This work examines the distribution of four euchromatic histone modification marks (H3K4me2, H3K4me3, H3K36me3, and H3K4K9ac) within rice centromeres and finds that these marks are almost exclusively associated with centromeric subdomains that contain actively transcribed genes.
Abstract
The presence of the centromere-specific histone H3 variant, CENH3, defines centromeric (CEN) chromatin, but poorly understood epigenetic mechanisms determine its establishment and maintenance. CEN chromatin is embedded within pericentromeric heterochromatin in most higher eukaryotes, but, interestingly, it can show euchromatic characteristics; for example, the euchromatic histone modification mark dimethylated H3 Lys 4 (H3K4me2) is uniquely associated with animal centromeres. To examine the histone marks and chromatin properties of plant centromeres, we developed a genomic tiling array for four fully sequenced rice (Oryza sativa) centromeres and used chromatin immunoprecipitation–chip to study the patterns of four euchromatic histone modification marks: H3K4me2, trimethylated H3 Lys 4, trimethylated H3 Lys 36, and acetylated H3 Lys 4, 9. The vast majority of the four histone marks were associated with genes located in the H3 subdomains within the centromere cores. We demonstrate that H3K4me2 is not a ubiquitous component of rice CEN chromatin, and the euchromatic characteristics of rice CEN chromatin are hallmarks of the transcribed sequences embedded in the centromeric H3 subdomains. We propose that the transcribed sequences located in rice centromeres may provide a barrier preventing loading of CENH3 into the H3 subdomains. The separation of CENH3 and H3 subdomains in the centromere core may be favorable for the formation of three-dimensional centromere structure and for rice centromere function.
INTRODUCTION
The centromere is the chromosomal domain that serves as the docking site for the assembly of the kinetochore for chromosome segregation. Centromeric (CEN) chromatin is marked by a centromere-specific histone variant, CENH3 (known as CENP-A in mammals and CID in Drosophila melanogaster) (Henikoff et al., 2001). Centromeres can be inactivated or activated in a noncentromeric genomic region without changing the underlying sequences (Sullivan and Schwartz, 1995; Han et al., 2006; Marshall et al., 2008b; Zhang et al., 2010). Thus, the establishment and maintenance of centromeres is not defined by the underlying DNA sequences but is determined by poorly understood epigenetic mechanisms (Allshire and Karpen, 2008).
It has been speculated that CEN chromatin may be modified differently from the flanking pericentromeric heterochromatin, and this may serve as an epigenetic mark for CENH3 loading and centromere identity (Sullivan and Karpen, 2004). Indeed, both DNA sequences and histone proteins associated with CEN chromatin can be epigenetically differentially modified compared with those associated with the flanking pericentromeric chromatin (Sullivan and Karpen, 2004; Cam et al., 2005; Zhang et al., 2008; Koo et al., 2011). In humans and D. melanogaster, blocks of CENP-A/CID nucleosomes are interrupted by nucleosomes containing dimethylated H3 Lys 4 (H3K4me2), a euchromatic mark associated with permissive transcription (Sullivan and Karpen, 2004). Depletion of H3K4me2 within the centromere of a human artificial chromosome (HAC) resulted in a failure to recruit HJURP, a CENP-A chaperone, and inactivation of the centromere (Bergmann et al., 2011), which demonstrated a functional link between epigenetic modification of CEN chromatin and the maintenance of centromere stability.
Endogenous human and D. melanogaster centromeres consist almost exclusively of satellite repeats (Henikoff et al., 2001); thus, in these species, the centromeric histone modification patterns can be analyzed only by low-resolution cytological methods. It remains unclear how the blocks of CENP-A/CID nucleosomes and H3 nucleosomes intermingle and what is the threshold of the modified centromeric histones that can be reliably detected by the cytological methods. By contrast, rice (Oryza sativa) provides an excellent model for studying epigenetic modifications of CEN chromatin. Four rice centromeres (Cen4, Cen5, Cen7, and Cen8) have been fully or nearly fully sequenced (Nagaki et al., 2004; Wu et al., 2004; Zhang et al., 2004; Matsumoto et al., 2005), representing the best sequenced and characterized endogenous centromeres from any multicellular eukaryote. We used this sequence resource to develop a genomic tiling array that contains these four centromeres and the entire rice chromosome 8. Here, we report high-resolution maps of histone modifications associated with endogenous centromeres. We demonstrate that the euchromatic characteristics of rice CEN chromatin are trademarks of the transcribed sequences embedded in the centromeric H3 subdomains.
RESULTS
A Genomic Tiling Array Covering Four Rice Centromeres
We developed a genomic tiling array that covers four rice centromeres (Cen4, Cen5, Cen7, and Cen8) using the NimbleGen 3x720K array based on the NimbleGen HD2 platform. Each of the four rice centromeres contains a crossing-over suppressed region (CSR), spanning 2.09 to 3.61 Mb of DNA. Each CSR contains a CENH3-associated core domain, spanning 420 to 820 kb (Yan et al., 2008) (Figure 1). The core domain contains several CENH3 subdomains, each flanked by CENH3-lacking subdomains (Yan et al., 2008) (Figure 1). The entire sequence of rice chromosome 8 was included in the tiling array to allow comparisons of centromeric and noncentromeric sequences. The NimbleGen HD2 platform has the maximum capacity of 2.1 million probes. It uses long oligo probes of variable lengths (50 to 75 bp) to allow similar Tm values for all probes, providing outstanding sensitivity, specificity, and reproducibility of the microarray data sets. The tiling array contained 682,368 probes, covering the CSRs of chromosomes 4 (2.18 Mb), 5 (2.09 Mb), 7 (3.61 Mb), and the entire chromosome 8 (28.31 Mb, including the CSR of 2.3 Mb; Figure 1) with a median probe spacing of 27 bp. The repetitive DNA sequences specific to the rice centromeres were excluded from the tilling array (see Methods). Each NimbleGen slide contains three replicates of the probe set located on separate blocks (see Supplemental Figure 1 online).
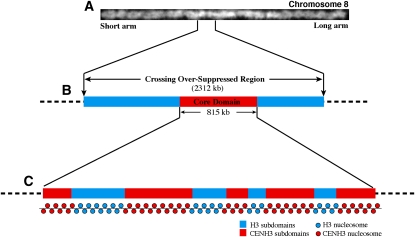
Figure 1.
