Abstract
In humans Uncoupling Proteins (UCPs) are a group of five mitochondrial inner membrane transporters with variable tissue expression, which seem to function as regulators of energy homeostasis and antioxidants. In particular, these proteins uncouple respiration from ATP production, allowing stored energy to be released as heat. Data from experimental models have previously suggested that UCPs may play an important role on aging rate and lifespan. We analyzed the genetic variability of human UCPs in cohorts of subjects ranging between 64 and 105 years of age (for a total of 598 subjects), to determine whether specific UCP variability affects human longevity. Indeed, we found that the genetic variability of UCP2, UCP3 and UCP4 do affect the individual's chances of surviving up to a very old age. This confirms the importance of energy storage, energy use and modulation of ROS production in the aging process. In addition, given the different localization of these UCPs (UCP2 is expressed in various tissues including brain, hearth and adipose tissue, while UCP3 is expressed in muscles and Brown Adipose Tissue and UCP4 is expressed in neuronal cells), our results may suggest that the uncoupling process plays an important role in modulating aging especially in muscular and nervous tissues, which are indeed very responsive to metabolic alterations and are very important in estimating health status and survival in the elderly.
Introduction
Un-Coupling Proteins (UCPs) belong to a family of anion transporters located in the inner membrane of mitochondria responsible for uncoupling substrate oxidation from ATP synthesis. As a consequence stored energy is released as heat. To date, five UCP homologues, UCP1 to UCP5, have been identified in mammals. UCP1 (4q28.31), the first member characterized, is predominantly expressed in brown adipose tissue (BAT), where it has a well-established role in cold- and diet-induced thermogenesis [1]. UCP2 and UCP3 share a common region on chromosome 11q13. UCP2 is widely expressed in many organs and tissues [2], while UCP3 is principally expressed in skeletal muscle, cardiac muscle, and BAT [3], [4]. Finally, UCP4 (6p12.3) and UCP5 (Xp24) are predominantly expressed in the central nervous system and at a lower level in other tissues [5], [6]. Unlike UCP1, the physiological role of the other UCPs still remains to be fully elucidated. It has been hypothesized that these proteins may provide protection from oxidative damage by preventing excessive production of mitochondrial Reactive Oxygen Species (ROS) [7], [8]. Indeed, Ucp-knockout mice have increased levels of ROS and show signs of increased oxidative damage [9]–[11]. Further evidence indicates that activation of UCP2 and UCP3 by ROS leads to a mild uncoupling and to diminished ROS formation [12], [13], while their inhibition by purine nucleotides increases membrane potential and mitochondrial ROS production [14].
The uncoupling activity of UCPs has also been linked to regulation of other more specific metabolic functions [15], [16] such as fatty acid oxidation [17], glucose-stimulated insulin secretion [18], whole body energy balance [19], and apoptosis [20]. Human aging is characterized by a gradual reduction in the ability to coordinate cellular energy expenditure and storage (crucial to maintain energy homeostasis), and by a gradual decrease in the in the ability to mount a successful stress response [21], [22]. These physiological changes are typically associated with changes in body composition (i.e increase in fat mass and the decline in fat-free mass), and with a chronic state of oxidative stress with important consequences on health status [23]–[25]. Mitochondrial function is crucial in these processes, being mitochondria the main cellular sites controlling energy metabolism and the redox state. Thus, by promoting fatty acid oxidation, and by reducing ATP and ROS production, the induction of mitochondrial uncoupling through UCPs may also be a critical pathway in the modulation of the rate of aging and lifespan [26]–[28]. This idea, first proposed by M. D. Brand [26] and termed “uncoupling-to-survival”, is now supported by several experimental evidences (for reviews see: [27], [29], [30]). Of interest, targeted expression of exogenous UCP extends lifespan of adult flies [31], [32], while mice with higher metabolism live longer and have higher uncoupling activity [33]. Transgenic and knockout mice for Ucp genes show alterations in lifespan [34], [35]. Interestingly, it has also been shown that in vivo uncoupling mimics metabolic and lifespan effects of calorie restriction (CR) [36]. Consistently, mice subjected to CR show an increased expression of Ucp2 and Ucp3 [37], [38].
