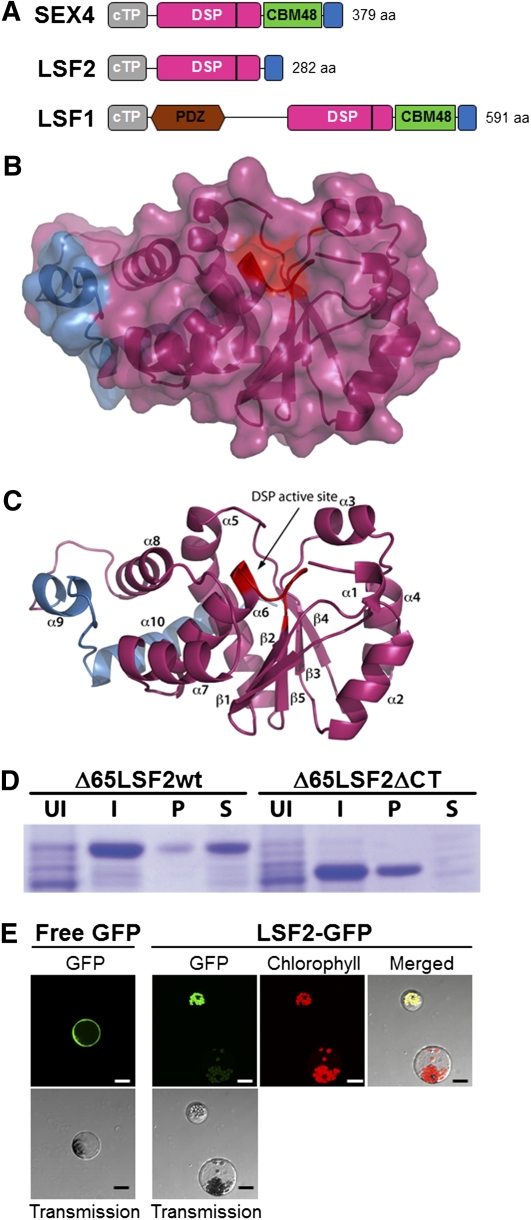
BLAST searches (http://blast.ncbi.nlm.nih.gov) of the Arabidopsis genome revealed two loci encoding proteins with high sequence similarity to SEX4: LSF1 and LSF2 (Figure 1A; see Supplemental Figures 1A and 1B online; Comparot-Moss et al., 2010). LSF2 encodes a 282–amino acid protein with a predicted molecular mass of 32.1 kD (http://www.isb-sib.ch/). The LSF2 protein contains a predicted 61–amino acid chloroplast transit peptide (cTP; ChlorP and TargetP prediction; http://www.cbs.dtu.dk/services/ChloroP; http://www.cbs.dtu.dk/services/TargetP/), a DSP domain (residues 85 to 247), and a C-terminal domain (CT; residues 248 to 282). The CT extension is not observed in other members of the DSP family but is conserved in LSF1 and LSF2 orthologs from other plant species. The DSP of LSF2 possesses the canonical DSP active-site signature residues HCxxGxxRA/T (see Supplemental Figure 1A online; Yuvaniyama et al., 1996). Interestingly, LSF2 does not possess the CBM (Figure 1A) located between the DSP and CT domains in both SEX4 and LSF1 nor does it possess the PDZ-like putative protein–protein interaction domain identified in LSF1 (Figure 1A; Fordham-Skelton et al., 2002).
Figure 1.
LSF2 Protein Structure, Heterologous Expression, and Subcellular Localization.
(A) Schematic representation of the domain topography of SEX4, LSF2, and LSF1. The cTP is in gray, the DSP domain in pink, the CBM in green, the PDZ-like domain in brown, and the CT domain in blue. The active site of the proteins is denoted with a black vertical line within the DSP domain. The lengths of the proteins are also indicated.
(B) Surface view of the LSF2 homology model based on the SEX4 crystal structure (Vander Kooi et al., 2010), showing the predicted integrated architecture between the DSP (pink) and CT domains (blue). The predicted active-site location is indicated in red.
(C) Ribbon diagram of the predicted LSF2 structure (B) with the active site labeled in red. Elements of secondary structure are numbered consecutively from the N to C termini.
(D) The CT domain is essential for soluble expression of LSF2. Coomassie blue–stained SDS-PAGE showing purification of Δ65LSF2 protein and Δ65LSF2ΔCT, which lacks the C-terminal 35 residues. I, cells induced with isopropyl β-d-1-thiogalactopyranoside; P, pellet of insoluble protein; S, soluble protein; UI, uninduced cells.
(E) Subcellular localization of transiently expressed LSF2-GFP fusion protein in Arabidopsis wild-type protoplasts. Green fluorescence of the control GFP protein is found in the cytosol, while LSF2-GFP fusion protein is located in the chloroplast. Transmission images of the same cells are also shown. Green fluorescence, transmission, and chlorophyll images were merged to show the accurate localization of GFP fluorescence in the chloroplast. Note how the untransformed protoplast only shows chlorophyll autofluorescence. Bar = 20 μm.
The recently determined structure of SEX4 provides a molecular basis for understanding its glucan phosphatase function (Vander Kooi et al., 2010). The DSP domain and CBM interact to form an integral structural unit. The CT domain contacts both the DSP and the CBM domain and is essential for the folding and solubility of recombinant SEX4 (Vander Kooi et al., 2010). The overall similarity between LSF2 and SEX4 sequences allowed us to model the structure of LSF2 (Figures 1B and 1C; see Supplemental Figure 1A online). Due to the absence of the CBM, the predicted LSF2 structure is more compact than SEX4. Like SEX4, the two α-helices in the CT domain are predicted to wrap around the DSP and cradle the final helix (α8), while making contact with multiple helices (α5, α6, and α7; Figure 1C). To determine whether the CT domain of LSF2 is important for its function, we generated a set of constructs to express LSF2 in Escherichia coli. We previously demonstrated that we could purify full-length SEX4, but it was far more soluble when we removed the cTP (Vander Kooi et al., 2010). Similarly, we found that we could purify full-length LSF2, but we obtained significantly more protein if we removed the cTP at residue Lys-65 (Δ65LSF2) (Figure 1D; see Supplemental Figure 1C online). However, when we deleted the CT domain from Δ65LSF2, the protein was entirely insoluble (Figure 1D, Δ65LSF2ΔCT). Similar results were obtained with LSF2ΔCT (see Supplemental Figure 1C online). Thus, the CT domain is necessary for soluble expression of LSF2, as for SEX4.
To discover whether LSF2 is chloroplastic, we examined the subcellular localization of the protein by transiently expressing an LSF2-green fluorescent protein (GFP) fusion in Arabidopsis protoplasts (Figure 1E). Free GFP was used as a control and was found in the cytoplasm, whereas the LSF2-GFP fusion protein was localized in the chloroplast. This is consistent with the prediction of a cTP in the LSF2 sequence and with a former localization study of putative Arabidopsis chloroplast protein kinases and phosphatases (among them LSF2; Schliebner et al., 2008).
Next, we investigated LSF2 expression patterns using a β-glucuronidase (GUS) transcriptional reporter construct in which 1.5 kb of the LSF2 promoter region upstream of the LSF2 translation start site was fused to the GUS gene, resulting in the LSF2pro:GUS fusion. The fusion construct was transformed into wild-type plants, and the GUS activity was analyzed in three independent transgenic T2 lines. GUS activity was found in all organs, especially in green tissues, which represent sites of starch storage, and in the vasculature (see Supplemental Figures 2A to 2G online). Analysis of publicly accessible transcriptome data was broadly consistent with the GUS activity results (see Supplemental Figure 2H online). Incubation of seedlings containing the LSF2pro:GUS construct in the dark for 72 h revealed decreased LSF2 expression (see Supplemental Figure 2G online). Again, this result was consistent with publicly accessible transcriptome data showing that LSF2 expression fluctuates diurnally, with transcript abundance declining gradually during the night to a low level, followed by a rapid increase during the first hours of the day (see Supplemental Figure 2I online). This expression pattern is similar to SEX4 (see Supplemental Figure 2I online) and other genes encoding enzymes of starch metabolism, suggesting they are coordinately regulated (Smith et al., 2004). Searching the AtProteome database (Baerenfaller et al., 2008; http://fgcz-atproteome.unizh.ch/) also provided evidence for the presence of LSF2 predominantly in green tissues (see Supplemental Figure 2J online).
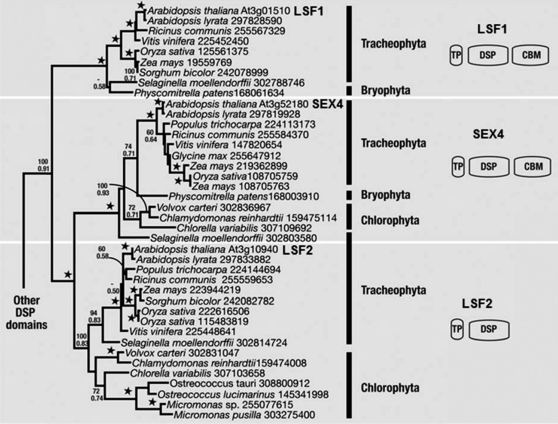
Homologs of LSF2 are found in vascular plants, mosses, and in green algae. Maximum likelihood (ML) and Bayesian analyses of 150 unambiguously aligned characters of the DSP domain support the relationship of SEX4, LSF1, and LSF2 (100% ML bootstrap and a posterior probability of 1.0; Figure 2; see Supplemental Data Set 1 online). The LSF1 proteins, which are absent from green algae, cluster at the base of the SEX4 and LSF2 sister clades (100% ML bootstrap and posterior probability of 1.0). So far, no phosphatase activity has been attributed to LSF1. The divergence of the DSP from SEX4 and LSF2 may suggest that it has acquired a novel function (Comparot-Moss et al., 2010; M. Umhang and S.C. Zeeman, unpublished data). Analysis of the Arabidopsis genes reveals distinct exon-intron structures (see Supplemental Figure 3 online). However, two exon-intron boundaries are conserved between members of the gene family: one within the DSP of all three genes and the other in the CBM domains of SEX4 and LSF1. Along with the similar domain organization in SEX4 and LSF1, this supports a common origin of the CBM in SEX4 and LSF1, indicating its presence in their common ancestor.
