Abstract
Qβ virus-like particles encapsulating multiple copies of fluorescent proteins were generated in high yields using a modular system enhanced by specific engineered RNA-protein interactions. The resulting particles were structurally indistinguishable from recombinant Qβ alone. The encapsidated proteins were nearly identical in photochemical properties to monomeric analogues, were more stable toward thermal degradation, and were protected from proteolytic cleavage. Residues on the outer capsid surface were chemically derivatized by acylation and azide-alkyne cycloaddition without affecting the fluorescence properties of the packaged proteins. A high affinity carbohydrate-based ligand of the CD22 receptor was thereby attached, and specific cell labeling by the particles was successfully detected by flow cytometry and confocal laser microscopy.
Keywords: Virus-like particles, fluorescent proteins, cell targeting
Introduction
Viruses and virus-like particles are process- and chemistry-friendly scaffolds for the display or encapsulation of functional molecules.1 The recombinant virus-like particle (VLP) derived from bacteriophage Qβ is comprised of 180 copies of a 14.2 kDa, 132 amino acid subunit.2 The X-ray crystal structure of the capsid is known to near-atomic resolution,2c and it is stable to extremes of temperature, pH, and chemical treatment.3 A wide variety of molecules have been conjugated to the outer surface of Qβ and related VLPs by chemical methods for the purpose of modifying the properties of the particle in vitro and in vivo.4 While dye molecules can be easily attached in this manner, their display on the exterior surface of the particle occupies connection points that could be used for other functional molecules and may alter the interactions of the particles by making the dye structures (which are often hydrophobic or charged) accessible to the solvent or potential binding agents.
Accordingly, we wished to put the dye marker for VLPs on the particle interior, and to do so by genetic means rather than by chemical modification. We have recently described the encapsidation of functional enzymes inside the Qβ VLP, promoted by an expressed RNA adapter containing binding motifs to the interior of the capsid shell and a peptide tag added to the cargo protein of interest.5 Here we describe the use of this modular methodology to package several fluorescent proteins commonly used as biomarkers in vitro and in vivo.6 We earlier neglected to cite the pioneering work of Stockley and coworkers, who first illuminated the potential of engineered modular packaging in the closely related MS2 particle.7 Vault,8 bacterial,9 and plant virus-derived10 shells have also previously been loaded with green fluorescent protein (GFP) using engineered peptide-protein interactions.
Materials and Methods
A. Cloning
All sequences were verified by direct sequencing of forward and reverse strands using unique primers at either ends (Retrogen). Plasmids were propagated in DH5a cells (BioPioneer) or One Shot Top10 (Invitrogen) and grown in SOB (Difco).
Mutations for superfolder GFP, blue fluorescent protein (BFP), and cyan fluorescent protein (CFP) were then installed by primer-directed mutagenesis. Each was prepared with a fused Rev peptide sequence as follows. The fluorescent protein gene (denoted XFP) was amplified by PCR from the pCDF-CP-XFP coding plasmids with primers XFP-F2 and XFP-R1, digested with NcoI and XhoI, gel purified and ligated into a similarly digested pCDF vector coding for the synthetic Rev-peptide in-frame and directly upstream from the NcoI site. For the free fluorescent proteins, separate coding plasmids were constructed to place a hexahistidine tag at the C-terminus. Amplification by PCR from these plasmids was performed with forward primer XFP-his-F1 (Table S3, Supporting Information) and XFP-R1. The resulting fragment was again digested and ligated into a similarly digested pCDF-1b vector, creating pCDF-XFP.
B. VLP Production and Purification
E. coli BL21 (DE3) (Invitrogen) cells harboring the appropriate plasmids were grown in either SOB (Difco) or MEM11 supplemented with carbenicilin and spectinomycin at 50 and 100 μg/mL, respectively. Starter cultures were grown overnight at 37 °C, and were used to inoculate larger cultures. Induction was performed with 1 mM IPTG at an OD600 of 1.0 in SOB or 2.0 in MEM for 4 hours at 37 °C for all XFP constructs. Cells were harvested by centrifugation in a JA-17 rotor at 13,700 g (10,000 rpm) and were either processed immediately or stored as a pellet at −80 °C. The cell lysate was prepared by resuspending the cell pellet with 5 mL Qβ buffer (20 mM Tris-HCl, pH 7.5, containing 10 mM MgCl2) or TBS and sonicating at 30W for 3 min with 5-second bursts and 5-second intervals. Cell debris was pelleted in a JA-17 rotor at 27000 g (14,000 rpm) and 2M ammonium sulfate was added to the supernatant to precipitate the VLPs. These were pelleted and resuspended in 0.5 mL of Qβ buffer or TBS. Lipids and membrane proteins were then extracted from particles with 1:1 n-butanol:chloroform; VLPs remain in the aqueous layer. Crude VLPs were further purified by sucrose density ultracentrifugation (10–40% w:v). Particles were either precipitated from the sucrose solution with 10% w:v PEG8000 or pelleted out by ultracentrifugation in a 70.1 Ti rotor (Beckman) at 450,000 g (70,000 rpm) for at least 2 hours. After assessment of purity, additional sucrose gradients were used to further purify VLPs to >95% if necessary.