In humans the correlation between mitochondrial uncoupling and aging emerges from association studies which support a role for UCPs in many age-related phenotypic traits involving alterations in cellular energy homeostasis such as obesity, diabetes and lipid-related diseases [39]–[41]. We recently performed a population-based study in a human cohort of elderly subjects which demonstrated that UCP3 variability has an impact on hand grip strength, one of the most important hallmarks of human aging [42] and more recently we also found that two variants (A-3826G and C-3740A) in the upstream enhancer region of human UCP1 gene affect the expression of the gene and are correlated with human longevity [43].
Given the tissue-specific expression of UCP genes, it seems likely that they may affect the senescence of different tissues with important effects on the overall aging process and consequently on lifespan. Thus, the goal of the present study was to assess both the primary effects of single loci and gene-gene interactions to test the hypothesis that other UCP genes may contribute to survival at very old age either independently and/or through complex interactions.
Methods
Ethical statement
Samples were collected within the framework of several recruitment campaigns carried out for monitoring the quality of aging in Calabria from 2002.The recruitment campaigns and subsequent analyses received the approval of the Ethical committee of the University of Calabria. All subjects provided written informed consent for studies on aging carried out by our research group. White blood cells (WBC) from blood buffy coats were used as a source of DNA.
The Sample
A total of 598 subjects (293 men and 305 women, age range 64–105 years; mean ages 82.74 (±11.66) and 85.23 (±10.84) years, respectively) participated in this study. All subjects were born in Calabria (southern Italy) and their ancestry in the region was ascertained up to the grandparents' generation. Younger subjects were contacted through family physicians. Subjects older than 90 years were identified through the population registers and then were contacted by specialized personnel and invited to join the study. Each subject underwent a medical visit carried out by a geriatrician who also conducted an interview including the administration of a structured questionnaire, validated at European level. The questionnaire collected socio-demographic information, evaluated physical and cognitive status, and self-reported health status. Subjects with dementia and/or neurologic disorders were not included.
The analyses were carried out by dividing the sample into two specific age classes obtained according to the survival function of the Italian population from 1890 onward [44]. The two “thresholds of longevity” used to define these age classes were 88 years for men and 91 years for women.
SNPs selection
Polymorphisms within the UCP genes were selected from literature data, and using information from online databases such as NCBI dbSNP and HapMap. SNP selection was based on allele frequency, position, and functional effects. For each gene the selected SNPs are reported in Table 1.
Table 1. Description and localization of selected SNPs in the UCP genes.
| Gene symbol | dbSNP ID | Physical location | Function annotation * | |
| UCP2 | rs660339 | C/T | exon 4 | Ala55Val |
| rs659366 | G/A | 5′-proximal region | −866 G/A | |
| UCP3 | rs15763 | C/T | 3′UTR | |
| rs1800849 | C/T | 5′-proximal region | −55 C/T | |
| UCP4 | rs9472817 | C/G | intron 8 | |
| rs10498769 | C/G | 5′UTR | ||
| UCP5 | rs2235800 | A/T | intron 7 | |
| rs5975178 | C/T | 5′-proximal region |
*Provided only for coding SNP (amino acid residues for two alleles) and for SNPs in the 5′ flanking region (the nucleotide positions relative to the transcription start sites in the promoter regions are indicated).
Genotyping
DNA was prepared from peripheral blood lymphocytes using standard techniques. Genotyping of the eleven polymorphic sites was carried out using a TaqMan Real Time PCR (SNP Genotyping kit, Applied Biosystems). In all assays the fluorescent FAM dye was used to label the wild-type allele, while the mutant allele was labeled by fluorescent VIC dye. PCR reactions were carried out in a total volume of 5 µl containing 20 ng of genomic DNA, 2.5 µl of TaqMan Universal Master mix (concentration of 2×), 0.25 µl of Custom TaqMan SNP Genotyping Assay (concentration of 20×) containing both primers and probes. The amplification protocol (60°C for 30 seconds, 95°C for 10 minutes followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute) was performed by using a StepOne thermal cycler (Applied Biosystems). Random regenotyping was conducted to confirm the results.
Genetic and statistical analyses
For each polymorphism of the UCP genes, allele frequencies were estimated by counting genes from the observed genotypes. The Hardy-Weinberg equilibrium (HWE) was tested using the exact test proposed by [45]. Standard errors for alleles were computed according to the hypothesis of the multinomial distribution.