Figure 2.
ML Phylogeny of the LSF1, LSF2, and SEX4 DSP Domain from Viridiplantae.
Numbers at nodes correspond to ML bootstrap support (top) and Bayesian posterior probabilities (bottom). Stars at nodes correspond to 100% bootstrap support and a posterior probability of 1.0. Major groups are bracketed and labeled to the right.
LSF2 Dephosphorylates Amylopectin and Soluble Phosphorylated Glucans
The expression of LSF2 in green tissues, its localization in the chloroplast, its similarity to SEX4, and its coordinated expression with other starch metabolizing enzymes all suggest that it may be a glucan phosphatase involved in transitory starch metabolism. To determine if LSF2 is a phosphatase, we tested whether the purified recombinant protein could dephosphorylate the artificial substrate para-nitrophenylphosphate (p-NPP; a universal chromogenic substrate for acid and alkaline phosphatases). LSF2 was active against p-NPP (Figure 3A) and had a similar specific activity and kinetic properties to SEX4 (Gentry et al., 2007). Within the DSP active-site motif, HCxxGxxRA/T (Yuvaniyama et al., 1996), the conserved Cys is essential for activity. Mutation of the corresponding Cys in LSF2 (Cys-193) to Ser abolished its activity against p-NPP (Figure 3A, LSF2 C/S), analogous to the C198S mutation in SEX4 (Gentry et al., 2007).
Figure 3.
LSF2 Is a Starch Binding Phosphoglucan Phosphatase Specific for C3-Bound Phosphate Esters in Starch.
(A) Specific activity of SEX4, LSF2, and the active-site mutant LSF2 C/S against the artificial substrate p-NPP at their optimal pH (6.5). Error bars indicate mean ± se (n = 3; P value < 0.05).
(B) Phosphate release measured by the malachite green assay using SEX4, LSF2, or the active-site mutant LSF2 C/S against solubilized amylopectin (left) and purified phospho-oligosaccharides (right) at their respective optimal pH (6.5). The amounts of the two phosphatases (SEX4 and LSF2) used in the assay were adjusted to equal hydrolytic activity on p-NPP, while the amounts of the two substrates were normalized to similar amounts of phosphate. Note the different scales on the y axes. Error bars indicate mean ± se (n = 3). Similar results were obtained using Δ78-LSF2 recombinant protein lacking the N-terminal 78 residues (corresponding to the cTP).
(C) Hydrolysis of C6- and C3-phosphate esters in native starch granules. Purified phosphate-free starch granules from GWD-deficient Arabidopsis sex1-3 mutants (Yu et al., 2001) were prelabeled with 33P at either the C6- or C3-positions and incubated with SEX4, LSF2, or active-site mutant LSF2 C/S recombinant proteins. Phosphate release over time was linear and is expressed relative to the total 33P incorporated into starch. The reaction time was 5 min. Each value is the mean ± se of four replicate samples.
(D) Binding of LSF2 to potato amylose-free (waxy) starch in vitro. SEX4, Δ78-LSF2, and AP (alkaline phosphatase from calf intestine) proteins were incubated with starch for 30 min at 20°C. The starch was pelleted by centrifugation. Proteins in the supernatant (S), in the pellet wash (W), and bound to the pellet (P) were visualized by SDS-PAGE and silver staining.
Next, we tested whether LSF2 was able to use phosphorylated glucans as substrates. Potato amylopectin is phosphorylated on ~1 in every 300 Glc residues (Blennow et al., 2002), while the soluble phospho-oligosaccharides that accumulate in sex4 (which have a degree of polymerization between 4 and 20) are singly or doubly phosphorylated (Kötting et al., 2009). We incubated recombinant LSF2 either with solubilized amylopectin or with purified phospho-oligosaccharides and quantified the amount of phosphate released using the malachite green assay (Werner et al., 2005). In each case, recombinant SEX4 was used as a positive control. As reported previously, SEX4 displayed robust phosphatase activity toward both substrates (Figure 3B; Gentry et al., 2007; Kötting et al., 2009). LSF2 could also liberate phosphate from both glucan substrates, although to a lesser extent than SEX4 (Figure 3B). As predicted, mutation of the active-site Cys to Ser abolished LSF2 activity. Collectively, these data show that LSF2 possesses a functional DSP domain and is capable of dephosphorylating glucan substrates even though it lacks a CBM.
LSF2 Specifically Dephosphorylates C3-Glucosyl Residues of Starch in Vitro
The two dikinases GWD and PWD phosphorylate the C6- and C3-positions of glucosyl units in amylopectin, respectively (Ritte et al., 2006). While SEX4 is able to hydrolyze both C6- and C3-bound phosphate, we considered the possibility that LSF2 might be specific for one or the other position. To test this, we phosphorylated purified sex1 starch granules (which are phosphate free; Yu et al., 2001) in vitro using recombinant potato GWD and recombinant Arabidopsis PWD sequentially. Using [β-33P]ATP as a substrate in one dikinase reaction and unlabeled ATP in the other, 33P-labeled phosphate was introduced at either the C6- or the C3-position. In all cases, the starch granules were phosphorylated at both locations (see Methods for details). After incubation of the recombinant proteins with the 33P-labeled starches, the released 33P was determined. The incubation times were short (i.e., 5 min), such that the rates of hydrolysis from both C6- and C3-positions were linear. The amounts of the two phosphatases were adjusted to equal hydrolytic activity on p-NPP (0.05 μg SEX4 and 0.2 μg LSF2). Recombinant SEX4 efficiently released phosphate from both positions, but twice as much from the C6- as from the C3-position (Figure 3C; Hejazi et al., 2010). This may reflect a preference for the C6-phosphate esters. However, as less 33P was incorporated into the C3-positions compared with the C6-positions (20.41 versus 47.72 pmol P mg−1 starch), it may also reflect a lower frequency of C3-bound phosphate groups on the granule surface. When LSF2 was incubated with starch granules labeled with 33P at the C6-position, no radioactivity was released. Conversely, LSF2 efficiently hydrolyzed the phosphate esters at the C3-position. Again, mutation of the conserved Cys residue abolished LSF2 activity (Figure 3C). When prelabeled starch was incubated with 25-fold more LSF2 protein for up to 2 h, a minimal amount of hydrolysis from the C6-positions was observed, although the rate was not linear (see Supplemental Figure 4 online). Thus, LSF2 is unique as it is highly specific for the C3-position of glucosyl residues of starch even if, under saturating conditions, it has a low capacity to dephosphorylate some C6-esters.
LSF2 Binds Starch Despite Lacking a CBM and Is Present inside Starch Granules
To test the starch binding capacity of LSF2, we performed a starch binding assay with purified recombinant LSF2 and SEX4 and a commercially available alkaline phosphatase as a nonbinding control (Figure 3D). The recombinant proteins were incubated with amylose-free potato starch, and the starch was washed with assay medium to remove all the unbound proteins (Figure 3D, lane W). All of the SEX4 protein bound to starch, as demonstrated by the absence of the protein in the soluble fraction after starch pulldown (Figure 3D). This result is consistent with previous reports that CBM48s have a high affinity for starch (Niittylä et al., 2006; Gentry et al., 2007). LSF2 was also able to bind to starch, but the affinity may be lower than that of SEX4, as demonstrated by the fact that some soluble LSF2 was still visible on silver-stained SDS-PAGE gels (Figure 3D). As expected, alkaline phosphatase did not bind to starch. These data show that despite lacking a CBM, recombinant LSF2 can still bind to starch, perhaps through secondary binding sites within or adjacent to the catalytic domain.
To confirm that endogenous LSF2 binds starch, we incubated potato starch with protein extracts from Arabidopsis leaves and analyzed the bound proteins by SDS-PAGE (see Supplemental Figure 5 online, Binding) and liquid chromatography–tandem mass spectrometry (MS/MS). Among the identified peptides derived from the bound proteins, one was unambiguously assigned to LSF2 in gel slices corresponding to the predicted molecular mass of 32 kD of LSF2 (Table 1). Next, we isolated starch from Arabidopsis leaves harvested at the end of the light period and analyzed proteins bound to the starch granule surface (External) and proteins encapsulated within the starch granule (see Supplemental Figure 5 online, Internal). One LSF2 peptide was identified in the external fraction and three in the internal fraction (Table 1). These proteomics data support our in vitro binding experiment with recombinant LSF2.
Table 1.
LSF2 Peptides Identified from Starch-Bound Proteins by Tandem Mass Spectrometry
| Sequence | Score | Expectation |
| Binding assay with Arabidopsis extract | ||
| DRDPLSLR | 32.04 | 0.00950 |
| External starch binding proteins | ||
| DFDPLSLR | 43.67 | 0.00065 |
| Internal starch proteins | ||
| AVSSLEWAVSEGK | 27.26 | 0.02100 |
| DELIVGSQPQKPEDIDHLK | 34.36 | 0.00260 |
| KLIQER | 27.81 | 0.04400 |
Proteins in the indicated gel slices (see Supplemental Figure 5 online) were in-gel digested with trypsin and analyzed by liquid chromatography–MS/MS. Mascot scores higher than 26 and expectation values below 0.05 were required for peptide identification.
Loss of LSF2 Results in Elevated Glucan-Bound Phosphate Levels
To study the function of LSF2 in vivo, we identified two independent Arabidopsis insertion mutants at the LSF2 locus (see Supplemental Figure 3 online) and designated them lsf2-1 (Sail_595F04) and lsf2-2 (GT10871). We obtained homozygous lines and confirmed the positions of the insertions by PCR and DNA sequencing (in exon 2 at bp +510 from the start codon and exon 4 at bp +1016 for lsf2-2 and lsf2-1, respectively). Quantitative RT-PCR revealed that both insertions prevented normal LSF2 expression relative to the corresponding wild types (see Supplemental Figure 6A online).