A Diagram of the Structure of Rice Cen8.
(A) A digitally straightened rice pachytene chromosome 8. Genetic crossing over is suppressed in a 2.3-Mb region (CSR) in the centromere, which accounts for ~10% of the chromosome (Yan et al., 2005).
(B) The CSR of chromosome 8 is shown as an example. The core domain of Cen8, which contains CENH3, is 815 kb and embedded within the CSR.
(C) The core domain is diagrammed, consisting of five blocks of CENH3 nucleosomes interspersed by four H3 subdomains.
Chromatin Immunoprecipitation–Chip Studies on Histone Modifications
We conducted chromatin immunoprecipitation (ChIP)-chip experiments using antibodies specific to four modified histones: dimethylated H3 Lys 4 (H3K4me2), trimethylated H3 Lys 4 (H3K4me3), trimethylated H3 Lys 36 (H3K36me3), and acetylated H3 Lys 4, 9 (H3K4K9ac). All four modifications are typically associated euchromatin and active transcription (Kouzarides, 2007). ChIP was conducted using leaf tissue from 2-week-old rice seedlings, and the immunoprecipitated DNA was amplified (see Methods). Before hybridizing the amplified DNA to the microarrays, we used ChIP-PCR to test the relative enrichment in the immunoprecipitated DNA of sequences from two genomic regions that were previously characterized as being associated with euchromatic histone marks (Yan et al., 2005) (see Supplemental Figure 2 online).
For the microarray analysis, three biological replicates were performed for each histone modification examined. We exchanged the positions of biological and technical replicates on the NimbleGen slides to remove any position effect. Thus, each histone modification included 18 hybridization experiments (see Supplemental Figure 1 online). Only two of the 72 (4 × 18) experiments showed a relatively low Pearson correlation coefficient (<0.9; see Supplemental Table 1 online). Most of the Pearson correlation coefficients were >0.95, indicating a high reproducibility and reliability in the data sets. We also compared the H3K4me2 ChIP-chip data with our pervious H3K4me2 results derived from ChIP-PCR (Yan et al., 2005). The tiling array includes probes associated with 51 rice genes that were analyzed by ChIP-PCR for their association with H3K4me2 (Yan et al., 2005). ChIP-chip and ChIP-PCR results were consistent for 49 (96%) of the 51 genes. The two remaining genes were not detected by ChIP-PCR previously but were detected by ChIP-chip, indicating that the HD2-based system is highly sensitive.
Euchromatic Histone Modifications Were Mostly Associated with Genic Regions
We used a sliding window method to identify genomic regions that showed significant enrichment of each of the four histone modifications. Each enriched region contained a minimum of five consecutive probes (see Methods). In general, most of the enriched regions on the tilling array were associated with genes (nontransposable element [TE] related), ranging from 74.2% H3K4me3- to 78.9% H3K4me2-enriched regions (Table 1). Similarly, most enriched regions in the four centromeres were associated with genes, ranging from 70.5% H3K36me3- to 86.0% H3K4K9ac-enriched regions. Overall, 5.8% of the DNA sequences (a total of 146,645 bp) within the core domains of the four centromeres (a total of 2545 kb) was associated with at least one of the four modifications.
Table 1.
Identification of Enriched Regions Associated with H3K4me3, H3K36me3, H3K4K9ac, and H3K4me2
| No. of Enriched Regions |
Covering Length (Mb) |
Location of Enriched Regions |
|||||||
| Modification | Total | CSRa | Chr. 8b | Total | CSRa | Chr. 8b | Gene Body | Gene Body and 1 kb Upstream | TE-Related |
| H3K4me3 | 3696 | 500 | 3296 | 7.27 | 0.88 | 6.57 | 74.2% (2742) | 78.2% (2890) | 4.1% (150) |
| H3K36me3 | 2222 | 374 | 1931 | 6.89 | 1.17 | 6.07 | 78.0% (1731) | 82.2% (1826) | 4.5% (101) |
| H3K4K9ac | 3204 | 492 | 2827 | 6.20 | 0.74 | 5.62 | 75.5% (2420) | 80.0% (2564) | 3.7% (118) |
| H3K4me2 | 3582 | 534 | 3170 | 7.79 | 1.32 | 6.83 | 78.9% (2826) | 85.8% (3075) | 4.2% (149) |
Includes the CSR of Cen4, Cen5, Cen7, and Cen8.
The entire chromosome 8, including the CSR.
We detected a total of 23 intergenic regions (at least 200 bp away from any annotated genes) in the four centromeres, which were associated with at least one of the four modifications. We manually examined the annotation of these 23 regions. We found that 15 of the 23 regions showed high similarity to proteins in the nonredundant database (BLASTx, P = 1e-5), and three additional regions were associated with noncoding transcripts. Thus, the four histone modifications were almost exclusively associated with transcription or putative transcription in the rice centromeres. However, these results do not exclude the possibility of euchromatic modifications associated with the repetitive DNA sequences in rice centromeres. The centromere-specific satellite repeat CentO arrays in rice centromere 11 were found to be hypomethylated using a cytology-based assay (Yan et al., 2010).
Association of Euchromatic Histone Modifications with Transcription
To further examine the relationship between transcription and euchromatic histone modifications, we conducted massively parallel pyrosequencing of mRNAs (RNA-Seq) using leaf tissue from rice seedlings at the same growth stage used for the ChIP experiments. We obtained 40.1 million RNA-Seq reads from two biological replicates (see Methods). Our RNA-Seq data showed a high correlation with the recently published rice RNA-Seq data that was also derived from 2-week-old seedling tissue (Lu et al., 2010) (Spearman’s rank correlation coefficient = 0.87). The RNA-Seq data were used to reveal if a specific region on the NimbleGen tilling array is associated with transcription.
For genes associated with all four histone modifications, 87.2% of them were transcribed (Figure 2A). By contrast, only 13.3% of the unexpressed genes were associated with all four histone modifications (Table 2). We divided the genes into six categories based on their expression level (see Methods). Genes with high expression levels had a high tendency to be associated with all four histone modifications (Figure 2B). When active genes were further divided into 20 groups based on their expression levels, the levels of all four histone modifications were positively correlated with the logarithmic average gene expression levels (see Supplemental Figure 3 online).
