To the Editor
To study proteins in the context of a cellular system, it is essential that the molecules with which a protein interacts are identified and the functional consequence of each interaction is understood. A plethora of resources now exist to capture molecular interaction data from the many laboratories generating such information, but whereas such databases are rich in information, the sheer number and variability of such databases constitutes a substantial challenge in both data access and quality assessment to the researchers interested in a specific biological domain.
Integrating data from these disparate resources remained a challenge until 2004, when the Human Proteome Organization Proteomics Standards Initiative (HUPO-PSI) released the PSI molecular interaction (MI) XML format, a community standard for the representation of molecular-interaction data. To concomitantly standardize annotation across the different databases, they also developed a controlled vocabulary enabling a detailed but consistent description of molecular interactions1. A simplified, standardized format for interaction data, the Molecular Interaction Tabular format (MITAB), is also available2. PSI-MI formats are now broadly accepted and widely implemented by over 30 databases and supported by key software tools.
The PSI-MI formats facilitate the integration of molecular interaction data from multiple sources, both by the user community and by dedicated software tools. However, users must still first collect data from each of the individual databases, which typically involves different queries at multiple websites or downloading data files from different web servers. Additionally, the retrieved data has then to be kept up to date with each release of the originating database. This challenge has led to the development of the PSI common query interface (PSICQUIC), a community standard for computational access to molecular-interaction data resources.
All data sources implementing PSICQUIC can be queried in the exact same way. Formulating the query once is sufficient to retrieve the relevant data from many interaction data sources. Independently published observations of an experimental system, curated by independent databases, are then integrated in response to a user query (Fig. 1). A PSICQUIC query can be a simple protein identifier or a complex construct using the syntax defined by the molecular interaction query language (MIQL) (Supplementary Note 1).
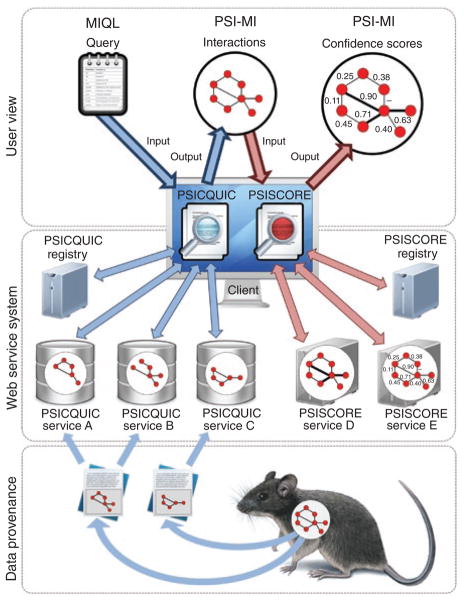
Figure 1.
PSICQUIC and PSISCORE architecture. A given biological system (sample) is observed by different experimental technologies, resulting in different publications reporting different, potentially partial, observations. A publication is potentially curated by more than one database. A PSICQUIC application sends a user query formulated in MIQL to all currently available PSICQUIC servers. Responses in unified PSI-MI format allow the PSICQUIC application to assemble a complete network view of the originally observed system. A given interaction network can be scored by multiple PSISCORE servers, each of them implementing one or more scoring methods (here symbolized by different line thickness (PSISCORE service D) and numbers (PSISCORE service E). The PSISCORE client application then presents the combined results to the user.
The existence of an open-source reference implementation for PSICQUIC allows the rapid setup of a local server for interaction data with limited effort. The PSICQUIC project site (http://psicquic.googlecode.com/) offers open-source client libraries and code examples, facilitating programmatic access to the PSICQUIC registry and services. Thus, PSICQUIC can be easily integrated with third-party applications. For instance, it is used by Cytoscape3 to query multiple web services at the same time for rendering the resulting interaction networks. PSICQUIC is also used by the International Molecular Exchange consortium (IMEx) to facilitate high-quality, nonredundant data sharing (unpublished data).
As a result, more than 16 million interactions are already accessible from 16 PSICQUIC services (Supplementary Table 1), which includes servers hosted by most major molecular interaction providers. All these services are listed in the PSICQUIC registry. Each service is classified by tags from a controlled vocabulary, which help the user to select the services of interest. The PSICQUIC architecture even allows seamless integration of commercial data sources with publicly available sources, based on access privileges of end users.
Another challenge in the field of molecular interactions is varying data quality. Owing to the diversity of techniques for experimental detection, computational prediction and curation of interaction data, adequate quality assessment methods have to account for the different evidence associated with each reported interaction. An interaction of two proteins can be supported, for example, by a single concurrent mention in a scientific publication or by multiple independent experimental observations, including details such as the protein-binding interface or assay parameters. Consequently, researchers require a system to retrieve confidence scores for user-defined sets of molecular interactions. This led to the development of the PSI confidence scoring system (PSISCORE) based on an earlier study4 (Supplementary Note 2).
Confidence measures for molecular interactions can use different, potentially complementary, properties of biological systems. Evidence-based confidence scores are commonly derived from the applied experimental detection technique or based on standard reference sets, functional annotations, evolutionary conservation, structural knowledge, literature support or network topology. The diversity of confidence measures raises questions about their comparison and combination. To date, the community has not agreed on a generally accepted common scoring scheme for molecular interactions5. Therefore, PSISCORE is based on the concept of decentralization, where individual scoring servers can apply different scoring methods for assessing diverse biological and methodological aspects of interaction data (Fig. 1).
The start and end point of a PSISCORE use case is a user-defined PSI-MI file that describes a set of molecular interactions. The interaction data can be the result of a previous PSICQUIC query (Supplementary Note 3) or contain publicly available experimental interactions and unpublished or computationally predicted results. PSISCORE can also be integrated into existing workflows as a quality filter to add the computed confidence scores to the PSI-MI file. It is easy to programmatically access PSISCORE or to incorporate the user’s own confidence scoring servers using the open-source libraries and the documentation at http://psiscore.googlecode.com/. All available scoring servers and their scoring methods are listed and described in the PSISCORE registry.
Supplementary Material
Acknowledgments
This study was supported by the European Commission under the Serving Life-science Information for the Next Generation contract 226073; Proteomics Standards Initiative and International Molecular Exchange contract FP7-HEALTH-2007-223411; Apoptosis Systems Biology Applied to Cancer and AIDS contract FP7-HEALTH-2007-200767; Experimental Network for Functional Integration contract LSHG-CT-2005-518254; German National Genome Research Network; German Research Foundation contract KFO 129/1-2; US National Institutes of Health grant R01GM071909; the Italian Association for Cancer Research; a Wellcome Trust Strategic Award to the European Molecular Biology Laboratory–European Bioinformatics Institute for Chemogenomics Databases; Grand Challenges in Global Health Research, the Canadian Institutes of Health Research, Foundation for the National Institutes of Health and Genome British Columbia; and a German Research Foundation–funded Cluster of Excellence for Multimodal Computing and Interaction. We thank organizers and sponsors of the Biohackathons 2008, 2009 and 2010 where part of the work was done.
Footnotes
Supplementary information is available on the Nature Methods website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Hermjakob H, et al. Nat Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 2.Kerrien S, et al. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shannon P, et al. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blankenburg H, et al. Bioinformatics. 2009;25:1321–1328. doi: 10.1093/bioinformatics/btp142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orchard S, et al. Proteomics. 2007;7:3436–3440. doi: 10.1002/pmic.200700658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.