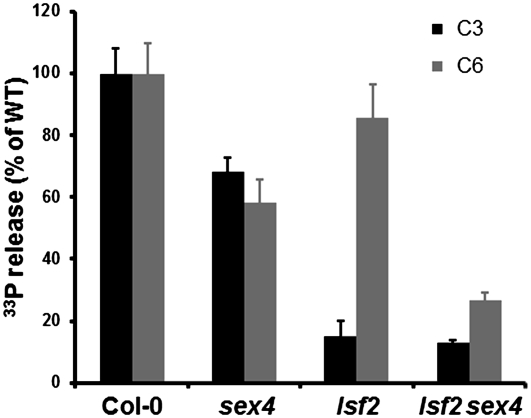
To determine the contribution of LSF2 to total glucan phosphatase activity in vivo, we incubated starch granules labeled with 33P at either the C6- or the C3-position with crude extracts from leaves of the lsf2 mutants or their respective wild types (Figure 4; see Supplemental Figure 6B online). Consistent with our in vitro assay, lsf2 extracts released 80% less phosphate from the C3-position than extracts of wild-type leaves, whereas phosphate release from the C6-position was unaltered. The residual C3-phosphatase activity of lsf2 extracts can be attributed to the activity of SEX4 or other phosphatases in lsf2 extracts.
Figure 4.
Hydrolysis of C6- and C3-Phosphate Esters from Starch Granules by Extracts of the Wild Type, lsf2, sex4, and lsf2 sex4.
Purified phosphate-free starch granules from GWD-deficient Arabidopsis sex1-3 mutants were prelabeled with 33P at either the C6- or C3-positions and were then incubated with desalted extracts from whole rosettes of wild-type (WT) Col-0, sex4, lsf2, and lsf2 sex4 plants harvested at the end of the light period. Phosphate release over time was linear under these conditions and was expressed relative to the phosphate released by wild-type extracts. Each value is the mean ± se of four replicate samples.
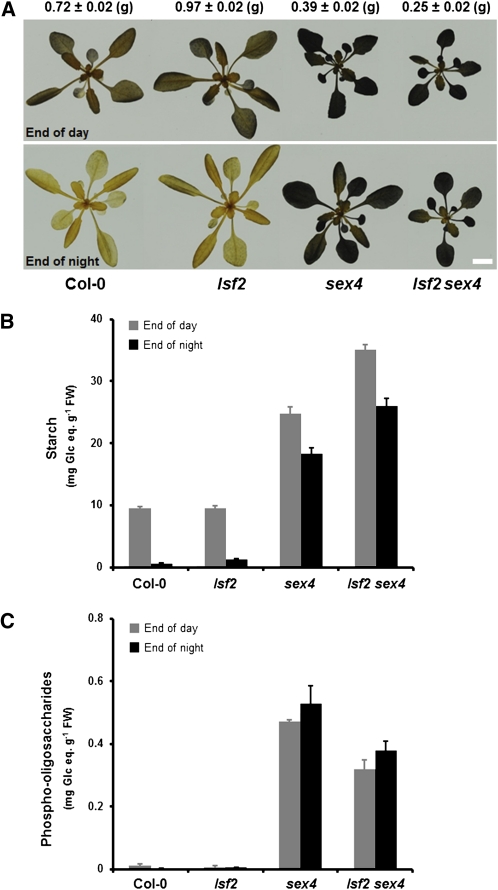
Leaves of lsf2-1 and lsf2-2 and their respective wild types were harvested at the end of the day and the end of the night. No differences in leaf starch content were revealed in either mutant compared with their wild types by qualitative iodine staining (Figure 5A) or by quantitative measurements after digestion of starch to Glc (Figure 5B; see Supplemental Figure 6C online). Thus, the loss of LSF2 does not prevent a normal rate of transitory starch degradation, at least under our growth conditions. We determined whether lsf2 mutants had altered glucan-bound phosphate by measuring total phosphate levels of leaf starch extracted at the end of the day and by measuring whether lsf2 plants contained soluble phospho-oligosaccharides. Total starch-bound phosphate was significantly elevated in both lsf2 alleles compared with their wild types (Table 2; see Supplemental Figure 6D online), supporting the idea that LSF2 dephosphorylates starch in vivo. However, no soluble phospho-oligosaccharides were detected in extracts of lsf2 leaves, whereas they were present in high amounts in sex4 extracts (Figure 5C; Kötting et al., 2009).
Figure 5.
Impact of the lsf2 Mutation on Starch Metabolism and Plant Growth.
(A) Photographs of wild-type Col-0 and lsf2, sex4, and lsf2 sex4 mutants harvested at the end of the day (top) and at the end of the night (bottom) after 4 weeks of growth. To visualize starch content, chlorophyll was cleared from the plants in 80% (v/v) ethanol and stained for starch with iodine solution. Representative plants were selected to show the reduced growth rate of the lsf2 sex4 mutant. Fresh weight average values are given above (g; n = 6). Bar = 1 cm.
(B) Leaf starch content at the end of the day (gray bars) and at the end of the night (black bars) in the wild-type Col-0 and lsf2, sex4, and lsf2 sex4 mutants. Each value is the mean ± se of nine replicate samples (P value < 0.05). FW, fresh weight.
(C) Phospho-oligosaccharides content at the end of the day (gray bars) and at the end of the night (black bars) in the wild-type Col-0 and lsf2, sex4, and lsf2 sex4 mutants. Each value is the mean ± se of nine replicate samples (P value < 0.05).
Table 2.
Glucan-Bound Phosphate Content in Wild-Type and Mutant Plants
| Starch | Amylopectin | |
| Genotype | nmol Phosphate μmol−1 Glc Equivalents | |
| Col-0 | 1.35 ± 0.037 | 1.47 ± 0.037 |
| lsf2 | 1.8 ± 0.04 | 1.97 ± 0.04 |
| sex4 | 1.09 ± 0.036 | 1.33 ± 0.036 |
| lsf2 sex4 | 1.3 ± 0.039 | 1.74 ± 0.039 |
Leaf starch was purified from pools of hundreds of 4-week-old plants harvested at the end of the light period. The amylopectin content was determined to be 92.6% ± 0.2% for the wild type, 91.4% ± 0.1% for lsf2, 79.1% ± 0.4% for sex4, and 75.5% ± 0.5% for lsf2 sex4. Starch-bound phosphate from the same preparation was determined using the malachite green assay (see Methods for details). The values show the results of one representative experiment with the se of three technical replicates (p value < 0.05). Similar results were obtained in a second independent experiment.
lsf2 Starch Contains High Levels of C3-Bound Phosphate
We reasoned that the elevated glucan-bound phosphate of the lsf2 starch could represent phosphate bound specifically to the C3-position. Therefore, we determined the chemical nature of the phosphate in starch isolated from leaves of wild-type and lsf2 plants by 31P NMR analysis (Figure 6; see Supplemental Figures 7 and 8 and Supplemental Table 1 online). Starch samples were digested with α-amylase and amyloglucosidase, and the products of digestion were subjected to matrix-assisted laser desorption ionization (MALDI)/MS/MS analysis prior to NMR analysis (see Methods). MALDI time-of-flight mass spectra revealed the presence of signals consistent with phospho-oligosaccharides, varying from three to 16 hexoses plus one or two phosphates (see Supplemental Figure 9 online). Although the phospho-oligosaccharide mixture is heterogeneous in terms of polymerization state, the 31P chemical shifts are mainly influenced by the local environment (e.g., formed by three consecutive glucoses) and are similar in phospho-oligosaccharides of different lengths.
Figure 6.
The lsf2 Mutation Causes Elevated C3-Bound Phosphate Levels.
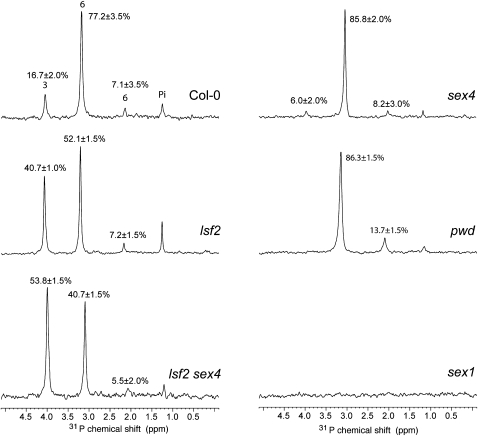
31P-NMR one-dimensional spectra of hydrolyzed starch of wild-type, lsf2, sex4, lsf2 sex4, sex1, and pwd plants harvested at the end of the light period recorded with between 9216 and 16,384 transients at 303K, pH 6.0. Peak areas are proportional to the relative amount of glucan-bound phosphate and are given as a percentage on top of each peak. Chemical shifts are referenced to external H3PO4 (85%).
A one-dimensional 31P spectrum of wild-type samples revealed four signals corresponding to four phosphate species. The type of linkage to Glc can be determined by analyzing through-bond long-range coupling constants (3JHP) between 1H and 31P with a 31P-1H heteronuclear single-quantum correlation spectrum (see Supplemental Figure 7, Supplemental Table 1, and Supplemental References online). In the case of O3 attachment, one signal correlating H3 and P as in Glc-3P is expected, whereas phosphate at O6 leads to two signals correlating H6 and H6′ with P. In the starch spectrum, signal 1 on the left shows one 1H-31P correlation (see Supplemental Figure 7C and Supplemental Table 1 online) and can thus be assigned as O3 attachment, and signals 2 and 3 show correlations to two protons and can be assigned as O6 attachment. Signal 4 does not show any 1H-31P correlation and likely originates from inorganic orthophosphate (see Supplemental Figure 7C and Supplemental Table 1 online). Our results build on previous NMR analyses (Ritte et al., 2006) but allow better separation of the signals at pH 6.0, enabling us to assign the previously unassigned signal 2 to a second C-6 phosphate species.