C. Characterization
Purity and Quantitation of Encapsidated Proteins
The purity of assembled VLPs was assessed by isocratic size-exclusion chromatography with a Superose 6 column on an Akta Explorer FPLC instrument. Non-aggregated Qβ particles elute approximately 3 mL after the void volume-associated peaks.
The protein content of each sample was analyzed with a Bioanalyzer 2100 Protein 80 microfluidics chip. The average number of encapsidated proteins was determined by normalizing the area integration of coat protein and cargo protein peaks to the calculated molecular weight of the proteins they signified, determining the molar ratio of coat protein to cargo protein and multiplying by 180 to obtain the number of cargo proteins loaded per VLP. Overall protein concentration was determined with a modified Lowry protein assay (Pierce).
Mass Spectrometry
Details of sample preparation and analysis by MALDI mass spectrometry are given in Supporting Information.
Electron Microscopy
TEM images were acquired with a HP CM100 electron microscope (HP) with 80kV, 1s exposure and Kodak SO163 film on carbon formavor grids stained with 2% uranyl acetate.
Hydrodynamic Radii
Purified particles were analyzed on a light-scattering plate reader (Wyatt DynaPro); typically at protein concentration of approximately 0.1 mg/mL, with 10 acquisitions of 10 seconds each per sample. All samples were found to have a radius of 14.2 ± 0.2 nm, showing that the packaging of different fluorescent proteins has no significant effect on the size of the VLPs.
D. Free XFP Production and Purification
The conditions used for expression of the free His6-tagged fluorescent proteins were the same as used for the VLPs. To isolate the desired material, the cleared cell lysate was passed through a cobalt-NTA Talon resin column (0.5 mL bed volume). The column was washed with 3 column volumes of T buffer (20 mM Tris-HCl pH 7.5), 3 volumes of T + 20 mM imidazole, 2 volumes of T + 100 mM imidazole and eluted with T + 300 mM imadazole. Fractions containing XFP were pooled and dialyzed against two changes of 2L of T and concentrated with an Amicon Ultra centrifugal filtration unit (10 kDa MWCO, Millipore). Purity was assayed by chip-based electrophoresis as above.
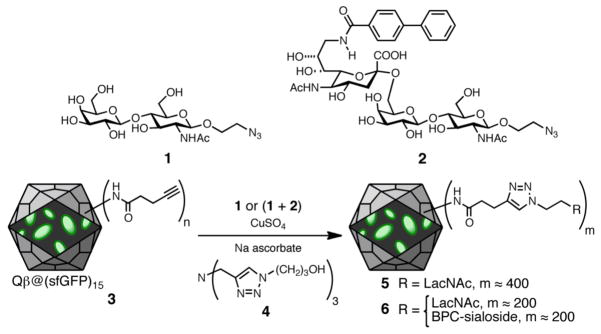
E. Preparation of Glycan-Decorated Particles
Particle 3 and compounds 1 and 2 were prepared as previously described (see Figure 4).12 CuAAC attachment to the particles was performed by the standard procedure employing ligand 4.13 Particles were purified by ultracentrifugation through 10–40% sucrose gradients, as is standard practice.11 Isolated yields of purified particles 5 and 6 were >70% relative to the amount of 3 used, and loadings of the LacNAc and BPC-sialoside moieties were determined by mass spectrometry.
Figure 4.
Derivatization of Qβ@GFP15 with glycan ligands LacNAc (using 1) and the BPC derivative of sialic acid (using 2) by Cu-catalyzed azide-alkyne cycloaddition chemistry.