Single-locus analysis
In recent years different robust tests have been proposed to test genotype-phenotype associations in case-control studies. One of these is the MAX3 test which assumes the maximum of three different Cochran-Armitage trend tests (CATTs) evaluated with respect to three different genetic models of inheritance: dominant, additive or recessive [46], [47]. The main drawback of such tests is represented by the fact that they could be seriously affected by confounding factors [48]. In this context, So and Sham recently proposed a robust association test allowing for quantitative or binary traits as well as covariates [49]. This test was based on the score test and has been implemented in the R package RobustSNP. In the present study the robust association test proposed by So and Sham was applied to estimate the association between the variability in the UCP genes and the probability of reaching advanced ages. In each test we included sex and Body Mass Index (BMI) as covariates.
Haplotypic analysis
Pairwise measures of linkage disequilibrium (LD) between the analyzed loci were calculated with the Haploview 4.2 [50]. The amount of LD was quantified by Lewontin's coefficient (D').
In order to model the effect of the UCP haplotypes on the probability to attain longevity we used the haplo.stats package of R. It implements a haplotype-based association analysis within the generalized linear model (GLM) framework that allows for ambiguous haplotypes. The haplo.score function of this package has been used to obtain the GLM-based score statistics for testing global and individual haplotype effects on the probability to attain longevity [51]. Permutation-based p-values were used to evaluate the significance of the scores obtained (10000 permutations). In this model the effect of the different haplotypes was assessed assuming a dominant model after adjusting for sex and BMI.
Interaction analysis
In order to explore the interaction effects between the analyzed polymorphisms on the probability to be part of the very old age group, the Model-Based Multifactor Dimensionality Reduction (MB-MDR) method was applied [52]. In this model only all possible second order interactions, after adjusting for main effects sex and BMI, were analyzed.
All statistical analyses were performed using genetics, RobustSNP, mbmdr and haplo.stats packages of R 2.10.1.
Results
Table 2 reports the socio-demographic characteristics of the sample analyzed according to age group. The observed genotype frequencies (Table S1) were in agreement with those expected at HWE (p>0.01). Table 3 reports the results of the association test for the three different genetic models (dominant, additive and recessive) obtained in the analyzed sample using the RobustSNP algorithm. Table 3 clearly shows that the variability of the UCP polymorphism was significantly associated with the analyzed phenotype. In particular, using the minor allele for each SNP as reference and after adjusting for BMI and sex, the dominant model for the rs660339 (UCP2) and the rs1800849 (UCP3) resulted to be significantly associated with the longevity phenotype (p = 0.001 in both cases), while the recessive model was the most likely for SNPs rs15763 (UCP3), rs9472817 (UCP4) and rs2235800 (UCP5) (p<0.05). However, after adjusting for multiple testing, all the previous associations remained statistically significant, except those for rs2235800 of the UCP5 gene (p = 0.058).
Table 2. Socio-demographic characteristics of the analysed sample according to sex and age group.
| Younger group | Males | Females | Total |
| N (%) | 155 (41.3) | 220 (58.7) | 375 (100) |
| Mean age (SD) | 72.86 (6.027) | 80.64 (9.127) | 77.42 (8.858) |
| BMI (SD) | 27.06 (4.122) | 26.38 (4.845) | 26.66 (4.566) |
| Older group | Males | Females | Total |
| N (%) | 138 (61.9) | 85 (38.1) | 223 (100) |
| Mean age (SD) | 93.85 (3.771) | 97.09 (3.235) | 95.09 (3.903) |
| BMI (SD) | 24.42 (3.701) | 22.50 (3.994) | 23.69 (3.918) |
Note: age-cut-offs to define the younger and older groups were 88 years for males and 91 years for females, as reported in Materials and Methods section.
SD: standard deviation.
Table 3. Results of the Robust SNP association test for the most likely genetic model (additive, recessive and dominant) obtained in the analyzed sample.