The ratio of C3- to C6-bound phosphate in wild-type starch was ~1:5 (Figure 6, Table 3), consistent with previous reports (Ritte et al., 2006; Haebel et al., 2008). We analyzed pwd, sex1, and sex4 starches as controls. As expected, C3-phosphate was reduced to the limits of detection in pwd starch, while sex1 starch was phosphate free (Figure 6; Ritte et al., 2006). In sex4 starch, the ratio of C3- to C6-bound phosphate was appreciably lower than the wild type (0.06 versus 0.20; Figure 6, Table 3; Comparot-Moss et al., 2010). Remarkably, in starch from both lsf2 mutant alleles, the proportion of C3-bound phosphate was increased to levels not previously reported (40.7 and 37.9% for lsf2-1 and lsf2-2, respectively; Figure 6, Table 3; see Supplemental Figure 8 online). These experiments suggest that the elevated levels of total glucan-bound phosphate in the lsf2 starch result from an accumulation of C3-bound phosphate. Thus, LSF2 makes a major contribution to dephosphorylation at the C3-position in vivo and appears to be only partially redundant with SEX4.
Table 3.
C3- and C6-Bound Phosphate Contents of Leaf Starch in Wild-Type and Mutant Plants
| C6-Bound | C3-Bound | C3/C6-Bound | |
| Genotype | nmol μmol−1 Glc Equivalents | ||
| Col-0 | 1.32 ± 0.05 | 0.27 ± 0.03 | 0.20 |
| sex4 | 1.24 ± 0.03 | 0.08 ± 0.03 | 0.06 |
| lsf2-1 | 1.25 ± 0.03 | 0.86 ± 0.02 | 0.69 |
| lsf2 sex4 | 0.79 ± 0.04 | 0.93 ± 0.03 | 1.16 |
| Landsberg erecta-0 | 1.47 ± 0.03 | 0.3 ± 0.04 | 0.2 |
| lsf2-2 | 1.47 ± 0.03 | 0.9 ± 0.04 | 0.61 |
Leaf starch was purified from plants harvested at the end of the light period, and total starch-bound phosphate was determined by the malachite green assay. The relative amounts of C3- and C6-bound phosphate were determined based on the peak areas of the corresponding 31P-NMR spectra (see Supplemental Figures 6 and 7 online).
lsf2 sex4 Double Mutants Show a Severe Starch Excess Phenotype and Growth Retardation
To uncover the functional relationship between SEX4 and LSF2, we generated the lsf2 sex4 double mutant using lsf2-1 (SAIL line 509F04) and sex4-3 (SALK line 102567; Niittylä et al., 2006) as parents. The ability of crude extracts of lsf2 sex4 leaves to act on sex1 starch granules prelabeled with 33P (see above) revealed that the dephosphorylating activity at both the C6- and the C3-position was significantly reduced compared with wild-type extracts (Figure 4). This suggests that SEX4 and LSF2 are the major glucan phosphatases in Arabidopsis leaves. However, some residual activity (18% at the C3-position and 30% at the C6-position) was still detected. This may mean that another phosphatase contributes to starch dephosphorylation (though we consider it unlikely to be LSF1; M. Umhang and S.C. Zeeman, unpublished data), but the activity may also be nonspecific, resulting from extrachloroplastic enzymes in the extract.
The loss of both glucan phosphatases impacts significantly on starch metabolism. lsf2 sex4 had significantly higher starch contents than sex4 at both the end of the day and the end of the night, although some daytime synthesis and nighttime degradation was still evident (Figure 5B). Thus, although lsf2 single mutants have normal starch levels, in the absence of SEX4, phosphate removal by LSF2 is important for starch degradation. The growth rate of lsf2 sex4 was reduced compared with both wild-type and sex4 plants (Figure 5A), which is expected as starch turnover is essential for normal growth in diurnal conditions (Zeeman et al., 2007). Intriguingly, total starch-bound phosphate levels in lsf2 sex4 were slightly lower than in lsf2 single mutants but higher than in sex4 and wild-type starches (Table 2). We used 31P-NMR spectroscopy to quantify the C3- and C6-bound phosphate (Figure 6). Interestingly, the ratio of C3- to C6-bound phosphate in lsf2 sex4 starch was even higher than in lsf2 starches (1.16 versus 0.69, respectively; Table 3). These data show that the glucan phosphatases have a major influence on both the amount and location of starch-bound phosphate.
To exclude the possibility that the changes in starch-bound phosphate are due to pleiotropic changes in the starch phosphorylating enzymes, we analyzed the amount of GWD and PWD proteins in the leaves of the phosphatase mutants. PWD protein amounts were the same in all lines (see Supplemental Figures 10A and 10B online). Slightly more GWD protein was detected in sex4 and lsf2 sex4 than in lsf2 and the wild type (see Supplemental Figure 10A and 10C online). Increased GWD could contribute to the elevated proportion of C6-bound phosphate in sex4 starch. However, the increase in C6-phosphate esters in sex4 is more likely due to the inability of LSF2 to act on C6-bound phosphate. The minor increase in GWD cannot explain the lsf2 sex4 phenotype.
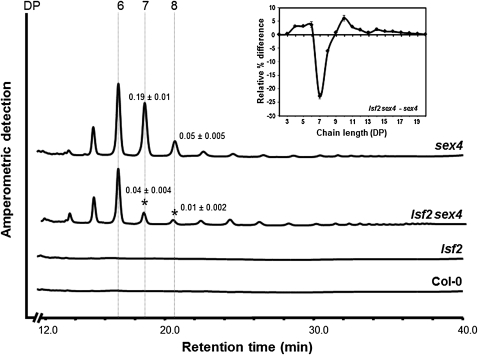
Most of the glucan-bound phosphate in sex4 is present as soluble phospho-oligosaccharides in the stroma. lsf2 sex4 leaves also contained phospho-oligosaccharides, but the amount was reduced by 30% relative to sex4 (Figure 5C). Analysis of the chain length distribution (Figure 7) revealed that the relative proportion of phospho-oligosaccharides with a degree of polymerization of seven and eight was significantly lower in lsf2 sex4 than in sex4. This suggests that different populations of phospho-oligosaccharides are released from the granule surface during starch degradation in the two lines. Alternatively, the way the released phospho-oligosaccharides are further metabolized differs in the two lines.
Figure 7.
Differences in the Chain Length Distribution of Phospho-Oligosaccharides Extracted from Leaves of sex4 and lsf2 sex4.
Soluble extracts from individual plants at the end of the night were dephosphorylated with 5 μg of recombinant SEX4 for 2 h. Released oligosaccharides were purified by ion-exchange chromatography and analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection. One representative chromatogram (from eight replicates of each) is shown. Numbered lines indicate the degree of polymerization (DP) of the detected oligosaccharides. Values above DP7 and DP8 indicate the amounts in milligram Glc equivalents g−1 fresh weight. Significant differences between sex4 and lsf2 sex4 are marked with asterisks (P value < 0.05). The inset shows the difference between the chain length distribution of the dephosphorylated oligosaccharides from lsf2 sex4 and sex4. Peak areas were summed, and the areas of the individual peaks were calculated as a percentage of the total ± se of at least four replicates. The difference plot was derived by subtracting the relative percentage values for sex4 from those of lsf2 sex4.
DISCUSSION
LSF2 Has Unique Biochemical Properties
This study identifies LSF2 as an enzyme involved in the transitory starch metabolism in Arabidopsis. It possesses a DSP domain highly similar to SEX4, has glucan phosphatase activity, and is able to dephosphorylate both soluble and insoluble substrates. Mutation of the active-site Cys conserved among different members of the DSP family abolished LSF2 activity. However, while SEX4 releases phosphate from both the C6- and the C3-positions of glucosyl residues of starch, we show that LSF2 has a strong preference for phosphate at the C3-position, although it also has a low capacity to hydrolyze the C6-bound phosphate.
It is interesting that LSF2 lacks a CBM domain (Figure 1A). Generally, CBMs function to increase the concentration of the enzyme on the substrate surface, to define affinity and ligand specificity, and potentially influence the substrate conformation prior to catalysis (Boraston et al., 2004). In SEX4, the DSP active site and the binding face of the CBM align to form a single continuous surface that is essential for glucan phosphatase activity (Vander Kooi et al., 2010). Mutations of conserved binding residues in the CBM abolish interaction with amylopectin and significantly reduce the ability of SEX4 to release phosphate from its glucan substrate (Gentry et al., 2007; Hejazi et al., 2010). By contrast, LSF2 does not require a CBM for its activity and presumably binds its substrate through secondary sites within or adjacent to the catalytic domain. Our structural modeling of LSF2 suggests that the absence of the CBM gives rise to a more compact protein structure compared with SEX4 (Figures 1B and 1C). It provides the basis for future mutagenic studies that will help define the structural features that allow substrate binding and confer specificity toward the C3-phosphate esters.
SEX4 and both LSF proteins possess a CT domain not found in other members of the DSP superfamily (Vander Kooi et al., 2010). In SEX4 and our structural model of LSF2, the CT domain interacts tightly with the DSP domain (Figures 1B and 1C). Truncation of the CT domain from either protein affects its solubility when expressed in a recombinant form in E. coli (Figure 1D; Vander Kooi et al., 2010). It is reasonable to hypothesize that the CT domain plays an important role in vivo for correct protein folding and/or stability. Further work will be required to determine whether it has an additional function specific to glucan phosphatases compared with other members of the DSP family. We are working to obtain the LSF2 crystal structure, which will help define the properties of the CT domain in LSF2 and understand how the enzyme interacts with its glucan substrates.
While SEX4, LSF1, and LSF2 share a common ancestry, the domains present in the common ancestor and the sequence of gene duplication events leading to the three gene classes are uncertain due to low phylogenetic signal from the short CBM domain and a lack of completed genomes. The most parsimonious explanation is that early in the lineage of green plants, a DSP domain from the eukaryotic host genome acquired a cTP and gained function in the site of starch storage, the plastid. Acquisition of a CBM domain gave rise to the common ancestor of LSF1, LSF2, and SEX4; phylogenetic analysis of the DSP domain as well as common exon-intron boundaries within the DSP and CBM-encoding sequences support a common ancestry for both domains. Duplication gave rise to the LSF1 class, which neofunctionalized (sequence divergence and acquisition of its PDZ-like domain), while a further duplication gave rise to SEX4 and LSF2, the CBM domain being lost in the latter (perhaps the result of imperfect gene duplication). As not all three genes were found in all taxa sampled (for example, the green algal genomes do not contain LSF1), this model implies some lineage-specific gene loss events. However, since relatively few green algal genomes are complete, and several of those are known to have reduced or divergent genomes (Derelle et al., 2006; Worden et al., 2009), gene loss might be limited to individual species.