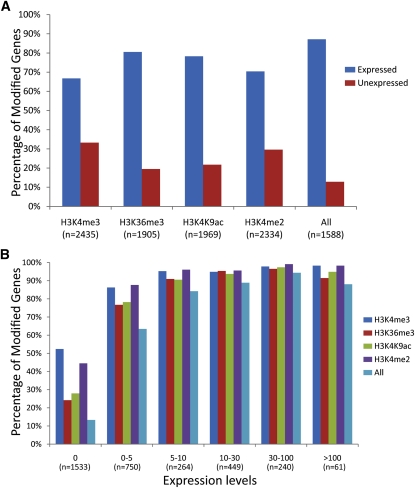
Figure 2.
Correlation between Gene Expression and Histone Modifications.
(A) Association of each of the four histone modifications (H3K4me3, H3K36me3, H3K4K9ac, and H3K4me2) with expressed and nonexpressed genes based on RNA-Seq data. Bars indicate the percentage of all expressed (blue) or unexpressed (red) genes enriched for this modification.
(B) Genes at different expression levels and their association with each of the four histone modifications. All genes were divided into six categories based their expression value FPKM (FPKM = 0, 0 to 5, 5 to 10, 10 to 30, 30 to 100 and >100). “All” indicates genes associated with H3K4me3, H3K36me3, H3K4K9ac, and H3K4me2 concurrently.
Table 2.
Histone Modifications Associated with Expressed and Nonexpressed Genes
| Gene Expression | H3K4me3 | H3K36me3 | H3K4K9ac | H3K4me2 | All Four Modifications | Total No. |
| Expressed gene | 1625 (92.1%) | 1534 (87.0%) | 1541 (87.4%) | 1643 (93.1%) | 1384 (87.2%) | 1764 |
| Nonexpressed gene | 810 (52.8%) | 371 (24.2%) | 428 (27.9%) | 691 (45.1%) | 204 (13.3%) | 1533 |
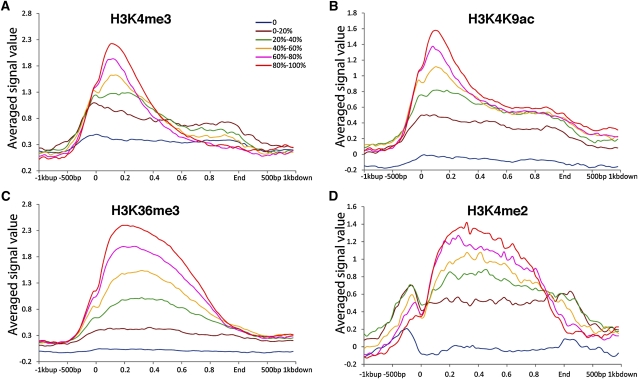
We aligned the histone modification data along rice genes, including 1 kb upstream of the transcription start sites (TSSs) and 1 kb downstream of the ends of the transcribed regions. H3K4me3 and H3K4K9ac peaked immediately after the TSS, tapered off within the first 20% of the gene, and then decreased sharply toward the 3′ end of the genes (Figure 3). H3K4me2 and H3K36me3 formed a broad plateau after the TSS, especially for highly expressed genes (Figure 3). Histone acetylation and methylation of H3K4 and K3K36 have been well documented to be associated with active genes. The patterns of the four histone modifications associated with rice genes were highly similar to those previously reported from both plant (Li et al., 2008; Wang et al., 2009; He et al., 2010) and mammalian species (Barski et al., 2007).
Figure 3.
Histone Modifications Associated with Rice Genes.
The distribution patterns of H3K4me3 (A), H3K4K9ac (B), H3K36me3 (C), and H3K4me2 (D) throughout the body and regulatory regions of rice genes. We divided the rice genes into six categories based on their expression levels; these are color coded and shown as separate lines (0, bottom 20%, 20 to 40%, 40 to 60%, 60 to 80%, top 20%). The y axis shows the relative signal strength for each modification. The x axis represents the genes, aligned by specific landmarks. For each gene, the gene body (including the coding sequence and 5′ and 3′ untranslated regions) was equally divided into 50 portions, which are plotted on the x axis as decimals from 0 to 1, with position 0 representing the TSS and position 1 representing the end of the gene. The 1 kb upstream and 1 kb downstream were also aligned based on the sequence positions.
The CENH3 Subdomains in Rice Centromeres Were Depleted of Histone H3–Associated Modifications
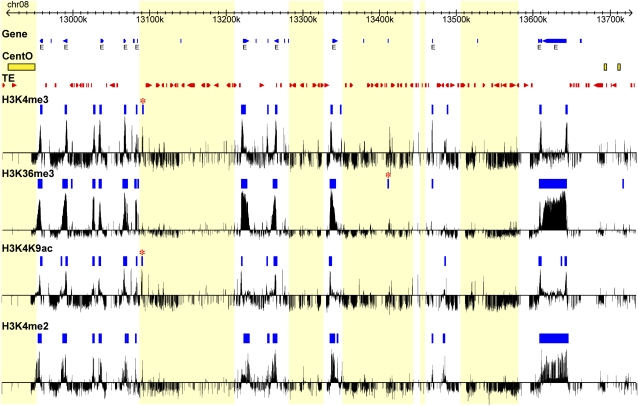
Each of the four rice centromeres (Cen4, Cen5, Cen7, and Cen8) consists of multiple subdomains associated with CENH3 (Yan et al., 2008). Each CENH3 subdomain is flanked by subdomains that lack CENH3 (Figure 1). The CENH3-lacking subdomains were assumed to contain only H3 nucleosomes; however, this was not supported by ChIP experiments using antibodies specific to H3 (Yan et al. 2005). The ChIP-chip results showed that the CENH3 subdomains in the four rice centromeres were depleted of all four histone modifications (Figure 4; see Supplemental Figure 4 online). The majority of the enriched regions were concurrently located in the CENH3-lacking subdomains (Table 3). Only 12 regions within the CENH3 subdomains, including three regions in Cen8 (Figure 4), were enriched with one to three of the four histone modifications. Manual annotation revealed that the DNA sequences of all 12 regions are related to TE, suggesting that these enriched regions may result from cross-hybridization of the ChIP DNA to partially homologous probes on the tiling array. These results confirm that the CENH3 subdomains largely do not contain any of these four types of modified histone H3. We refer to the CENH3-lacking subdomains as centromeric H3 subdomains hereafter.