| Gene | SNP | MAF | Z(ADD) | Z(REC) | Z(DOM) | P(ADD) | P(REC) | P(DOM) | P-value* |
| UCP2 | rs659366 | A (0.295) | 0.936 | 0.338 | 0.978 | 0.349 | 0.735 | 0.328 | 0.555 |
| rs660339 | T (0.341) | 3.049 | 1.477 | 3.190 | 0.002 | 0.140 | 0.001 | 0.003 | |
| UCP3 | rs15763 | T (0.247) | 2.163 | 2.468 | 1.454 | 0.031 | 0.014 | 0.146 | 0.029 |
| rs1800849 | T (0.099) | 2.788 | −1.197 | 3.186 | 0.005 | 0.231 | 0.001 | 0.003 | |
| UCP4 | rs9472817 | G (0.498) | −1.984 | −3.607 | 0.316 | 0.047 | <0.001 | 0.752 | 0.001 |
| rs10498769 | G (0.190) | −0.085 | −0.660 | 0.130 | 0.933 | 0.509 | 0.897 | 0.761 | |
| UCP5 | rs2235800 | A(0.387) | 1.554 | 2.120 | 0.849 | 0.120 | 0.034 | 0.396 | 0.058 |
| rs5975178 | C (0.474) | 0.203 | 0.210 | 0.170 | 0.839 | 0.833 | 0.865 | 0.959 |
Z(ADD), Z(REC) and Z(DOM) are the z-statistics under the additive, recessive and dominant models respectively. P(ADD), P(REC) and P(DOM) are the p-values under the three genetic models.
*p-value was adjusted for multiple testing of different genetic models obtained by the proposed analytic approach.
MAF: Minor Allele Frequency.
From the z-statistics of these models we observed that carriers of the T allele at rs660339 and at rs1800849 (dominant models) significantly influenced the probability to be part of the oldest group (positive scores). For the recessive models, homozygous subjects for the T allele at rs15763 variation also increased the probability to be part of the oldest group, while this probability was significantly decreased (negative score) for homozygous subjects with the T allele at rs9472817.
By using the MB-MDR approach we did not find any significant interaction effect among the analyzed polymorphisms (data not shown).
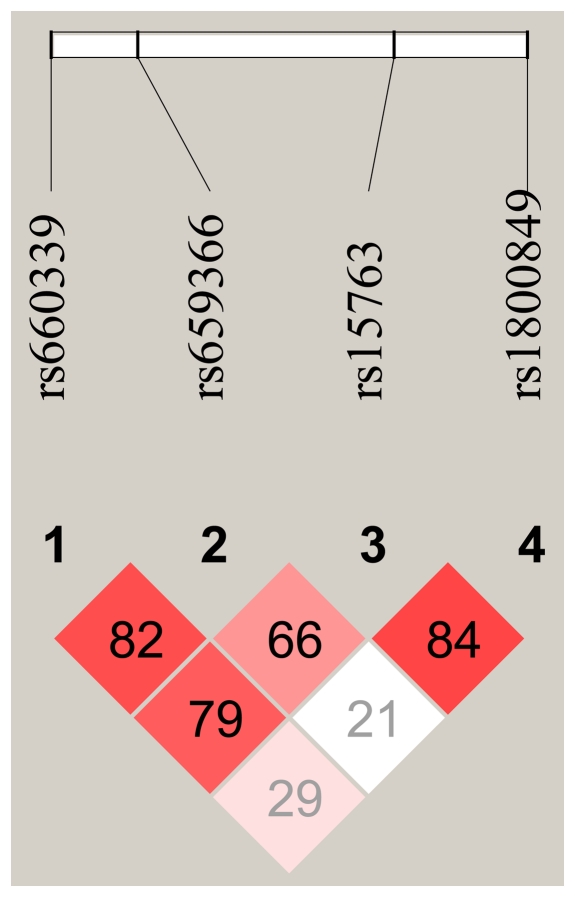
Subsequently, using the aforementioned GLM-based algorithm, we evaluated the effect of the UCP haplotypes on the probability to attain longevity. LD analysis showed that among the four SNPs in the UCP2-UCP3 gene region rs660339, rs659366 and rs15763 were in moderate LD (see Fig. 1), while rs1800849 was in LD with rs15763 (D' = 0.83), but it was virtually unlinked with the others (D'<0.30). Finally, we found a weak LD between rs9472817 and rs10498769 in the UCP4 gene (D' = 0.47), and between rs2235800 and rs5975178 in the UCP5 gene (D' = 0.44). Based on these LD patterns, and using a D' cutoff equal to 0.6, haplotypes were reconstructed at the UCP2-UCP3 and UCP3 loci.
Figure 1. Schematic representation of linkage disequilibrium (D' coefficient) among the four SNPs of the UCP2-UCP3 gene cluster.