The Function of LSF2 in Starch Metabolism
The evidence that LSF2 functions in the metabolism of transitory starch in Arabidopsis chloroplasts is compelling. First, LSF2 has a high degree of sequence similarity with SEX4, is expressed in starch-containing tissues, and shares a similar diurnal expression pattern to many genes involved in starch metabolism (see Supplemental Figure 2 online; Smith et al., 2004). Second, we show that LSF2 is localized in the chloroplasts (Figure 1E), which confirms an earlier finding (Schliebner et al., 2008). Third, we show that LSF2 can release phosphate bound to the C3-positions of the glucosyl residues of starch (Figure 3C). Although disruption of LSF2 does not in itself affect diurnal starch levels, it causes a dramatic increase in the proportion of C3-bound phosphate in the starch and a modest increase in total starch phosphate content (Tables 2 and 3, Figure 6).
Transient glucan phosphorylation is required for starch degradation, as indicated by the starch excess phenotypes of sex1, pwd, and sex4 mutants. Ritte et al. (2004) demonstrated that the rate of phosphate incorporation into starch was higher during periods of starch breakdown than during periods of starch synthesis. Since the phosphorylated glucans are themselves degraded during starch breakdown, the increased incorporation of phosphate is only transient (Ritte et al., 2004). However, there is an important difference between the rate of phosphorylation during starch degradation and the amount of starch-bound phosphate we measure. The latter reflects primarily the degree to which phosphorylation proceeds concurrently with starch biosynthesis (Nielsen et al., 1994). As lsf2 single mutants do not have a starch-excess phenotype, the starch we analyzed was synthesized during a single day. Thus, the increased amount of starch-bound phosphate must reflect the increased rate of phosphate incorporation during daytime biosynthesis. This in turn implies that LSF2 acts during the day to reduce C3-bound phosphate to the levels observed in the wild type and that net phosphate incorporation is a balance between the phosphorylation and dephosphorylation reactions. This hypothesis is also supported by the finding that LSF2 protein is encapsulated within starch granules in vivo (Table 1).
Unlike SEX4, LSF2 is not essential for starch breakdown in chloroplasts (Figure 5). Although our data show that SEX4 cannot fully compensate for the absence of LSF2, its activity on the C3-bound phosphate (Figure 3C) may be sufficient to mediate a normal rate of starch breakdown under the growth conditions we used. However, when SEX4 is missing, LSF2 is important in starch degradation because the severity of the starch excess phenotype in lsf2 sex4 is increased compared with sex4 (Figures 5A and 5B). Phosphate removal is important for the action of glucan-degrading enzymes. For example, β-amylases are unable to degrade chains that carry a phosphate close to the nonreducing end (Takeda and Hizukuri, 1981). It is also possible that other glucan hydrolytic steps might be prevented by phosphate groups. For example, while debranching enzymes can release phospho-oligosaccharides, their action might be inhibited if a phosphate group is located too close to their target branch points.
In sex4 mutants, the decrease in glucan phosphatase activity results in a large increase in glucan-bound phosphate, most of which is present as soluble phospho-oligosaccharides. These are released from the granule surface at night by debranching (ISA3) and endoamylolysis (AMY3; Kötting et al., 2009). Interestingly, no phospho-oligosaccharides accumulate in lsf2 mutants, probably because any that are released are dephosphorylated by SEX4. Phospho-oligosaccharides accumulate in lsf2 sex4, but unexpectedly there were less than in sex4 (Figure 5C). Furthermore, compared with sex4, the phospho-oligosaccharides of lsf2 sex4 had an altered chain length distribution (Figure 7). To explain this phenotype, we propose that the loss of both SEX4 and LSF2 results in a greater degree (and possibly a different pattern) of phosphorylation at the granule surface. We suggest that this inhibits glucan hydrolases: both β-amylases, which would release maltose, and debranching enzymes/α-amylase, which could release phospho-oligosaccharides. This explains both the increased starch content and the lower level of phospho-oligosaccharides. It is notable that phospho-oligosaccharide chains with degree of polymerization of seven and eight were significantly lower in lsf2 sex4 compared with sex4 (Figure 7), perhaps because fewer chains of this length are released from the granule surface. However, phospho-oligosaccharides may be further metabolized by stromal enzymes (such as AMY3 and DPE1; Critchley et al., 2001; Kötting et al., 2009), and the chain length profile could reflect the enzymatic rearrangements possible, depending on the phosphorylation pattern.
A further consideration is the degree of interdependence between phosphorylation and glucan hydrolysis. Edner et al. (2007) demonstrated not only that starch hydrolysis is stimulated by phosphorylation, but also that the reverse is true, as phosphorylation is stimulated by prior hydrolysis (Edner et al., 2007). Thus, if the loss of the glucan phosphatases prevents hydrolysis, this may in turn inhibit the further phosphorylation of the starch granule surface. This could account for the overall lower degree of glucan-bound phosphate in lsf2 sex4 compared with sex4.
Potential Biotechnological Applications of lsf2-Like Starches
The discovery that mutation of LSF2 results in starch with elevated amounts of phosphate specifically bound to the C3-position (Figure 6) creates options to produce novel starches for industrial applications. The degree of phosphorylation considerably influences the physico-chemical properties of starch granules, especially the hydration status (Muhrbeck and Eliasson, 1991). The presence of phosphate induces structural changes in amylopectin, promoting the solubility of the glucan chains (Blennow and Engelsen, 2010; Hejazi et al., 2010) and confers a high swelling power to starch gels (Blennow et al., 2001). Moreover, it has recently been proposed that, while C6-phosphorylation causes only minor changes in amylopectin structure, phosphorylation at the C3-position imposes significant steric effects and is predicted to induce conformational changes in the glucan backbone that can disrupt starch crystallinity (Hansen et al., 2009). Given that C3-phosphate esters are scarce in nature, the lsf2-like starches with elevated amounts of C3-bound phosphate may potentially have interesting biotechnological applications.
Thus far, only GWD has been manipulated in crop plants. Antisense inhibition of GWD in potato tubers resulted in a starch that has low phosphate content and low paste viscosity. It also prevents unwanted starch degradation in stored tubers (Lorberth et al., 1998). Overexpressing GWD in cereal endosperms is claimed to result in starches with very high swelling power when heated in water (Zeeman et al., 2010, and references therein). The impact of manipulating PWD, SEX4, and LSF2 in starch crops has yet to be determined. Potentially, the coordinated manipulation of both phosphorylating and dephosphorylating enzymes may allow both the amount of starch-bound phosphate and the ratio at the C3- or C6-positions to be controlled, leading to novel starches with further improved functionalities.
METHODS
Plant Materials and Growth Conditions
Plants for metabolite measurements were grown in a controlled environment chamber (Percival AR-95L; CLF Plant Climatics) in a 12-h-light/12-h-dark cycle with a constant temperature of 22°C, 65% relative humidity, and a uniform illumination of 150 μmol photons m−2 s−1. Plants used for the preparation of leaf starch granules were grown in a climate chamber (Weiss Umwelttechnik) with 16-h-light/8-h-dark regime and a constant temperature of 21°C and 60% relative humidity. Light intensity was between 120 and 140 μmol photons m−2 s−1. To promote uniform germination, imbibed seeds were stratified for 3 d at 4°C in the dark.
The following Arabidopsis thaliana T-DNA insertion mutants were used in this study: sex4-3 (Salk_102567; Niittylä et al., 2006), sex1-3 (Yu et al., 2001), pwd (SALK_110814; Kötting et al., 2005), lsf2-1 (Sail_595_F04; this work), and lsf2-2 (GT10871; this work). Arabidopsis ecotype Columbia-0 (Col-0) was used in all experiments, except for experiments involving lsf2-2, which was in a Landsberg erecta ecotype background. The lsf2 sex4 double mutant was obtained by crossing lsf2-1 and sex4-3 single mutants. Homozygous double mutants were identified by PCR-based screening and DNA sequencing, using gene-specific primers alone or in combination with a T-DNA left border-specific primer (see Supplemental Table 2 online for primer sequences).
LSF2 Subcellular Localization
To localize LSF2, its coding sequence was amplified from a full-length cDNA obtained from the Riken Bioresource Center (stock number pda16983) and cloned in frame with the N terminus of GFP in the vector pGFP2 (Haseloff and Amos, 1995). The LSF2-GFP fusion protein was transiently expressed in isolated Arabidopsis mesophyll protoplasts as described previously (Fitzpatrick and Keegstra, 2001). GFP fluorescence and chlorophyll autofluorescence were monitored using a confocal laser scanning microscope (TCS-NT; Leica Microsystems) with excitation windows of 507 to 520 nm and 620 to 700 nm, respectively. TCS-NT software version 1.6.587 was used for image acquisition and processing.
Quantitative RT-PCR
Total RNA was extracted from leaves using an RNeasy mini kit (Qiagen). Following DNase-I treatment, 1 μg of total RNA of each sample was used to produce cDNA using oligo(dT) primer (18-mer) and the SuperScript III first-strand synthesis system (Invitrogen). Quantitative PCR was performed using the SYBR Green Supermix (Eurogentec) with an iCycler (Applied Biosystems). Primer sequences are indicated in Supplemental Table 2 online. Reactions were run in triplicate with three different cDNA preparations, and the iQ5 Optical System Software (Applied Biosystems) was used to determine the threshold cycle (Ct) when fluorescence significantly increased above background. Gene-specific transcripts were normalized to PP2A and quantified by the ΔCt method (Ct of gene of interest − Ct of PP2A gene). Real-time SYBR green dissociation curves showed one species of amplicon for each primer combination.