Figure 4.
Histone Modifications Associated with Rice Cen8.
The sequence coordinates are shown at the top, with blue triangles representing genes, yellow rectangles representing CentO satellite repeats, and red triangles representing TE insertions. Expressed genes are marked with an “E” (RNA-Seq expression value > 0 FPKM). Enriched regions associated with H3K4me3, H3K36me3, H3K4K9ac, and H3K4me2 are marked by blue boxes; the bars underneath represent the normalized log2 value of the intensity of ChIP-chip hybridization relative to the reference. Three enriched regions within the CENH3 subdomains are marked by an asterisk. The light-yellow boxes across all tracks indicate the CENH3 binding subdomains.
Table 3.
Histone Modifications Associated with CENH3 Subdomains
| No. of Modified Regions in Centromere/CENH3 Subdomainsa | Length of Modified Regions in Centromere/CENH3 Subdomains (bp)b | ||||||||
| Centromere | H3K4me3 | H3K36me3 | H3K4K9ac | H3K4me2 | Allc | H3K4me3 | H3K36me3 | H3K4K9ac | H3K4me2 |
| Cen4 | 19/2 | 17/2 | 20/3 | 16/1 | 10/0 | 24,506/2,204 | 60,725/1,191 | 25,948/2,965 | 57,518/533 |
| Cen5 | 10/1 | 9/1 | 8/0 | 13/1 | 6/0 | 24,984/635 | 26,048/158 | 14,620/0 | 31,030/1,694 |
| Cen7 | 2/2 | 3/3 | 3/3 | 0/0 | 0/0 | 454/454 | 666/666 | 969/969 | 0/0 |
| Cen8 | 17/1 | 15/1 | 19/1 | 18/0 | 10/0 | 27,460/962 | 71,534/764 | 24,708/1,216 | 65,219/0 |
| Total | 48/6 | 44/7 | 50/7 | 47/2 | 26/0 | 77,404/4,255 | 158,973/2,779 | 66,245/5,150 | 153,767/2,227 |
The total number of enriched regions/the number of regions within the CENH3 subdomains in the same centromere.
The total length of enriched regions/the length of regions within the CENH3 subdomains in the same centromere.
Regions with all four modifications concurrently.
Similar Histone Modification and Expression Patterns Were Associated with Genes Located in Different Regions of Rice Chromosome 8
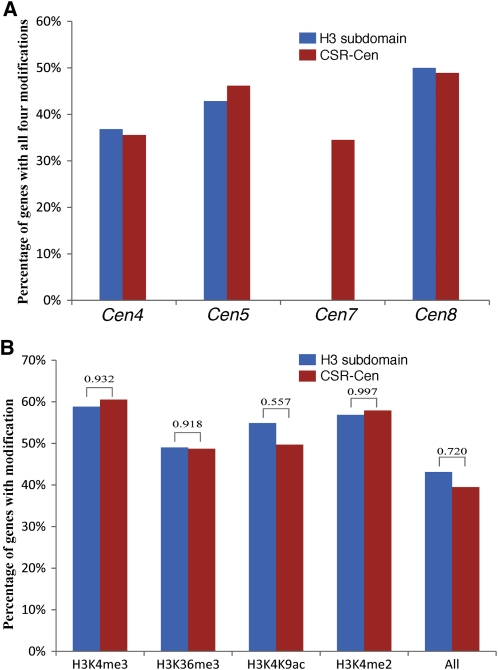
We compared the histone modification patterns associated with genes located in the H3 subdomains with those located in the rest of the CSR (CSR-Cen, all CSR genes excluding those in the core centromere) (Figure 5A). Although the percentage of genes associated with all four histone modifications varied from ~35 to 50% among the four centromeres, genes located in the H3 subdomains and those located within CSR-Cen showed similar modification patterns (Figure 5A). Approximately 43% of the genes located in the H3 subdomains were associated with all four modifications, which is not significantly different from that (39%) of the genes located in CSR-Cen (P = 0.72). Similar conclusions were obtained when the same two sets of genes were analyzed for individual histone modifications (P > 0.5; Figure 5B).
Figure 5.
Histone Modifications Associated with Genes Located in Different Subdomains within Rice Centromeres.
(A) Percentages of genes associated with all four histone modifications in four rice centromeres. H3 subdomain, H3 subdomains within the core of the centromere; CSR-Cen, CSR excluding the centromere core. Note: Rice Cen7 does not contain any known genes.
(B) Percentages of genes associated with H3K4me3, H3K36me3, H3K4K9ac, and H3K4me2 in H3 subdomains and CSR-Cen regions. The P values of χ2 tests are shown on the top of the bars.
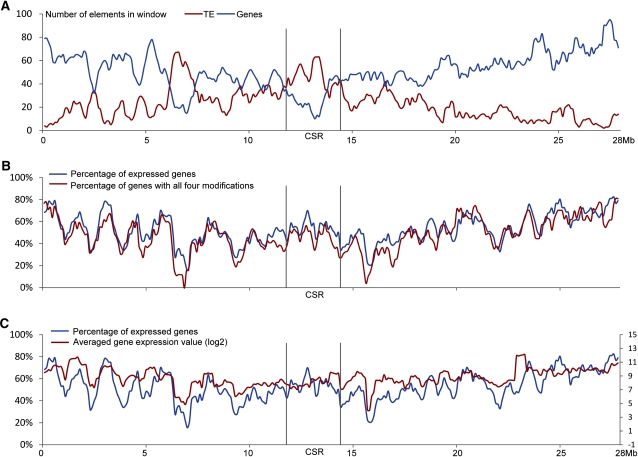
We then extended the same comparison to include all of chromosome 8 by analyzing the variance in percentage of genes associated with specific modifications using a sliding window method. The distal ends of rice chromosome 8 showed a higher density of genes and a lower density of TEs compared with the pericentromeric region (Figure 6A), which correlates with the brighter 4′,6-diamidino-2-phenylindole staining pattern in the pericentromeric region, as seen on the pachytene chromosome 8 (Figure 1). The CSR of chromosome 8 showed a higher density of TEs and a lower density of genes than the rest of the chromosome. Although the density of genes in the CSR is low, the percentage of CSR genes associated with all four histone modifications was not significantly different from the percentage of genes located in the rest of the chromosome 8 (Kolmogorov-Smirnov test, P = 0.36) (Figure 6B).