As shown in Table 4, the analysis of the UCP2-UCP3 haplotypes revealed that the CAC haplotype, consisting of the minor allele for rs660339, major allele for rs659366, and minor allele rs15763, was associated with a decreased probability of attaining longevity (P = 0.003), while the TGT haplotype (opposite combination of alleles) acted in a reverse fashion by increasing this probability (p = 0.046). However, the strength of these associations was influenced by the allelic status at rs659366 of the relevant haplotype. Indeed, the CGC and TAT haplotypes, when compared to the above, differ only for rs659366 and did not show any effect. For the UCP3 haplotypes involving rs15763 and rs1800849, we found that carriers of the CC haplotype have a decreased probability of achieving longevity (P<0.001). Conversely, as previously reported in the single-locus analysis, the presence of a T allele at either rs15763 or rs1800849 increased such probability, although this positive effect was more pronounced when the T allele at rs1800849 was involved (p = 0.044 vs P = 0.006).
Table 4. Estimation of haplotype frequencies in the UCPs SNPs and association with longevity in the analyzed sample.
| Gene | SNPs | Freqa | Score | P-valueb | ||
| UCP2-UCP3 | rs660339 | rs659366 | rs15763 | |||
| C | A | C | 0.029 | −2.876 | 0.003 | |
| C | G | C | 0.596 | −1.747 | 0.086 | |
| C | G | T | 0.028 | 0.317 | 0.754 | |
| C | A | T | 0.004 | 0.877 | 0.400 | |
| T | A | C | 0.077 | 0.953 | 0.347 | |
| T | G | C | 0.050 | 0.971 | 0.339 | |
| T | A | T | 0.184 | 1.390 | 0.165 | |
| T | G | T | 0.030 | 1.985 | 0.046 | |
Estimated haplotype frequency.
Monte-Carlo p-value from 104 replications.
Note: p-values of global score statistics based on 104 replications were 0.047 for UCP2-UCP3 haplotypes, 0.0028 for UCP3 haplotypes.
Discussion
Substantial evidence suggests that the ability of UCPs to reduce ROS and regulate energy utilization underpins the ability of UCPs to promote lifespan in various experimental models [27], [28], [29], [30], [35]. In the present study we found that variants in UCP2, UCP3, and UCP4 significantly affect an individual's chances of becoming ultra-nonagenarians. The different localization of the proteins we found associated with longevity allows us to predict the areas where the uncoupling process may play an important role in survival at very old age.
UCP2 and UCP3 genes cluster on chromosome 11q13 in a region linked to lower resting metabolic rate in humans [53], which is also syntenic to a region of mouse chromosome 7 linked to hyperinsulinemia and obesity [54]. Their chromosomal location and their association with fatty acid transport across mitochondrial inner membrane and β-oxidation, support the hypothesis that, in addition to functioning as regulators of oxidative stress, both UCP2 and UCP3 can modulate cellular energy metabolism [16], [55]. While mounting evidence has documented the importance of both UCP2 and UCP3 variability in the pathophysiology of different chronic metabolic diseases, such as diabetes and obesity [41], [56], relatively little is known of their implication in human aging and longevity. In this study we found that the Ala55Val (rs660339) of UCP2, and -55 C/T (rs1800849) and rs15763 of UCP3 were significantly associated with survival at very old age after Bonferroni correction.
Ala55val SNP in exon 4 causes a conservative amino acid change that does not seem to cause a functional change in the protein. However, it has been reported that the Val/Val genotype causes a lower degree of uncoupling, an enhanced metabolic efficiency, and a lower fat oxidation than the Ala/Ala and Ala/Val genotypes [57]. In addition this genotype has been associated to higher exercise energy efficiency [58], higher risk in obesity, higher incidence of diabetes [59], [60], and a higher acute insulin response to glucose [61]. Moreover, individuals with the Val/Val genotype had greater weight loss and a higher BMI [62]. Nevertheless, other studies have produced conflicting results [63]–[66]. Here we show that this polymorphism also contributes to extend lifespan. Given the wide distribution of UCP2, it is likely that systemic effects on global energy metabolism and redox state underlie the association with longevity. However, UCP2 also exhibits tissue-specific regulation suggesting tissue specific physiological effects. In this regard, evidence has been provided that in the brain, liver and other tissues UCP2 functions as regulator of oxidative stress [67]–[69], in the heart as a regulator of energy availability [70], in white adipose tissue and skeletal muscle as a regulator of fatty acid metabolism [71], [72], and as a regulator of insulin dynamics in pancreatic β and α-cells [73], [74]. In this scenario, variants of UCP2 may have positive and negative effects depending on the tissue and on the fine balance between energy production/consumption and mitochondrial ROS generation.