Construction of the LSF2pro:GUS Fusion Gene and Arabidopsis Transformation
A DNA fragment corresponding to 1.5 kb of genome sequence upstream of the LSF2 start codon was amplified from Arabidopsis genomic DNA by PCR and sequenced to confirm that no spontaneous mutation was introduced. The DNA fragment was inserted into the pMDC163 binary vector (Curtis and Grossniklaus, 2003) upstream of the GUS reporter gene to create a recombinant unit LSF2pro:GUS. The reporter gene fusion was introduced into wild-type Arabidopsis plants (Col-0) through Agrobacterium tumefaciens–mediated transformation using the floral dip method (Clough and Bent, 1998). The independent transformants were selected on half-strength Murashige and Skoog media (Dufecha Biochemie) supplemented with hygromycin (50 μg mL−1) and transferred to soil after 2 to 3 weeks. T2 plants (i.e., progeny of transgenic generation 1) were used for the GUS staining. The primers used to confirm the recombinant transgene in transgenic plants are listed in Supplemental Table 2 online.
Histochemical GUS Staining and Microscopy
Seedlings were immersed in GUS staining solution (50 mM sodium phosphate buffer, pH 7.0, 0.05% [w/v] 5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid, cyclohexylammonium salt, 1 mM K3[Fe(CN)6], 1 mM K4[Fe(CN)6], and 0.05% [v/v] Triton X-100) and infiltrated under vacuum for 30 min. Staining proceeded for 4 or 16 h at 37°C. Chlorophyll was removed with 70% (v/v) ethanol, and the plant tissues were examined using conventional light microscopy. Images of GUS staining patterns are representative of at least three independent transgenic lines.
Homology Modeling of LSF2
HHpred (Söding, 2005; Söding et al., 2005) and InterPro Domain Scan (Zdobnov and Apweiler, 2001) were used to determine which DSP structure was the best template to model LSF2. The top hits were aligned with LSF2 using profile multiple alignment with predicted local structure 3D (PROMALS3D; Zdobnov and Apweiler, 2001). These alignments were the inputs in the alignment mode of Swiss-Model from Swiss PDB viewer version 8.05 (Arnold et al., 2006). Multiple homology models were inspected manually and by Anolea, Gromos, and Verify3d (Lüthy et al., 1992; Melo et al., 1997; Christen et al., 2005). The model using SEX4 (Protein Data Bank code 3nme) as the template was the best model.
Phylogenetic Analysis
Genomic sequences and gene models of Arabidopsis lyrata, Arabidopsis thaliana, Ricinus communis, Vitis vinifera, Oryza sativa, Zea mays, Sorghum bicolor, Selaginella moellendorffii, Physcomitrella patens, Populus trichocarpa, Glycine max, Zea mays, Volvox carteri, Chlamydomonas reinhardtii, Chlorella variabilis, Ostreococcus tauri, Osterococcus lucimarinus, Micromonas sp Micromonas pusilla, Mus musculus, Gallus gallus, Homo sapiens, Paramecium tetraurelia, Tetrahymena thermophila, Cyanidioschyzon merolae, Thalassiosira pseudonana, Porphyra yezoensis, Caenorhabditis elegans, Plasmodium falciparum, Guillardia theta, as well as all completed bacterial genomes were mined using BLASTx and tBLASTn (cutoff of e−4) with LSF1, LSF2, and SEX4 from Arabidopsis. Results were subject to reciprocal BLAST against the Arabidopsis genome, proteins with a different top hit were noted, and the corresponding Arabidopsis sequences were added to the results. All protein sequences were aligned using ClustalX (Thompson et al., 1997), and the alignment was imported into MacClade (Maddison and Maddison, 1992) for refinement. All proteins of bacterial origin as well as proteins with reciprocal results other than LSF1, LSF2, and SEX4 were easily aligned within the DSP domain. However, they generally encoded additional domains not present in LSF1, LSF2, or SEX4, which severely compromised the inclusion set within DSP and, after distance analysis of the DSP domain, were confirmed to be more related to other proteins. These were excluded from further analysis, leaving only proteins from eukaryotic species, the majority of which were from plants, green algae, and metazoans. The DSP domains of the remaining proteins were realigned with ClustalX. Ambiguously aligned characters were excluded in MacClade, and any sequences of the same species that were identical after exclusion of ambiguous characters were also collapsed to a single taxon, resulting in a matrix of 65 taxa and 150 characters. The alignment of these sequences is showed in Supplemental Data Set 1 online. ML phylogenies were inferred using (1) PhyML (Guindon and Gascuel, 2003) with the Dayoff substitution matrix and eight categories of substitution rates and (2) RAxML7.04 software (Stamakis, 2006) using the GTR+GAMMA model of evolution. The alpha value and number of invariable sites were calculated from the data sets. The branching support was assessed using ML bootstrap analysis (PhyML with four rate categories and 100 replications, RAxML, GTR+GAMMA, and 1000 replications) and Bayesian posterior probability values based on 1,000,000 generations and priors set to default using MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003).
Cloning, Expression, and Purification of Recombinant Proteins
The full-length cDNA of LSF2 was cloned into the pProEXHT vector (Invitrogen) according to standard protocols. Additional pET28b LSF2 constructs were generated where the first 78 or 65 amino acids (pET28b Δ78-LSF2 and pET28b Δ65-LSF2, respectively) or the last 35 amino acids (pET28b LSF2ΔCT and pET28b Δ65LSF2ΔCT) were deleted. pET21 Δ52-SEX4 has been previously described (Gentry et al., 2007). A point mutation in the LSF2 gene resulting in the C193S substitution was generated with the QuickChange site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions and cloned into the pProEXHta vector. Recombinant proteins were expressed with an N- or C-terminal hexahistidine tag in Escherichia coli BL21 (DE3) CodonPlus cells (Stratagene). Fusion proteins were expressed and purified from soluble extracts of E. coli using Ni2+-NTA agarose affinity chromatography as described previously (Kötting et al., 2005). Primer sequences used for recombinant cloning are listed in Supplemental Table 2 online.
Measurement of Phosphatase Activity
Phosphatase activity of recombinant enzymes against p-NPP (Fluka), solubilized amylopectin (Sigma-Aldrich), and purified phospho-oligosaccharides was measured using modifications of previously described methods (Worby et al., 2006). In all assays, the amount of SEX4 and LSF2 recombinant enzymes used was 0.05 and 0.2 μg, respectively. For p-NPP hydrolysis, each enzyme was incubated with 50 mM p-NPP at 37°C in 50-μL reactions with SEX4 assay medium containing 100 mM sodium acetate, 50 mM bis-Tris, 50 mM Tris, and 2 mM DTT, pH 6.5. Reactions were stopped at specific times by the addition of 200 μL 250 mM NaOH. The amount of released p-NPP was quantified by measuring absorbance at 410 nm. Activity against solubilized potato (Solanum tuberosum) amylopectin or purified phospho-oligosaccharides was determined by measuring released orthophosphate using the malachite green reagent. Phospho-oligosaccharides were isolated as previously described (Kötting et al., 2009). Recombinant enzymes were incubated with solubilized amylopectin (equivalent to 45 μg dry weight) or purified phospho-oligosaccharides (equivalent to 2 nmol phosphate) at 37°C in 20-μL reactions with assay medium (as above). Reactions were stopped with 20 μL of N-ethylmaleimide (250 mM) after the indicated incubation times. Subsequently, 80 μL of the malachite green reagent [2.5% (w/v) (NH4)6Mo7O24 and 0.15% (w/v) malachite green in 1 M HCl] was added, and the color allowed to develop at 20°C for 10 min. Absorbance at 660 nm was used to determine the phosphate groups released against a standard curve prepared with K2HPO4. Assays were performed in triplicate.
Starch Binding Assays
The starch binding capacity of recombinant LSF2 and SEX4 (Gentry et al., 2007) was determined in vitro. Calf intestine alkaline phosphatase (Fermentas) served as a nonstarch binding control. Each enzyme (5 μg) was incubated on a rotating wheel with prehydrated amylose-free potato starch (equivalent to 30 mg dry weight) at room temperature for 30 min in a final volume of 250 μL with assay medium (as above). Starch was pelleted by centrifugation, and unbound proteins remained in the supernatant. The pellet was washed once in 250 μL assay medium. Then, bound proteins were eluted by resuspending the starch pellet in 100 μL total protein extraction buffer (40 mM Tris-HCl, pH 6.8, 5 mM MgCl2, 4% SDS, and Complete Protease Inhibitor Cocktail [Roche]) for 30 min at 37°C. The soluble fraction and the supernatant from the wash were concentrated to 100 μL in Amicon Ultra spin concentrators (molecular mass cutoff of 10 kD; Millipore). Equal volumes of the concentrated unbound fraction and the eluted bound fraction were subjected to SDS-PAGE and visualized by silver staining. The activity of unbound proteins in the supernatant was measured against p-NPP (see above), and compared with control reactions that contained no starch.
Starch Granule Protein Analysis by MS/MS
Arabidopsis proteins were extracted from rosettes in 40 mM Tris-HCl, pH 6.8, 5 mM MgCl2, 1 mM DTT, and 1 mM phenylmethylsulfonyl fluoride. Extracted proteins (45 mg) were incubated for 4 h with 6 g of potato starch in a final volume of 50 mL at 4°C. The starch was collected by centrifugation and washed once with the same medium, and bound proteins were eluted with protein extraction buffer (see above).