Figure 6.
Histone Modification and Expression Patterns of Rice Genes Located along Chromosome 8.
(A) Distribution of genes and TEs along rice chromosome 8 (window size = 500 kb; step = 100 kb). The CSR of Cen8 contains a lower density of genes than the rest of the chromosome (except for the ~6- to 7.5-Mb domain on the short arm).
(B) Percentage of genes within sliding windows that are associated with all four histone modifications (H3K4me3, H3K4me2, H3K36me3, and H3K4K9ac).
(C) Percentages of expressed genes with the sliding window and their average expression levels along chromosome 8.
A sliding window method was also used to examine the percentages of expressed genes and the gene expression levels (displayed by log2 averaged gene expression value) along rice chromosome 8. Similarly, these values within CSR were not statistically different from those associated with the rest of the chromosome (Kolmogorov-Smirnov test: P = 0.76 for percentage of expressed genes, and P = 0.16 for gene expression level) (Figure 6C).
DISCUSSION
Euchromatic Histone Modifications Associated with Rice CEN Chromatin
The histone modifications associated with CEN chromatin have mostly been examined in humans. In endogenous human centromeres, CENP-A nucleosomes are interspersed with H3-associated nucleosomes marked by H3K4me2 (Sullivan and Karpen, 2004; Mravinac et al., 2009). Interestingly, these centromeric H3 nucleosomes were neither associated with the euchromatic marks H3K4me3 and acetylated H3/H4 nor with the heterochromatic marks H3K9me2 and H3K9me3 (Sullivan and Karpen, 2004). A recent HAC-based study suggested that the presence of H3K4me2 is essential for the proper centromere function of the artificial chromosome (Bergmann et al., 2011). However, ChIP-chip analysis revealed that H3K4me2 is not a prominent mark in three human neocentromeres (Alonso et al., 2010). H3K4me2 was only detected in a <2-kb region within a 75- to 90-kb CENP-A domain in the neocentromeres. In addition, these small H3K4me2 pockets were not caused by neocentromere activation, as they existed in the same region on the corresponding normal human chromosome. Thus, this minimum amount of H3K4me2 is unlikely to play a critical role in neocentromere function (Alonso et al., 2010). Similarly, H3K4me2 is depleted in the centromeres of maize (Zea mays; Shi and Dawe, 2006; Jin et al., 2008) and Neurospora crassa (Smith et al., 2011).
We demonstrate that H3K4me2 is distributed throughout the centromeric H3 subdomains of rice Cen4, Cen5, and Cen8 (Figure 4; see Supplemental Figure 4 online). However, H3K4me2 is neither the sole euchromatic mark in these centromeres nor ubiquitous in rice CEN chromatin. The majority of H3K4me2 regions were associated with transcription, and the same regions were also enriched for the transcription-related histone marks H3K4me3, H3K36me3, and H3K4K9ac. Genes located in the centromeric H3 subdomains showed histone modification patterns similar to genes located throughout chromosome 8 of rice (Figure 6). Thus, the euchromatic histone modifications associated with rice Cen4, Cen5, and Cen8 are not specific to rice CEN chromatin but are hallmarks of the DNA sequences, including active genes, embedded within the centromeric H3 subdomains. In comparison, none of these four histone modifications were detected in Cen7, which contains only a small centromeric H3 subdomain that lacks any transcribed sequences (see Supplemental Figure 4 online). Most intergenic sequences within the centromeric H3 subdomains were not associated with the euchromatic histone modifications (Figure 4; see Supplemental Figure 4 online). A ChIP-PCR–based analysis showed that at least some of these intergenic sequences in Cen8 are associated with the heterochromatic histone modification mark H3K9me2 (Yan et al., 2005).
Transcription and Centromere Function
Depletion of H3K4me2 at the centromere of a HAC caused a loss of transcription of the underlying centromeric satellite DNA and ultimately the inactivation of the centromere (Bergmann et al., 2011). The impact of H3K4me2 and transcription on centromere function demonstrated in this HAC-based system contrasts with the fact that centromeres are always associated with gene-poor regions, which may lack transcription. Human neocentromeres and evolutionarily new centromeres are often associated with gene deserts (Lomiento et al., 2008; Alonso et al., 2010). Similarly, induced neocentromeres in Candida albicans were preferentially associated with gene-poor regions (Ketel et al., 2009).
CENH3-associated nucleosomes appear to be incompatible with transcription because insertion of a marker gene in CEN chromatin of Schizosaccharomyces pombe resulted in silencing of the transgene (Allshire et al., 1995). Neocentromere formation in S. pombe can negatively impact the transcription of the underlying endogenous genes (Ishii et al., 2008). Similarly, neocentromere formation can cause silencing of the underlying transgene in C. albicans (Ketel et al., 2009). Transcripts derived from centromeric repetitive DNA were reported in a number of plant and animal species (Topp et al., 2004; May et al., 2005; Wong et al., 2007; Carone et al., 2009). However, the transcription of centromeric DNA is likely associated with H3 nucleosomes within the centromeric cores.
We propose that transcription within the H3 subdomains of rice centromeres may provide a barrier for the loading of CENH3. Transcription in the centromeric H3 subdomains may assist in evicting any misloaded CENH3. Such a loading barrier would help maintain the separation of H3 nucleosome blocks from CENH3 nucleosome blocks. We analyzed the expression patterns of all rice chromosome 8 genes based on the microarray-based gene expression data that were compiled from 32 stages/organs of rice reproductive development using a total of 95 microarray hybridizations (Fujita et al., 2010). Expression data were found for eight Cen8 genes. Interestingly, the expression of five of the eight genes was detected in all 95 microarray experiments, and one was detected in 94 experiments. The remaining two genes (LOC_Os08g21720 and LOC_Os08g22250) were only detected in two or three experiments, respectively. In addition, LOC_Os08g21720 showed sequence similarity with Ty1-copia retrotransposons (e value = 2E-36). LOC_Os08g22250 is a hypothetical protein without any similarity to known proteins in the nonredundant database. These results show that most Cen8 genes express constitutively, which may be favorable for their potential role in separating the H3 and CENH3 subdomains.