F. Cell culture
All cell culture reagents were purchased from Invitrogen, unless noted otherwise. CHO cells stably expressing human CD22 (designated CD22-CHO) and Flp-In™-CHO cells (designated WT-CHO) were maintained in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12), supplemented with 10% Newborn Calf Serum (NCS) (Omega Scientific, Inc.), 1 mM sodium pyruvate, 2.5 mM L-Glutamine, 100 units/mL penicillin, 100 μg/mL streptomycin, and either 30 μg/mL Hygromycin B (CHO-CD22+) or 25 μg/mL Zeocin (Flp-In™-CHO). Cells were grown at 37°C in a humidified 5% CO2/95% air atmosphere.
G. Flow cytometry
Cell monolayers were detached using Accutase (Innovative Cell Technologies, Inc.) and rinsed twice with PBS. Approximately 1 × 105 cells were aliquoted into Titertube Micro Test Tubes (Bio-Rad). Qβ particles were added directly to the cell suspensions to a final concentration of 25 μg/mL (10 nM in particles) and incubated for 1 hr at 37°C in a humidified 5% CO2/95% air atmosphere. Cells were then washed three times with FACS buffer (PBS, 1% NCS, 2 mM EDTA) and fixed with 1% paraformaldehyde (Electron Microscopy Sciences) for 10 min at RT. After two additional washings, cells were resuspended and stored in FACS buffer at 4°C until analysis. Flow cytometric data was obtained on a Digital LSR II flow cytometer (BD Biosciences). In addition to a live scatter gate, the population was also gated for doublet discrimination, and at least 10,000 events were collected. The data was analyzed using FlowJo software (Tree Star Inc.). Experiments were performed in triplicate and repeated at least twice.
H. Confocal microscopy
Approximately 3 × 104 cells were seeded on glass coverslips and allowed to adhere for 48 hours. Qβ particles were prepared in complete growth media. Cells were rinsed once with PBS, before the addition of Qβ particles to a final concentration of 2.5 μg/mL (1.0 nM) or 25 μg/mL (10 nM). The treated cells were then incubated at 37°C in a humidified 5% CO2/95% air atmosphere for 1 hr. After the hour incubation period, cells were rinsed twice with PBS, fixed with 2% paraformaldehyde for 20 min at RT, and washed again two times with PBS. Cellular membranes were stained with wheat germ agglutinin, Alexa Fluor® 555 conjugate and nuclei were stained with DAPI (10 μg/mL in PBS) (Biotium, Inc.), washed twice with PBS, and mounted on glass slides (Propper Manufacturing Co. Inc.) using ImmunO-Fluore mounting media (MP Biomedicals LLC). Images were acquired on a Bio-Rad (Zeiss) Radiance 2100 Rainbow laser scanning confocal microscope equipped with a 60x oil immersion objective, and analyzed using ImageJ software.
RESULTS AND DISCUSSION
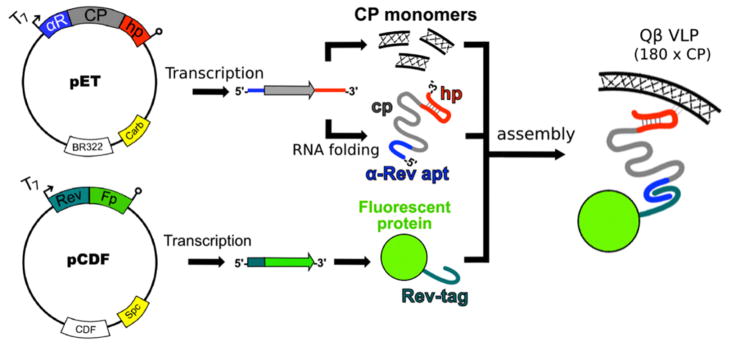
The gene for an engineered “superfolder” (sf) variant of green fluorescent protein (sfGFP)14 was encoded into a dual-plasmid expression system for Qβ VLP expression and packaging, as shown schematically in Figure 1 and described previously.5 Two binding domains were introduced to the T7 polymerase-transcribed coat protein (CP) mRNA, carried on a ColE1-group plasmid. The packaging construct employs an RNA aptamer that binds an arginine-rich peptide (Rev) derived from HIV-1,15 inserted just upstream of the ribosome binding site, and another RNA sequence (the “Qβ packaging hairpin”) that binds to the interior of the assembled coat protein, positioned immediately downstream of the stop codon. The cargo fluorescent protein was N-terminally tagged with the Rev peptide and inserted into a compatible CloDF13-group plasmid. Transformation with both plasmids and expression in BL21(DE3) E. coli yielded VLPs encapsidating the Rev-tagged protein. Such species are designated Qβ@(protein)n, where n = the average number of proteins packaged per particle. Superfolder variants of blue and cyan fluorescent proteins (BFP and CFP) were generated by making mutations analogous to those made in GFP to confer folding stability (Y66H for BFP, Y66W for CFP),14,16 and these proteins plus the red fluorescent mCherry protein were each incorporated separately into Qβ VLPs by the same procedure.