Arabidopsis starch with bound proteins was isolated as described previously (Ritte et al., 2000). To extract proteins bound to the surface of the granules, the starch was incubated with the total protein extraction buffer described above for the starch binding assays. Protein encapsulated inside the starch granules were subsequently isolated by boiling the granules in a buffer containing SDS as described by Boren et al. (2004), except with omission of DTT (Boren et al., 2004). Extracted proteins were separated and visualized by Coomassie Brilliant Blue–stained SDS-PAGE. For proteomics analysis, gel slices were diced into small pieces and in-gel digestion was performed as describe previously (Shevchenko et al., 1996). After digestion, peptides were dried in a SpeedVac (Eppendorf AG) and subsequently dissolved in 3% (v/v) acetonitrile 0.2% (v/v) trifluoretic acid. Peptides were then desalted using Sepak C18 cartridges (Waters), redried in the SpeedVac, and then redissolved in 3% (v/v) acetonitrile and 0.2% (v/v) formic acid and analyzed on a Fourier transform ion cyclotron resonance mass spectrometer (Thermo Fisher Scientific) according to previously described methods (Agne et al., 2010). MS/MS spectra were searched with Mascot version 2.2.04 (Matrix Science) against the Arabidopsis TAIR10 protein database (downloaded on January 17, 2011) with a concatenated decoy database supplemented with contaminants. The search parameters were as follows: requirement for tryptic ends, one missed cleavage allowed, mass tolerance = ±5 ppm. Besides carbamidomethylation of Cys residues as fixed modification, oxidation of Met was included as variable modification. Peptide identification was accepted with a minimal Mascot ion score of 26 and a Mascot expectation value below 0.05, resulting in a false positive rate at the peptide level of below 1% for all measured samples.
Iodine Staining
Four-week-old Arabidopsis rosettes were harvested at the end of day or end of night and were incubated in 80% (v/v) ethanol for 12 h to remove the chlorophyll. The cleared plants were rinsed in water and stained in Lugol solution (Sigma-Aldrich) for 10 min.
Starch and Phospho-Oligosaccharide Extraction and Quantification
For starch content and phospho-oligosaccharide measurements, whole rosettes from 4-week-old Arabidopsis plants were harvested at the end of day or end of night, weighed, and snap frozen in liquid N2. Subsequent analyses were performed as previously described (Kötting et al., 2009).
GWD and PWD Protein Quantification
To extract total proteins, whole rosettes of 4-week-old Arabidopsis plants were harvested and immediately snap frozen in liquid N2. The entire rosette was homogenized in total protein extraction buffer as described in the starch binding assays section. Extracted proteins were quantified using the Pierce BCA protein assay kit (Thermo Fisher Scientific). Equal amounts of protein were separated by SDS-PAGE, electroblotted onto polyvinylidene fluoride, and probed with an antibody raised against potato GWD (Ritte et al., 2000) or Arabidopsis PWD (Kötting et al., 2005). The GWD and PWD bands, detected by chemiluminescence using a ChemiGlow West kit (Cell Biosciences), were quantified using gel analysis tools on ImageJ software (v1.42q; National Institutes of Health).
Quantification of Total Phosphate on Starch
Starch was isolated from whole Arabidopsis rosettes as described previously (Kötting et al., 2005). Starch granules (5 mg) were acid hydrolyzed in 50 μL 2 M HCl for 2 h at 95°C. The reaction was neutralized with 100 μL 1 M NaOH, and 50 μL was incubated with 15 units of Antarctic Phosphatase (New England Biolabs) for 2 h at 37°C in a final volume of 100 μL with assay medium (see above). Released orthophosphate was determined using the malachite green reagent, as above.
Phosphate Release from 33P-Labeled Granules
Phosphate-free starch granules isolated from the Arabidopsis sex1-3 mutant (Yu et al., 2001) were prephosphorylated with 33P at the C6- or C3-position as described by Hejazi et al. (2010). In both cases, the starch granules were phosphorylated at both locations, but the 33P-label was only at one or the other position. Recombinant potato GWD and recombinant Arabidopsis PWD were generated as described elsewhere (Ritte et al., 2002; Kötting et al., 2005). [β-33P]ATP was from Hartmann Analytic. Recombinant SEX4, LSF2, or LSF2 C/S (50 ng in each case) was incubated in dephosphorylation medium (100 mM sodium acetate, 50 mM bis-Tris, 50 mM Tris-HCl, pH 6.5, 0.05% [v/v] Triton X-100, 1 μg/μL [w/v] BSA, and 2 mM DTT) with 4 mg mL−1 starch prelabeled at either the C6- or the C3-position (see above) in a final volume of 150 μL on a rotating wheel for 5 min at 20°C.
Crude extracts of soluble protein were produced from 4-week-old Arabidopsis plants by homogenizing whole rosettes in a medium containing 50 mM HEPES-KOH, pH 7.5, 1 mM EDTA, 5 mM DTT, 10% (v/v) glycerol, and Complete Protease Inhibitor Cocktail. Extracts were desalted using NAP-5 Sephadex G-25 columns (GE Healthcare). Protein (37.5 μg) from these extracts was incubated with 0.75 mg of either C3- or C6-33P-labeled granules at 20°C for 20 min in reaction medium containing 50 mM HEPES-KOH, pH 7.0, 5 mM MgCl2, 5 mM CaCl2, 0.1% (w/v) BSA, 2 mM DTT, and 0.025% (v/v) Triton X-100 at a final reaction volume of 150 μL. Reactions were stopped by adding 50 μL of 10% (w/v) SDS, and the starch was pelleted by centrifugation. The amount of 33P released into the medium was quantified by scintillation counting. The starch pellet in blank reactions was also counted, and the data are expressed as the percentage 33P released into the supernatant. Phosphate release over time was linear under these conditions.
NMR Analysis of Starch-Bound Phosphate
Starch was isolated from Arabidopsis wild-type and mutant lines as described above, and 50 mg was suspended in 500 μL of medium containing 3 mM NaCl, 1 mM CaCl2, and 60 μg of α-amylase from pig pancreas (Roche). The suspension was shaken vigorously at 95°C for 5 min until the starch had gelatinized. A further 50 μg α-amylase and 450 μg amyloglucosidase from Aspergillus niger (Roche) was added, and digestion was performed at 37°C for 12 h with shaking, after which the solution was clear and nonviscous. All NMR measurements were performed on an Avance III 600-MHz spectrometer equipped with a QCI CryoProbe (Bruker) at 303K. Prior to analysis, the pH was adjusted to 6.0 with 0.2 M NaOH, and 5% (v/v) D2O was added to all samples. One-dimensional 31P spectra were recorded with 9200 to 16,400 transients, a recycle delay of 3.8 s, and 1H WALTZ16 decoupling (Shaka et al., 1983) at a field strength of 2.8 kHz. Spectra were indirectly referenced to H3PO4 (85% weight solution in water; AppliChem) using a Ξ value of 0.404807356 (Maurer and Kalbitzer, 1996). All spectra were processed with Topspin 2.1 (Bruker). Glucose-3-phosphate (Glycoteam) and glucose-6-phosphate (Roche) were used as a reference for peak identification.
MALDI/MS/MS Analysis of Phosphorylated Oligosaccharides
One milligram of wild-type digested starch (as prepared for NMR analysis) was dissolved in 25 μL of 10% (v/v) acetonitrile. Samples were spotted on the target by mixing 1 μL of matrix (40 mg mL−1 2,5-dihydroxybenzoic acid in 50% [v/v] acetonitrile, 1% [w/v] H3PO4 for wild-type starch, and 15 mg mL−1 2,5-dihydroxybenzoic acid in 30% [v/v] acetonitrile and 0.1% [v/v] trifluoroacetic acid for the rest) with 1 μL of each sample and then analyzed by MALDI (Ultraflex II TOF/TOF equipped with a smart beam laser) in the reflector negative ion mode. The analysis mass-to-charge range was 500 to 5000.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PP2A, At1g13320; LSF2, At3g10940; SEX4, At3g52180; LSF1, At3g01510; GWD, At1g10760; and PWD, At5g26570.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Structural Elements and Sequence Similarities between SEX4 and LSF2.
Supplemental Figure 2. Temporal and Spatial Expression Pattern of the LSF2 Gene.
Supplemental Figure 3. The Intron-Exon Structure of the Homologous Genes LSF2, LSF1, and SEX4.
Supplemental Figure 4. LSF2-Mediated Hydrolysis of C6- and C3-Phosphate Esters at Native Starch Granules.
Supplemental Figure 5. SDS-PAGE of Proteins Binding to Starch Granules.
Supplemental Figure 6. Phenotypic Characterization of lsf2 Mutant Alleles.
Supplemental Figure 7. Assignment of 31P Signals Using 2D NMR Spectroscopy.
Supplemental Figure 8. 31P-NMR Analysis of Arabidopsis Starch-Bound Phosphate.
Supplemental Figure 9. MALDI/MS/MS Analysis of Phosphorylated Oligosaccharides.
Supplemental Figure 10. GWD and PWD Protein Levels in Leaves of Wild-Type Col-0, lsf2, sex4, and lsf2 sex4 Plants.
Supplemental Table 1. Acquisition Parameters for 2D NMR Spectra.
Supplemental Table 2. Sequence of Primers Used in This Work.
Supplemental Data Set 1. DSP Domain Protein Sequences and Unambiguously Aligned Character Set Used as Input for Phylogenetic Analyses.
Supplemental References. Software Tools and Literature for NMR Spectroscopy.
Acknowledgments
We thank Simona Eicke, Tina Marthaler, Nadine Müller, Mena Nater, and André Imboden for technical help; the Functional Genomics Center Zürich for technical support in the MALDI/MS/MS analysis of phosphorylated oligosaccharides; Kuan-Jen Lu for assisting with the transcriptional data analysis; Klara Simkova for assisting with real-time PCR data analysis; and Shaun Peters for providing the pProEXHT vector collection and E. coli BL21 (DE3) CodonPlus competent cells. This work was supported by ETH Zurich (to O.K. and S.C.Z.), by the Swiss-South African Joint Research Program (to D. Santelia and S.C.Z.; Grant IZ LS Z3122916), by the Swiss National Foundation Sinergia Program (to M.S. and F.H.-T.A.; Grant CRSII3_127333), by the National Center for Research Resources of the National Institutes of Health (P20RR0202171), by the University of Kentucky College of Medicine startup funds, and by National Institutes of Health Grants R00NS061803 and R01NS070899 to D.A.M. and M.S.G.
AUTHOR CONTRIBUTIONS
D. Santelia, S.C.Z., O.K., M.S.G., and F.H.-T.A designed the research. D. Santelia, O.K., D. Seung, M.S., M.T., S.B., D.A.M., A.L., and N.P. performed the research. D. Santelia and S.C.Z. wrote the article.
