Understanding the development of a complex structure such as a fruit provides both an interesting developmental model and an important task for agriculture, holding the potential of improving both product quality and human nutrition (Klee, 2010). For this task, the tomato (Solanum lycopersicum) fruit has proven a tractable model system, with complex metabolism and accessible genetics and genomics (Carrari and Fernie, 2006). For fruit quality, characterizing the complex mixture of sugars, acids, and volatiles that contribute to tomato flavor and the alterations of cell wall structure that contribute to tomato texture will be essential for improving the quality of tomatoes that have been bred for yield and stability during postharvest handling. For nutrition, characterizing the important phytonutrients, such as flavonols, vitamin A precursors, and antioxidants, will be essential to improve human health in both over- and undernourished populations.
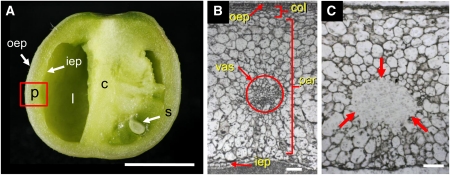
In addition to its tremendous metabolic complexity, there is also a great spatial complexity to a developing tomato fruit. Because most previous studies have characterized tomato fruit development using homogenized pericarp tissues, this spatial complexity has been lost. To remedy this, Matas et al. (pages 3893–3910) combined laser capture microdissection of specific fruit cell types with high-throughput pyrosequencing to examine the transcriptomes of key tomato tissues. They characterized the outer and inner epidermis, the collenchyma, the parenchyma, and the vascular tissues of the developing tomato pericarp in tomato fruits at the maximal expansion stage (see figure). Although this preripening stage does not show the complexities associated with ripening, this proof-of-concept study does allow examination of key factors in tomato development.
Laser capture microdissection of tomato fruit. Tomato fruit in cross section (A). Labels indicate pericarp (p), empty locule (l), columella (c), seed (s), outer epidermis (oep), and inner epidermis (iep). Cryosection of the tomato fruit pericarp before microdissection (B). Labels indicate outer epidermis (oep), collenchyma (col), vascular bundle (vas), parenchyma (par), and inner epidermis (iep) of the pericarp. Cryosection after microdissection of the vascular bundle; dissected area is indicated by arrows (C). Bars = 5 mm in (A) and 100 μm in (B) and (C). (Reprinted from Figures 1A, 1D, and 1G of Matas et al. [2011].)
After optimizing the fixation and sectioning methods to get the best recovery of sequences from each tissue, the authors examined the distribution of over 20,000 unigenes by both hierarchical clustering and pairwise comparisons. They found that 821 transcription factors showed different expression patterns in the different tissues, demonstrating that this method can help reveal the regulation of tissue differentiation. They also examined the implementation of tissue differentiation by profiling genes with functions involved in energy metabolism, cell wall dynamics, and cuticle formation. Interestingly, in the examination of the tissue specificity of genes involved in cuticle formation, they found that the inner epidermis of the pericarp also develops a cuticle. Indeed, they verified the properties of this inner cuticle by characterizing its structure and lipid composition. Thus, this method has proven useful for elucidating the spatial complexities of tomato fruit differentiation as well as for the examination of cell types and for gene discovery.
References
- Carrari F., Fernie A.R. (2006). Metabolic regulation underlying tomato fruit development. J. Exp. Bot. 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Klee H.J. (2010). Improving the flavor of fresh fruits: Genomics, biochemistry, and biotechnology. New Phytol. 187: 44–56 [DOI] [PubMed] [Google Scholar]
- Matas A.J., et al. (2011). Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 187: 3893–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]