Figure 1.
Schematic representation of the technique used to package protein inside Qβ VLPs. Compatible T7 expression vectors drive expression of capsid protein (CP), Rev-tagged cargo enzyme, and bifunctional mRNA. The Rev-tag binds to the α-Rev aptamer (apt) and Qβ genome packaging hairpin (hp) binds to the interior of the CP monomers, tethering the enzyme to the interior of the VLP.
Yields of purified particles were usually in excess of 50 mg per liter of culture. To determine the number of encapsidated fluorescent proteins, the integration of electrophoretic peaks corresponding to denatured Qβ capsid and fluorescent protein were corrected for the relative sizes of the proteins to determine the relative amounts of each. Under standard conditions (using Qβ hairpin, α-Rev aptamer, and the Rev peptide tag, with expression in SOB media), an average of 10–11 sfGFP proteins were encapsidated per particle, with some preparations giving an average of up to 15 per particle. Omission of one or more of the components cut the average by 40–60% (Supporting Information), showing that the RNA adapter interactions are important, but that undirected packaging occurs fairly well with this cargo. We suggest that nonspecific association of the highly positively-charged Rev tag with random RNA inside the particle may be responsible, as discussed in Supporting Information. Expression yields and encapsidation of the other fluorescent proteins used here were very similar to sfGFP under the same conditions.
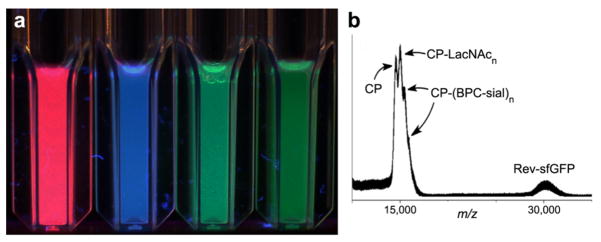
The particles containing each of the fluorescent proteins were indistinguishable from monodisperse 28-nm-diameter wild-type VLPs by all techniques that report on the exterior surface or size of the particles, including size-exclusion FPLC, dynamic light scattering, and transmission electron microscopy (Supporting Information). MALDI-mass spectrometry of denatured particles showed both the coat protein and the Rev-tagged encapsidated fluorescent protein (Figure 2).
Figure 2.
a) Photograph of solutions of Qβ VLPs containing 10±2 copies of the indicated fluorescent proteins, irradiated at 365 nm. b) MALDI-TOF of denatured particle 6 (see Figure 4).
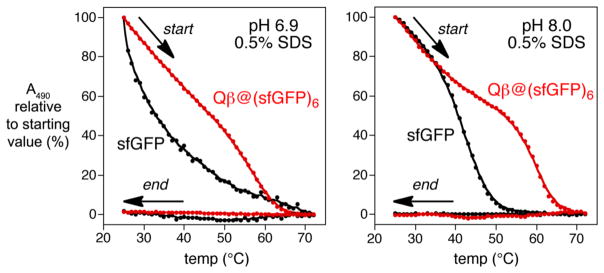
The encapsidated proteins were found to exhibit nearly identical absorbance and emission wavelengths, and rates of photobleaching, as the free fluorescent proteins. These properties did not depend on the number of cargo proteins in the particle. The excitation and emission intensities of the packaged proteins were somewhat more intense than their free analogues on a per-fluorescent-protein basis (Supporting Information), allaying concerns about self-quenching when confined at high local concentration inside the capsid (approximately 1.5–5 mM, depending on the number of fluorescent proteins encapsidated). In addition, the protein cargo was somewhat more resistant to thermal denaturation, as illustrated in Figure 3 for GFP. When heated in the presence of 0.5% sodium dodecyl sulfate (SDS), the GFP absorbance from Qβ@sfGFP6 was preserved to a significantly greater degree above 50°C than for the free protein. The capsid structure was not disrupted at these temperatures, as shown by dynamic light scattering and size-exclusion FPLC measurements, which were identical to the wild-type particle. In each case, cooling did not regenerate the chromophore fluorescence, consistent with irreversible disruption of the packaged protein tertiary structure. Lastly, the packaged proteins were completely resistant to protease digestion under conditions at which the free fluorescent proteins were fully degraded, (Supporting Information), demonstrating the physical protection offered by the nanoparticle container.