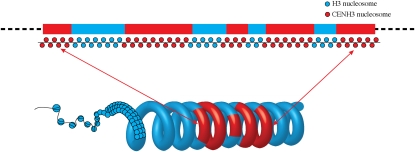
A centromere consisting of alternating CENH3 and H3 nucleosome blocks would be ideal for its three-dimensional structure: folding or looping of the nucleosome blocks will move the CENH3 subdomains to the outside of the chromatin to provide accessibility for kinetochore assembly and move the centromeric H3 subdomains to an interior position (Marshall et al., 2008a) (Figure 7). The centromeric H3 subdomains positioned on the interior of the chromatin could then be modified properly for other centromeric functions, such as chromatid cohesion. The structure of alternating CENH3 and H3 nucleosome blocks was reported in a human neocentromere (Chueh et al., 2005) and is also likely associated with the maize B chromosome centromere (Koo et al., 2011). Other epigenetic modifications of CEN chromatin, such as differential cytosine methylation of centromeric DNA (Zhang et al., 2008; Koo et al., 2011; Zakrzewski et al., 2011), could also serve as barriers for the separation of H3 and CENH3 nucleosome blocks.
Figure 7.
A Model of a Potential Three-Dimensional Structure of Rice centromeric.
Coiling or folding of the nucleosome blocks within the centromeric core moves the CENH3 subdomains to the inner kinetochore and the centromeric H3 subdomains to an interior position. The coiling model followed Marshall et al. (2008a).
METHODS
Development of a Rice CEN Genomic Tiling Array
We designed a customized NimbleGen 3x720K tiling microarray that covered four rice (Oryza sativa) centromeres (Cen4, Cen5, Cen7, and Cen8) and the entire chromosome 8 (Chr8). Each slide contained three replicates of 696,675 probes, with each replicate including 682,368 experimental probes and 4769 control probes, which are in triplicate. The probes were selected from the rice version 4 pseudomolecules (http://rice.plantbiology.msu.edu/downloads.shtml) on the basis of synthesis cycles, Tm, and sequence uniqueness. The resulting probes were then compared with the rice genome via BLASTn, where probes with more than 10 copies in the genome (≥90% similarity and ≥90% coverage) were excluded. The final list of experimental probes consisted of 30,706 probes from Cen4 (2.54 Mb, 7,860,001 to 10,400,000 bp), 28,183 probes from Cen5 (2.1 Mb, 11,370,001 to 13,470,000 bp), 53,404 probes from Cen7 (3.62 Mb, 9,100,001 to 12,720,000 bp), and 570,075 probes from Chr8 (28.31 Mb). The probes were between 50 and 75 bp in length, with a median spacing of 27 bp (the distance between the start positions of adjacent probes).
In addition, we also designed four sets of control probes, 4769 in total. The first set of 335 probes was derived from 55 genes in a 3.5-Mb region spanning rice Cen8; these genes were found to be enriched for both H4 acetylation and H3K4me2 (35), H4 acetylation only (4), H3K4me2 only (4), or neither of them (12) (Yan et al., 2005). The second set of 1878 probes (negative controls) was selected from four 400-kb regions, one each from Chr1 (1,600,001 to 2,000,000 bp), Chr2 (4,000,001 to 4,400,000 bp), Chr3 (1,550,001 to 1,950,000 bp), and Chr4 (34,600,001 to 35,000,000 bp). These 400-kb sequences were split into 60-bp bins, sliding every 20 bp, and via BLASTn were compared against the rice genome. Adjacent unique bins (no other hits with ≥40 bp alignment except to itself) were merged into subsequences, and reconstructed bins of 160 bp or longer were retained. For each of the four 400-kb regions, we split the retained subsequences into nonoverlapping 15-mer oligonucleotides and concatenated all the 15-mer oligonucleotides into a single pseudomolecule, requiring that the coordinates of any two neighboring 15-mer oligonucleotides be separated by least 30 kb. The four pseudomolecules were finally used for probe selection. The third set of 994 probes (negative controls) was selected from 90 Arabidopsis thaliana genes whose transcription was supported by the existence of cognate full-length cDNAs. These Arabidopsis genes have homologs in the rice genome but lack sequence similarity at the DNA level. The last set of 1562 control probes was designed from the 490,520-bp rice mitochondrial genome (BA000029.3), which was masked for regions that matched elsewhere in itself or the nuclear genome sequence. The array was manufactured by Roche NimbleGen using Maskless Array Synthesizer technology.
ChIP
ChIP was performed as previously described with only minor modifications (Nagaki et al., 2003). Approximately 5 g of leaf tissue from 2-week-old seedlings of rice variety Nipponbare was used in each ChIP experiment. Rabbit polyclonal antibodies against H3K4me2 (Upstate 07-030), H3K4me (Abcam ab8580), H3K36me3 (Abcam ab9050,) and H3K4K9Ac (Upstate 06-599) were used in the ChIP and rabbit normal serum in mock experiments. We also took an aliquot from the chromatin solution as input DNA. The input DNA was not immunoprecipitated. All DNA from ChIP experiments was amplified using the GenomePlex whole-genome amplification kit (Sigma-Aldrich) with a fragmentation reaction. To reduce the bias of the amplification, amplifications were performed in eight tubes and upon amplification completion were mixed into one tube. The amplified DNA was purified using a PCR purification kit (Qiagen).
We conducted ChIP-PCR to verify the relative enrichment of each histone modification. ChIP-PCR was performed in triplicate in a 20-μL PCR mixture (1× PCR buffer (without Mg), 1.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphate, 0.2 μM primers, 1 ng ChIP, mock or input DNA, and 0.5 units of Platinum Taq DNA polymerase (Invitrogen) and run at 95°C for 5 min, 32 cycles of 95°C for 30 s, 65°C for 30 s, and 72°C for 30 s. The primers used were from rice centromeres, as previously described (Yan et al., 2005). PCR products were electrophoresed on an agarose gel and stained with ethidium bromide.