References
- Agne B., Andrès C., Montandon C., Christ B., Ertan A., Jung F., Infanger S., Bischof S., Baginsky S., Kessler F. (2010). The acidic A-domain of Arabidopsis TOC159 occurs as a hyperphosphorylated protein. Plant Physiol. 153: 1016–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Baerenfaller K., Grossmann J., Grobei M.A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008). Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Baunsgaard L., Lütken H., Mikkelsen R., Glaring M.A., Pham T.T., Blennow A. (2005). A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J. 41: 595–605 [DOI] [PubMed] [Google Scholar]
- BeMiller J.N. (1997). Starch modification: Challenges and prospects. Starch-Stärke 49: 127–131 [Google Scholar]
- Blennow A., Bay-Smidt A.M., Olsen C.E., Moller B.L. (1998). Analysis of starch-bound glucose 3-phosphate and glucose 6-phosphate using controlled acid treatment combined with high-performance anion-exchange chromatography. J. Chromatogr. A 829: 385–391 [Google Scholar]
- Blennow A., Bay-Smidt A.M., Olsen C.E., Møller B.L. (2000). The distribution of covalently bound phosphate in the starch granule in relation to starch crystallinity. Int. J. Biol. Macromol. 27: 211–218 [DOI] [PubMed] [Google Scholar]
- Blennow A., Engelsen S.B. (2010). Helix-breaking news: Fighting crystalline starch energy deposits in the cell. Trends Plant Sci. 15: 236–240 [DOI] [PubMed] [Google Scholar]
- Blennow A., Mette Bay-Smidt A., Bauer R. (2001). Amylopectin aggregation as a function of starch phosphate content studied by size exclusion chromatography and on-line refractive index and light scattering. Int. J. Biol. Macromol. 28: 409–420 [DOI] [PubMed] [Google Scholar]
- Blennow A., Nielsen T.H., Baunsgaard L., Mikkelsen R., Engelsen S.B. (2002). Starch phosphorylation: A new front line in starch research. Trends Plant Sci. 7: 445–450 [DOI] [PubMed] [Google Scholar]
- Boraston A.B., Bolam D.N., Gilbert H.J., Davies G.J. (2004). Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 382: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boren M., Larsson H., Falk A., Jansson C. (2004). The barley starch granule proteome - Internalized granule polypeptides of the mature endosperm. Plant Sci. 166: 617–626 [Google Scholar]
- Christen M., et al. (2005). The GROMOS software for biomolecular simulation: GROMOS05. J. Comput. Chem. 26: 1719–1751 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Comparot-Moss S., et al. (2010). A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol. 152: 685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley J.H., Zeeman S.C., Takaha T., Smith A.M., Smith S.M. (2001). A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 26: 89–100 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E., et al. (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA 103: 11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edner C., Li J., Albrecht T., Mahlow S., Hejazi M., Hussain H., Kaplan F., Guy C., Smith S.M., Steup M., Ritte G. (2007). Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial beta-amylases. Plant Physiol. 145: 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick L.M., Keegstra K. (2001). A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J. 27: 59–65 [DOI] [PubMed] [Google Scholar]
- Fordham-Skelton A.P., Chilley P., Lumbreras V., Reignoux S., Fenton T.R., Dahm C.C., Pages M., Gatehouse J.A. (2002). A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J. 29: 705–715 [DOI] [PubMed] [Google Scholar]
- Fulton D.C., et al. (2008). Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry M.S., Dowen R.H., III, Worby C.A., Mattoo S., Ecker J.R., Dixon J.E. (2007). The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J. Cell Biol. 178: 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Haebel S., Hejazi M., Frohberg C., Heydenreich M., Ritte G. (2008). Mass spectrometric quantification of the relative amounts of C6 and C3 position phosphorylated glucosyl residues in starch. Anal. Biochem. 379: 73–79 [DOI] [PubMed] [Google Scholar]
- Hansen P.I., Spraul M., Dvortsak P., Larsen F.H., Blennow A., Motawia M.S., Engelsen S.B. (2009). Starch phosphorylation--maltosidic restrains upon 3′- and 6′-phosphorylation investigated by chemical synthesis, molecular dynamics and NMR spectroscopy. Biopolymers 91: 179–193 [DOI] [PubMed] [Google Scholar]
- Haseloff J., Amos B. (1995). GFP in plants. Trends Genet. 11: 328–329 [DOI] [PubMed] [Google Scholar]
- Hejazi M., Fettke J., Kötting O., Zeeman S.C., Steup M. (2010). The Laforin-like dual-specificity phosphatase SEX4 from Arabidopsis hydrolyzes both C6- and C3-phosphate esters introduced by starch-related dikinases and thereby affects phase transition of alpha-glucans. Plant Physiol. 152: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O., Pusch K., Tiessen A., Geigenberger P., Steup M., Ritte G. (2005). Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 137: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O., Santelia D., Edner C., Eicke S., Marthaler T., Gentry M.S., Comparot-Moss S., Chen J., Smith A.M., Steup M., Ritte G., Zeeman S.C. (2009). STARCH-EXCESS4 is a laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell 21: 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorberth R., Ritte G., Willmitzer L., Kossmann J. (1998). Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat. Biotechnol. 16: 473–477 [DOI] [PubMed] [Google Scholar]
- Lüthy R., Bowie J.U., Eisenberg D. (1992). Assessment of protein models with three-dimensional profiles. Nature 356: 83–85 [DOI] [PubMed] [Google Scholar]
- Maddison W.P., Maddison D.R. (1992). MacClade: Analysis of phylogeny and character evolution. Version 3. (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Maurer T., Kalbitzer H.R. (1996). Indirect referencing of 31P and 19F NMR spectra. J. Magn. Reson. B. 113: 177–178 [DOI] [PubMed] [Google Scholar]
- Melo F., Devos D., Depiereux E., Feytmans E. (1997). ANOLEA: A www server to assess protein structures. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5: 187–190 [PubMed] [Google Scholar]
- Muhrbeck P., Eliasson A.C. (1991). Influence of the naturally-occurring phosphate-esters on the crystallinity of potato starch. J. Sci. Food Agric. 55: 13–18 [Google Scholar]
- Nielsen T.H., Wischmann B., Enevoldsen K., Moller B.L. (1994). Starch phosphorylation in potato tubers proceeds concurrently with de novo biosynthesis of starch. Plant Physiol. 105: 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T., Comparot-Moss S., Lue W.L., Messerli G., Trevisan M., Seymour M.D., Gatehouse J.A., Villadsen D., Smith S.M., Chen J., Zeeman S.C., Smith A.M. (2006). Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J. Biol. Chem. 281: 11815–11818 [DOI] [PubMed] [Google Scholar]
- Ritte G., Lloyd J.R., Eckermann N., Rottmann A., Kossmann J., Steup M. (2002). The starch-related R1 protein is an alpha-glucan, water dikinase. Proc. Natl. Acad. Sci. USA 99: 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G., Lorberth R., Steup M. (2000). Reversible binding of the starch-related R1 protein to the surface of transitory starch granules. Plant J. 21: 387–391 [DOI] [PubMed] [Google Scholar]
- Ritte G., Heydenreich M., Mahlow S., Haebel S., Kötting O., Steup M. (2006). Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 580: 4872–4876 [DOI] [PubMed] [Google Scholar]
- Ritte G., Scharf A., Eckermann N., Haebel S., Steup M. (2004). Phosphorylation of transitory starch is increased during degradation. Plant Physiol. 135: 2068–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Santelia D., Zeeman S.C. (2011). Progress in Arabidopsis starch research and potential biotechnological applications. Curr. Opin. Biotechnol. 22: 271–280 [DOI] [PubMed] [Google Scholar]
- Schliebner I., Pribil M., Zühlke J., Dietzmann A., Leister D. (2008). A survey of chloroplast protein kinases and phosphatases in Arabidopsis thaliana. Curr. Genomics 9: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaka A.J., Keeler J., Freeman R. (1983). Evaluation of a new broad-band decoupling sequence: Waltz-16. J. Magn. Reson. 53: 313–340 [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Smith S.M., Fulton D.C., Chia T., Thorneycroft D., Chapple A., Dunstan H., Hylton C., Zeeman S.C., Smith A.M. (2004). Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 136: 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J. (2005). Protein homology detection by HMM-HMM comparison. Bioinformatics 21: 951–960 [DOI] [PubMed] [Google Scholar]
- Söding J., Biegert A., Lupas A.N. (2005). The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33(Web Server issue): W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamakis A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Takeda Y., Hizukuri S. (1981). Studies on starch phosphate. Part 5. Reexamination of the action of sweet-potato beta-amylase on phosphorylated (1->4)-α-D-glucan. Carbohydr. Res. 89: 174–178 [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Kooi C.W., Taylor A.O., Pace R.M., Meekins D.A., Guo H.F., Kim Y., Gentry M.S. (2010). Structural basis for the glucan phosphatase activity of Starch Excess4. Proc. Natl. Acad. Sci. USA 107: 15379–15384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T.P., Amrhein N., Freimoser F.M. (2005). Novel method for the quantification of inorganic polyphosphate (iPoP) in Saccharomyces cerevisiae shows dependence of iPoP content on the growth phase. Arch. Microbiol. 184: 129–136 [DOI] [PubMed] [Google Scholar]
- Worby C.A., Gentry M.S., Dixon J.E. (2006). Laforin, a dual specificity phosphatase that dephosphorylates complex carbohydrates. J. Biol. Chem. 281: 30412–30418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden A.Z., et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324: 268–272 [DOI] [PubMed] [Google Scholar]
- Yu T.S., et al. (2001). The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13: 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaniyama J., Denu J.M., Dixon J.E., Saper M.A. (1996). Crystal structure of the dual specificity protein phosphatase VHR. Science 272: 1328–1331 [DOI] [PubMed] [Google Scholar]
- Zdobnov E.M., Apweiler R. (2001). InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]
- Zeeman S.C., Kossmann J., Smith A.M. (2010). Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61: 209–234 [DOI] [PubMed] [Google Scholar]
- Zeeman S.C., Smith S.M., Smith A.M. (2007). The diurnal metabolism of leaf starch. Biochem. J. 401: 13–28 [DOI] [PubMed] [Google Scholar]