Figure 3.
Estimation of the thermal stabilities of free (black) and packaged (red) sfGFP at two different pH values, by following absorbance at 490 nm, characteristic of the properly folded protein. Bufers: pH 6.9 = 25 mM Na phosphate; pH 8.0 = 25 mM Tris-HCl.
Protein nanoparticles have demonstrated promise as tools for chemical biology. For example, we have previously demonstrated that conjugation of the 9-biphenylcarbonyl (BPC) derivative of the sialoside Siaα2-6Galβ1-4GlcNAc (2)17 to Qβ endows the particle with strong and selective affinity for cells bearing the lectin CD22.12 To test the practicality of packaged fluorescent proteins for tracking such particles, Qβ@sfGFP15 was decorated with a short alkyne linker by acylation of surface amino groups (giving 3, Figure 4). The resulting particle was addressed by Cu-catalyzed azide-alkyne cycloaddition (CuAAC) under the influence of the accelerating ligand 4.13 The azide component was either the Galβ1-4GlcNAc (LacNAc) disaccharide azide (1) alone as a negative control, or a 1:1 mixture of 1 and 2 at the same overall concentration. In this way, the resulting particles 5 and 6 bore identical numbers of triazole-linked glycans, but only one (6) displayed the high-affinity CD22 ligand. MALDI-MS analysis showed coat protein subunits bearing 0, 1, 2, and 3 glycans (one example is shown in Figure 2B). Estimation of their relative amounts18 indicated an average loading of approximately 400 glycans per particle.
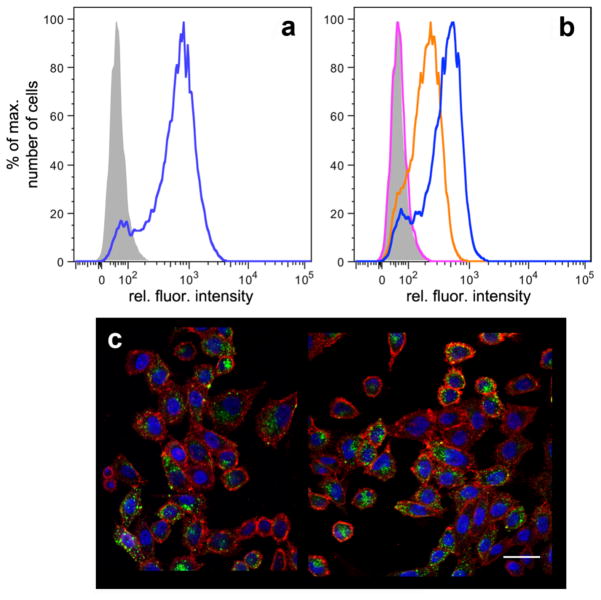
The binding of these particles with Chinese hamster ovary (CHO) cells expressing recombinant CD22 (CD22-CHO) and wild type CHO (WT-CHO) cells lacking the receptor was measured by flow cytometry. As shown in Figure 5A,B and Supporting Information, the CD22+ cells were bound by BPC-sialoside-bearing particles with signal strength similar to that of a commercial anti-CD22 antibody. Confocal microscopy confirmed the selective entry of BPC-sialoside-modified Qβ particles into CD22+ cells, as shown in Figure 5C, but not into WT-CHO cells, nor into either cell line when the BPC-sialoside was missing (Supporting Information). These observations illustrate that the encapsidated GFP was sufficient for flow cytometry and fluorescence microscopy operations, giving signal intensities comparable to particles chemically labeled with AlexaFluor-488.12
Figure 5.