It is widely accepted that damage by free radicals to the molecules with which they come into contact are the underlying cause of aging, therefore the ability of UCP2 to attenuate steady-state levels of ROS strongly suggests an essential role in lifespan extension. Recently, Andrews proposed that UCP2 promotes longevity by shifting a cell towards fatty acid fuel utilization thus pointing to a major role of UCP2 in modulating metabolism [75]. This hypothesis is somewhat supported by Barbieri and colleagues [76] who analyzed the Ala55val polymorphism in a human cohort of elderly subjects from an Italian population. Although the authors did not find an association with longevity, they found that individuals with the Val/Val genotype had significantly higher energy expenditure parameters. Interestingly, the A-IGF1R/Asp-IRS2/Val-UCP2 allele combination was associated with a better metabolic profile, higher energy expenditure parameters, and lower mortality rates in longevity, thus indicating a contribution of Ala55val polymorphism to the regulation of energy balance and survival [76]. The latter and our finding suggest that the 55Val allele might promote longevity by conferring protection towards age-related decline of metabolic rate. It is indeed well-known that human aging is accompanied by a decrease in resting metabolic rate, the largest component of total energy expenditure, which significantly affects disability and morbidity among the elderly [24], [77]. On the other hand, long-lived subjects show a preserved metabolic profile being less prone to the metabolic deregulation normally occurring with aging [78]. Accordingly, Rizzo and colleagues [79] reported that compared to aged subjects, healthy long-lived subjects display energy expenditure parameters that are closer to the values of healthy middle-aged adults. In a recent study we demonstrated a correlation between the activity of UCP1 and human survival, and hypothesized a slower decline of energy expenditure with age at the basis of the correlation [43].
Skeletal muscle, where UCP3 is mainly expressed, contributes considerably to the basal metabolic rate [80]. We found that carriers of the TT genotype (rs15763), and carriers of the -55 T allele (rs180084) were significantly over-represented among long-lived subjects. The rs15763 is located in the 3′UTR of the gene and has no clear functional role so far; on the contrary, the -55 T allele has been associated with a significantly increased gene expression and has been shown to positively modulate the resting metabolic rate of skeletal muscle [81], [82]. In addition, the over-expression of UCP3 in muscle cells has been shown to be associated with decreased production of ROS as well as with facilitating fatty acid oxidation [17], [83]. The physiological status of the muscle mass reflects a complex equilibrium between nutrition, metabolism and the response to stress. Aging muscle is characterized by a progressive loss of mass and a gradual increase of weakness leading to sarcopenia, a condition associated with physical disability, and with an increased risk of developing disorders such as atherosclerosis, type II diabetes and hypertension [84]. Sarcopenia is closely linked to a decrease in resting metabolic rate as well as to mitochondrial dysfunction and oxidative stress [85]. Therefore, in the context of the proposed functions of UCP3, the physiological consequence of an increased UCP3 activity in skeletal muscle might be to slow down the age related decline of muscle performance as a result of decreased ROS production, an increased protection of mitochondria from lipid peroxidation, and better metabolic efficiency [28], [83]. Accordingly, it has been found that mild uncoupling has an impact on cellular aging in human muscles in vivo [86]. This is in keeping with our previous work, where we have shown that the hand grip strength, the most effective death predictor in the elderly, was higher in the carriers of rs1800849-T allele than in the remaining the population [42].
Haplotype analysis confirmed the single SNP analyses, and highlighted the importance of the entire UCP2-UCP3 locus in human aging and longevity. In fact, one finding of interest emerging from this analysis is that the UCP2-rs659366 (-866 G/A), which was uninformative to single-locus analysis, seemed instead to be effective in modulating the aging process. In particular the rs659366G allele appeared to exert a modest beneficial effect on human survival. Indeed, the presence of the rs659366G allele reduced the negative effect of the rs659366C-rs15763C haplotype, while it increased the positive effect of the rs659366T-rs15763T haplotype. The rs659366 (-866G/A) variant changes promoter activity, affects metabolic rate and oxidative stress, and has been associated with metabolic traits in several, although not all, studies [87].