Microarray Hybridization
The ChIP and input DNA were labeled according to published protocols (Selzer et al., 2005) and Roche NimbleGen’s ChIP-chip user’s guide v6.1. In short, 1 μg of amplified ChIP DNA and 1 μg of input DNA was labeled using 5′ Cy3- or Cy5-labeled random nonamers, respectively (NimbleGen dual-color DNA labeling kit). DNA was incubated for 3 h at 37°C with 100 units of (exo-) Klenow fragment and 10 μL of 10 mM deoxynucleotide triphosphate mix (NimbleGen dual-color DNA labeling kit). The labeled samples were then precipitated with NaCl and isopropanol and rehydrated in 25 μL of water. The immunoprecipitated and input samples were combined in a 1.5-mL tube and dried down using a SpeedVac. Samples were resuspended in 5.6 μL of water and 14.4 μL of Roche NimbleGen hybridization buffer (NimbleGen hybridization and sample tracking control kit) and incubated at 95°C. The combined and resuspended samples were then hybridized to the array for 16 to 20 h at 42°C with mixing. Arrays were washed using the Roche NimbleGen wash buffer system and dried using the NimbleGen microarray dryer (Roche NimbleGen). Arrays were scanned at 5-μm resolution using the GenePix4000B scanner (Axon Instruments). Data were extracted from scanned images using NimbleScan v2.4 data extraction software, which allows for extraction, grid alignment, and generation of data files.
RNA-Seq
Rice seedlings at the same growth stage and under the same growing condition as those for ChIP experiments were used for RNA-Seq analysis. Total RNA was extracted using the RNeasy plant kit (Qiagen). Two biological replicates were prepared for RNA-Seq. RNA quality was checked with a Bioanalyzer 2100 (Agilent Technologies). High-quality RNA (RNA integrity number >8) was used for downstream DNase I treatment to completely remove any genomic DNA contamination. About 10 μg total RNA was converted to cDNA using the mRNA-seq kit from Illumina, and the barcoded cDNA library was sequenced on an Illumina Genome Analyzer 2 platform. We used TopHat software (Trapnell et al., 2009) to align the sequence reads to the reference genome and used Cufflink (Roberts et al., 2011) to call the expression values (fragments per kilobase of exon model per million mapped fragments [FPKM]) of annotated rice genes. We obtained 40.1 millions of RNA-Seq reads, and 89.2% of them were mapped to the rice genome (TIGR 5). Two biological replicates showed a high correlation (Spearman's rank correlation coefficient = 0.964). Thus, the data from the two replicates were merged for further analyses. We also downloaded recently published rice RNA-Seq data that were also derived from 2-week-old rice seedlings (Lu et al., 2010) for a comparative analysis with our RNA-Seq data sets. We filtered out TE-related genes in association analysis between gene expression and histone modifications. We divided the genes into six categories based their expression values (FPKM = 0, 0 to 5, 5 to 10, 10 to 30, 30 to 100, and >100) or divided active genes in 20 groups with same amount from the top 5% to the bottom 5%.
Data Analysis
Raw data for H3K4me3, H3K36me3, and H3K4K9ac were normalized by the biweight method. Raw data for H3K4me2 were normalized by the MA2C method (Song et al., 2007). All normalized data followed a Gaussian distribution, which was required to use ChIPOTle (Buck et al., 2005), the method we used to identify enriched regions. To remove the variance of replicates, the data were processed with Quantile normalization and then averaged. Regions enriched with one of the four histone modifications were identified by ChIPOTle (Buck et al., 2005) (version 1.11, sliding window = 600 bp, step = 150 bp, α = 0.01 after Benjamini and Hochberg multiple comparison correction [Benjamini and Hochberg, 1995] to control false discovery rate < 0.01). A minimum of five consecutive probes was required to be classified as an enriched region. The rice annotation information from TIGR database (release 5) was used in the analysis. The information of CSR, centromere regions, and the information of CENH3 binding domains in rice genome were defined by our lab previously (Yan et al., 2005, 2008). The intergenic transcripts used in this study were identified in rice previously (Li et al., 2007).
Accession Number
The ChIP-chip and RNA-Seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE29597 and GSE33265, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ChIP-Chip Experimental Design Using the NimbleGen 3x720K Tiling Array.
Supplemental Figure 2. Relative Enrichment Tests Using ChIP-PCR of Genes Associated with Rice Cen8.
Supplemental Figure 3. Correlation between the Expression Levels of the Genes and Each of the Four Euchromatic Histone Modifications.
Supplemental Figure 4. Histone modifications associated with rice Cen4, Cen5, and Cen7.
Supplemental Table 1. Correlation of ChIP-Chip Hybridization Experiments.
Acknowledgments
We thank Zhukuan Cheng for providing the pachytene chromosome 8 image in Figure 1, Robin Buell for helping with the rice RNA-Seq experiments, and the Dale Bumpers National Rice Research Center for providing the Nipponbare seeds. This research was supported by Grants DBI-0603927 and DBI-0923640 from the National Science Foundation to J.J.
AUTHOR CONTRIBUTIONS
J.J. and A.L.I. designed the research. S.K., W.Z., and H.R. performed the research. Y.W. and H.Y. analyzed data. J.J., Y.W., and A.L.I. wrote the article. J.J. and A.L.I. are joint senior authors who contributed equally.