Packaged GFP-dependent analysis of cell binding and internalization. a) Flow cytometry of CD22-CHO cells treated with buffer alone (grey) or fluorescently labeled anti-CD22 antibody (blue); 4°C for 1 h, followed by washing. b) As in panel (a); cells treated with buffer (grey), 5 (25 μg/mL, 10 nM in particles, pink), 6 (2.5 μg/mL, 1 nM in particles, orange), or 6 (25 μg/mL, 10 nM in particles, blue). c) Representative confocal laser microscopy image of CD22-CHO cells treated with 6 for 1 h at 37°C. Blue = DAPI stained nuclei, red = cell membrane (wheat germ agglutinin AlexaFluor® 555 conjugate), green = encapsidated GFP, scale bar = 30 μm. Negative control images showing no detectable binding or internalization of GFP-containing particles in the absence of CD22 on the cells or ligand 2 on the particles are included in Supporting Information.
CONCLUSIONS
The packaging technique used here represents a convenient and modular way to encode proteins inside Qβ virus-like particles, without sacrificing the advantageous properties of easy purification and high stability that characterize these versatile nanoparticles. Furthermore, the labeling of capsid amino groups (and perhaps some amino groups on the fluorescent protein) with an alkynyl-N-hydroxysuccinimide ester followed by a Cu-catalyzed click reaction (Figure 5) resulted in no change in fluorescence properties compared to the starting particle (data not shown), demonstrating that the particles retain their chemical tailorability without affecting the cargo.
Supplementary Material
Acknowledgments
This work was supported by the NIH (CA112085 and GM60938), the W.M. Keck Foundation, and the Postdoc-Programme of the German Academic Exchange Service (DAAD) (fellowship for F.M.). DNA encoding the mCherry protein was a kind gift from the Tsien laboratory at UCSD.
Footnotes
SUPPORTING INFORMATION. Experimental details and additional characterization data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Strable E, Finn MG. Curr Top Microbiol Immun. 2009;327:1–22. doi: 10.1007/978-3-540-69379-6_1. [DOI] [PubMed] [Google Scholar]; (b) Destito G, Schneemann A, Manchester M. Curr Top Microbiol Immunol. 2009;327:95–122. doi: 10.1007/978-3-540-69379-6_5. [DOI] [PubMed] [Google Scholar]; (c) Jennings GT, Bachmann MF. Biol Chem. 2008;389:521–536. doi: 10.1515/bc.2008.064. [DOI] [PubMed] [Google Scholar]
- 2.(a) Brown SD, Fiedler JD, Finn MG. Biochemistry. 2009;48:11155–7. doi: 10.1021/bi901306p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Vasiljeva I, Kozlovska T, Cielens I, Strelnikova A, Kazaks A, Ose V, Pumpens P. FEBS Lett. 1998;431:7–11. doi: 10.1016/s0014-5793(98)00716-9. [DOI] [PubMed] [Google Scholar]; (c) Golmohammadi R, Fridborg K, Bundule M, Valegard K, Liljas L. Structure. 1996;4:543–54. doi: 10.1016/s0969-2126(96)00060-3. [DOI] [PubMed] [Google Scholar]
- 3.(a) Overby LR, Barlow GH, Doi RH, Jacob M, Spiegelman S. J Bacteriol. 1966;91:442–8. doi: 10.1128/jb.91.1.442-448.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ashcroft AE, Lago H, Macedo JMB, Horn WT, Stonehouse NJ, Stockley PG. J Nanosci Nanotechnol. 2005;5:2034–2041. doi: 10.1166/jnn.2005.507. [DOI] [PubMed] [Google Scholar]; (c) Manzenrieder F, Luxenhofer R, Retzlaff M, Jordan R, Finn MG. Angew Chem Int Ed. 2011;50:2601–2605. doi: 10.1002/anie.201006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Udit AK, Everett C, Gale AJ, Reiber Kyle J, Ozkan M, Finn MG. ChemBioChem. 2009;10:503–10. doi: 10.1002/cbic.200800493. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Strable E, Prasuhn DE, Jr, Udit AK, Brown S, Link AJ, Ngo JT, Lander G, Quispe J, Potter CS, Carragher B, Tirrell DA, Finn MG. Bioconjugate Chem. 2008;19:866–75. doi: 10.1021/bc700390r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Prasuhn DE, Jr, Yeh RM, Obenaus A, Manchester M, Finn MG. Chem Commun. 