UCP4 and UCP5, together with UCP2, are known as neuronal UCPs due to their widespread distribution in the brain [6]. Neurons have a very high metabolic rate and consequently a high production of ROS. Andrews and co-workers observed that neuronal uncoupling activity leads to decreased ROS levels, a decreased Ca2+ voltage-dependent influx and increased local temperature in neuronal microenvironment [88]. We found that only the UCP4 rs9472817-GG genotype negatively impacts on the probability to attain longevity. It has been shown that UCP4 activity can induce an adaptive shift in energy metabolism, from mitochondrial respiration to glycolysis, that helps sustain neurons under conditions of metabolic and oxidative stress [89], [90]. Therefore, by decreasing free radical production and stabilizing cellular calcium homeostasis, UCPs expressed in neurons may positively influence neuronal function (synaptic transmission and plasticity) and retard the cellular deterioration associated with aged-related neurological disorders. Accordingly, the expression of UCP4 has been found significantly reduced in brains of subjects affected by Alzheimer disease [91]. Moreover, it was found that CC genotype for rs10807344 of UCP4 gene, not found to be associated in the present study with the longevity phenotype, exerts a protective effect on the occurrence of multiple sclerosis and of leukoaraiosis, a vascular demyelinization of the white matter of the brain [92], [93]. Thus, it is likely that variants affecting the UCP4 function might have consequences on the aging of the nervous system, and then might have a potential impact on health and longevity. Due to the fact that rs9472817 is located in intron 8, the exact molecular mechanism responsible for the effect on survival remains to be elucidated. Although it has been reported that intronic SNPs may have an effect on gene expression [94], it is more plausible for rs9472817 to be in LD with some other functional genetic alterations affecting the function of UCP4 and this has consequences on the aging nervous system, leading to the observed detrimental effect of the GG genotype. Further studies are needed to clarify this point.
In conclusion, we found that the genetic variability of UCP genes affects human longevity. This finding is in agreement with previous data showing that energy storage and expenditure have a key role in survival at old age and support the Uncoupling-to-survive hypothesis. Although this hypothesis was initially based on the possible effects of UCPs on the oxidative stress, the most recent data have emphasized the impact of UCP variations on the metabolic efficiency. It is then likely that the impact of UCPs' gene variation on longevity is due to the complexity of these functions rather than the sole effect of oxidative stress. On the other hand, due to the presence of different UCP genes, each active in specific tissues, further analyses will be necessary to understand the specific role of the uncoupling process in different tissues and, consequently, in correlation with differing metabolism and, possibly, nutrients.
Supporting Information
Genotypic and allelic frequencies in the analyzed sample by group.
(PDF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Fondi di Ateneo Unical (ex 60%) to GP and GR. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.Pecqueur C, Couplan E, Bouillaud F, Ricquier D. Genetic and physiological analysis of the role of uncoupling proteins in human energy homeostasis. J Mol Med. 2001;79:48–56. doi: 10.1007/s001090000150. [DOI] [PubMed] [Google Scholar]
- 3.Boss O, Samec S, Paoloni-Giacobino A, Rossier C, Dulloo A, et al. Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408:39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- 4.Vidal-Puig A, Solanes G, Grujic D, Flier JS, Lowell BB. UCP3: an uncoupling protein homologue expressed preferentially and abundantly in skeletal muscle and brown adipose tissue. Biochem Biophys Res Commun. 1997;235:79–82. doi: 10.1006/bbrc.1997.6740. [DOI] [PubMed] [Google Scholar]
- 5.Yang X, Pratley RE, Tokraks S, Tataranni PA, Permana PA. UCP5/BMCP1 transcript isoforms in human skeletal muscle: relationship of the short-insert isoform with lipid oxidation and resting metabolic rates. Mol Genet Metab. 2002;75:369–373. doi: 10.1016/S1096-7192(02)00008-2. [DOI] [PubMed] [Google Scholar]