References
- Allshire R.C., Karpen G.H. (2008). Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R.C., Nimmo E.R., Ekwall K., Javerzat J.P., Cranston G. (1995). Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9: 218–233 [DOI] [PubMed] [Google Scholar]
- Alonso A., Hasson D., Cheung F., Warburton P.E. (2010). A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K.R., Roh T.Y., Schones D.E., Wang Z.B., Wei G., Chepelev I., Zhao K.J. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Methodol. 57: 289–300 [Google Scholar]
- Bergmann J.H., Rodríguez M.G., Martins N.M.C., Kimura H., Kelly D.A., Masumoto H., Larionov V., Jansen L.E.T., Earnshaw W.C. (2011). Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 30: 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.J., Nobel A.B., Lieb J.D. (2005). ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 6: R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H.P., Sugiyama T., Chen E.S., Chen X., FitzGerald P.C., Grewal S.I.S. (2005). Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Carone D.M., Longo M.S., Ferreri G.C., Hall L., Harris M., Shook N., Bulazel K.V., Carone B.R., Obergfell C., O’Neill M.J., O’Neill R.J. (2009). A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma 118: 113–125 [DOI] [PubMed] [Google Scholar]
- Chueh A.C., Wong L.H., Wong N., Choo K.H.A. (2005). Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum. Mol. Genet. 14: 85–93 [DOI] [PubMed] [Google Scholar]
- Fujita M., et al. (2010). Rice expression atlas in reproductive development. Plant Cell Physiol. 51: 2060–2081 [DOI] [PubMed] [Google Scholar]
- Han F.P., Lamb J.C., Birchler J.A. (2006). High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 103: 3238–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G.M., et al. (2010). Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Malik H.S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Ishii K., Ogiyama Y., Chikashige Y., Soejima S., Masuda F., Kakuma T., Hiraoka Y., Takahashi K. (2008). Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321: 1088–1091 [DOI] [PubMed] [Google Scholar]
- Jin W.W., Lamb J.C., Zhang W.L., Kolano B., Birchler J.A., Jiang J.M. (2008). Histone modifications associated with both A and B chromosomes of maize. Chromosome Res. 16: 1203–1214 [DOI] [PubMed] [Google Scholar]
- Ketel C., Wang H.S.W., McClellan M., Bouchonville K., Selmecki A., Lahav T., Gerami-Nejad M., Berman J. (2009). Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 5: e1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo D.-H., Han F.P., Birchler J.A., Jiang J.M. (2011). Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res. 21: 908–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Li L., et al. (2007). Global identification and characterization of transcriptionally active regions in the rice genome. PLoS ONE 2: e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.Y., et al. (2008). High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 20: 259–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomiento M., Jiang Z.S., D’Addabbo P., Eichler E.E., Rocchi M. (2008). Evolutionary-new centromeres preferentially emerge within gene deserts. Genome Biol. 9: R173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T.T., Lu G.J., Fan D.L., Zhu C.R., Li W., Zhao Q.A., Feng Q., Zhao Y., Guo Y.L., Li W.J., Huang X.H., Han B. (2010). Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 20: 1238–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O.J., Chueh A.C., Wong L.H., Choo K.H.A. (2008b). Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82: 261–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O.J., Marshall A.T., Choo K.H.A. (2008a). Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J. Cell Biol. 183: 1193–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T., et al. ; International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- May B.P., Lippman Z.B., Fang Y.D., Spector D.L., Martienssen R.A. (2005). Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 1: e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravinac B., Sullivan L.L., Reeves J.W., Yan C.M., Kopf K.S., Farr C.J., Schueler M.G., Sullivan B.A. (2009). Histone modifications within the human X centromere region. PLoS ONE 4: e6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K., Cheng Z.K., Ouyang S., Talbert P.B., Kim M., Jones K.M., Henikoff S., Buell C.R., Jiang J.M. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36: 138–145 [DOI] [PubMed] [Google Scholar]
- Nagaki K., Talbert P.B., Zhong C.X., Dawe R.K., Henikoff S., Jiang J.M. (2003). Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163: 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pimentel H., Trapnell C., Pachter L. (2011). Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27: 2325–2329 [DOI] [PubMed] [Google Scholar]
- Selzer R.R., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L. (2005). Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer 44: 305–319 [DOI] [PubMed] [Google Scholar]
- Shi J., Dawe R.K. (2006). Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics 173: 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.M., Phatale P.A., Sullivan C.M., Pomraning K.R., Freitag M. (2011). Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol. Cell. Biol. 31: 2528–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.S., Johnson W.E., Zhu X.P., Zhang X.M., Li W., Manrai A.K., Liu J.S., Chen R.S., Liu X.S. (2007). Model-based analysis of two-color arrays (MA2C). Genome Biol. 8: R178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A., Karpen G.H. (2004). Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A., Schwartz S. (1995). Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet. 4: 2189–2197 [DOI] [PubMed] [Google Scholar]
- Topp C.N., Zhong C.X., Dawe R.K. (2004). Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA 101: 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.F., Elling A.A., Li X.Y., Li N., Peng Z.Y., He G.M., Sun H., Qi Y.J., Liu X.S., Deng X.W. (2009). Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L.H., Brettingham-Moore K.H., Chan L., Quach J.M., Anderson M.A., Northrop E.L., Hannan R., Saffery R., Shaw M.L., Williams E., Choo K.H.A. (2007). Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 17: 1146–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.Z., et al. (2004). Composition and structure of the centromeric region of rice chromosome 8. Plant Cell 16: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.H., Kikuchi S., Neumann P., Zhang W.L., Wu Y.F., Chen F., Jiang J.M. (2010). Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J. 63: 353–365 [DOI] [PubMed] [Google Scholar]
- Yan H.H., Jin W.W., Nagaki K., Tian S., Ouyang S., Buell C.R., Talbert P.B., Henikoff S., Jiang J.M. (2005). Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell 17: 3227–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.H., Talbert P.B., Lee H.R., Jett J., Henikoff S., Chen F., Jiang J.M. (2008). Intergenic locations of rice centromeric chromatin. PLoS Biol. 6: e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewski F., Weisshaar B., Fuchs J., Bannack E., Minoche A.E., Dohm J.C., Himmelbauer H., Schmidt T. (2011). Epigenetic profiling of heterochromatic satellite DNA. Chromosoma 120: 409–422 [DOI] [PubMed] [Google Scholar]
- Zhang W.L., Friebe B., Gill B.S., Jiang J.M. (2010). Centromere inactivation and epigenetic modifications of a plant chromosome with three functional centromeres. Chromosoma 119: 553–563 [DOI] [PubMed] [Google Scholar]
- Zhang W.L., Lee H.R., Koo D.H., Jiang J.M. (2008). Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell 20: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang Y.C., Zhang L., Li Y., Lu T.T., Lu Y.Q., Feng Q., Zhao Q., Cheng Z.K., Xue Y.B., Wing R.A., Han B. (2004). Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 32: 2023–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]