2007:1269–71. doi: 10.1039/b615084e. [DOI] [PubMed] [Google Scholar]; (d) Banerjee D, Liu A, Voss N, Schmid S, Finn MG. Chembiochem. 2010;11:1273–1279. doi: 10.1002/cbic.201000125. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Prasuhn DE, Jr, Singh P, Strable E, Brown S, Manchester M, Finn MG. J Am Chem Soc. 2008;130:1328–34. doi: 10.1021/ja075937f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Hooker JM, Kovacs EW, Francis MB. J Am Chem Soc. 2004;2004:3718–3719. doi: 10.1021/ja031790q. [DOI] [PubMed] [Google Scholar]; (g) Kovacs EW, Hooker JM, Romanini DW, Holder PG, Berry KE, Francis MB. Bioconjugate Chem. 2007;18:1140–1147. doi: 10.1021/bc070006e. [DOI] [PubMed] [Google Scholar]; (h) Stephanopoulos N, Tong GJ, Hsiao SC, Francis MB. ACS Nano. 2010;4:6014–6020. doi: 10.1021/nn1014769. [DOI] [PubMed] [Google Scholar]
- 5.Fiedler JD, Brown SD, Lau J, Finn MG. Angew Chem, Int Ed. 2010;49:9648–9651. doi: 10.1002/anie.201005243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Tsien RY. Annu Rev Biochem. 1998;67:509–44. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]; (b) Labas YA, Gurskaya NG, Yanushevich YG, Fradkov AF, Lukyanov KA, Lukyanov SA, Matz MV. Proc Natl Acad Sci USA. 2002;99:4256–61. doi: 10.1073/pnas.062552299. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Shaner NC, Steinbach PA, Tsien RY. Nature Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 7.(a) Wu M, Brown WL, Stockley PG. Bioconjugate Chem. 1995;6:587–595. doi: 10.1021/bc00035a013. [DOI] [PubMed] [Google Scholar]; (b) Brown WL, Mastico RA, Wu M, Heal KG, Adams CJ, Murray JB, Simpson JC, Lord JM, Taylor-Robinson AW, Stockley PG. Intervirology. 2002;45:371–380. doi: 10.1159/000067930. [DOI] [PubMed] [Google Scholar]
- 8.Kickhoefer VA, Garcia Y, Mikyas Y, Johansson E, Zhou JC, Raval-Fernandes S, Minoofar P, Zink JI, Dunn B, Stewart PL, Rome LH. Proc Natl Acad Sci USA. 2005;102:4348–4352. doi: 10.1073/pnas.0500929102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seebeck FP, Woycechowsky J, Zhuang W, Rabe JP, Hilvert D. J Am Chem Soc. 2006;128:4516–4517. doi: 10.1021/ja058363s. [DOI] [PubMed] [Google Scholar]
- 10.(a) Minten IJ, Hendriks LJA, Nolte RJM, Cornelissen JJLM. J Am Chem Soc. 2009;131:17771–17773. doi: 10.1021/ja907843s. [DOI] [PubMed] [Google Scholar]; (b) Minten IJ, Nolte RJM, Cornelissen JJLM. Macromol Biosci. 2010;10:539–545. doi: 10.1002/mabi.201000030. [DOI] [PubMed] [Google Scholar]
- 11.Studier FW. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 12.Kaltgrad E, O’Reilly MK, Liao L, Han S, Paulson JC, Finn MG. J Am Chem Soc. 2008;130:4578–9. doi: 10.1021/ja077801n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong V, Presolski SI, Ma C, Finn MG. Angew Chem, Int Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 15.Xu W, Ellington AD. Proc Natl Acad Sci USA. 1996;93:7475–7480. doi: 10.1073/pnas.93.15.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heim R, Prasher DC, Tsien RY. Proc Natl Acad Sci USA. 1994;91:12501–4. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Zaccai NR, Maenaka K, Maenaka T, Crocker PR, Brossmer R, Kelm S, Jones EY. Structure. 2003;11:557–567. doi: 10.1016/s0969-2126(03)00073-x. [DOI] [PubMed] [Google Scholar]; (b) Collins BE, Blixt O, Han S, Duong B, Li H, Nathan JK, Bovin N, Paulson JC. J Immunol. 2006;177:2994–3003. doi: 10.4049/jimmunol.177.5.2994. and references therein. [DOI] [PubMed] [Google Scholar]
- 18.Patel MK, Vijayakrishnan B, Koeppe JR, Chalker JM, Doores KJ, Davis BG. Chem Commun. 2010;46:9119–9121. doi: 10.1039/c0cc03420g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.