- 6.Smorodchenko A, Rupprecht A, Sarilova I, Ninnemann O, Bräuer AU, et al. Comparative analysis of uncoupling protein 4 distribution in various tissues under physiological conditions and during development. Biochim Biophys Acta. 2009;1788:2309–2319. doi: 10.1016/j.bbamem.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Nègre-Salvayre A, Hirtz C, Carrera G, Cazenave R, Troly M, et al. A role for uncoupling protein-2 as a regulator of mitochondrial hydrogen peroxide generation. FASEB J. 1997;11:809–815. [PubMed] [Google Scholar]
- 8.Mailloux RJ, Harper ME. Uncoupling proteins and the control of mitochondrial reactive oxygen species production. Free Radic Biol Med. 2011;51:1106–1115. doi: 10.1016/j.freeradbiomed.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Arsenijevic D, Onuma H, Pecqueur C, Raimbault S, Manning BS, et al. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Genet. 2000;26:435–439. doi: 10.1038/82565. [DOI] [PubMed] [Google Scholar]
- 10.Vidal-Puig AJ, Grujic D, Zhang CY, Hagen T, Boss O, et al. Energy metabolism in uncoupling protein 3 gene knockout mice. J Biol Chem. 2000;275:16258–16266. doi: 10.1074/jbc.M910179199. [DOI] [PubMed] [Google Scholar]
- 11.Brand MD, Pamplona R, Portero-Otin M, Requena JR, Roebuck SJ, et al. Oxidative damage and phospholipids fatty acyl composition in skeletal muscle mitochondria from mice underexpressing or overexpressing uncoupling protein 3. Biochem J. 2002;368:597–603. doi: 10.1042/BJ20021077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Echtay KS, Roussel D, St-Pierre J, Jekabsons MB, Cadenas S, et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 13.Brand MD, Buckingham JA, Esteves TC, Green K, Lambert AJ, et al. Mitochondrial superoxide and ageing: uncoupling-protein activity and superoxide production. Biochem Soc Symp. 2004;71:203–213. doi: 10.1042/bss0710203. [DOI] [PubMed] [Google Scholar]
- 14.Brand MD, Esteves TC. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab. 2005;2:85–93. doi: 10.1016/j.cmet.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Nübel T, Ricquier D. Respiration under control of uncoupling proteins: Clinical perspective. Horm Res. 2006;65:300–310. doi: 10.1159/000092847. [DOI] [PubMed] [Google Scholar]
- 16.Echtay KS. Mitochondrial uncoupling proteins--what is their physiological role? Free Radic Biol Med. 2007;43:1351–1371. doi: 10.1016/j.freeradbiomed.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 17.MacLellan JD, Gerrits MF, Gowing A, Smith PJ, Wheeler MB, et al. Physiological increases in uncoupling protein 3 augment fatty acid oxidation and decrease reactive oxygen species production without uncoupling respiration in muscle cells. Diabetes. 2005;54:2343–2350. doi: 10.2337/diabetes.54.8.2343. [DOI] [PubMed] [Google Scholar]
- 18.Chan CB, Kashemsant N. Regulation of insulin secretion by uncoupling protein. Biochem Soc Trans. 2006;34:802–805. doi: 10.1042/BST0340802. [DOI] [PubMed] [Google Scholar]
- 19.Bézaire V, Seifert EL, Harper ME. Uncoupling protein-3: clues in an ongoing mitochondrial mystery. FASEB J. 2007;21:312–324. doi: 10.1096/fj.06-6966rev. [DOI] [PubMed] [Google Scholar]
- 20.Mattson MP, Kroemer G. Mitochondria in cell death: novel targets for neuroprotection and cardioprotection. Trends Mol Med. 2003;9:196–205. doi: 10.1016/s1471-4914(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 21.Wilson MM, Morley JE. Invited review: Aging and energy balance. J Appl Physiol. 2003;95:1728–1736. doi: 10.1152/japplphysiol.00313.2003. [DOI] [PubMed] [Google Scholar]
- 22.Frisard M, Ravussin E. Energy metabolism and oxidative stress: impact on the metabolic syndrome and the aging process. Endocrine. 2006;29:27–32. doi: 10.1385/ENDO:29:1:27. [DOI] [PubMed] [Google Scholar]
- 23.Fried LP, Tangen CM, Walston J, Newman AB, Hirsch C, et al. Cardiovascular Health Study Collaborative Research Group. Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci. 2001;56:146–56. doi: 10.1093/gerona/56.3.m146. [DOI] [PubMed] [Google Scholar]
- 24.Roberts SB, Rosenberg I. Nutrition and ageing: changes in the regulation of energy metabolism with ageing. Physiol Rev. 2006;86:651–667. doi: 10.1152/physrev.00019.2005. [DOI] [PubMed] [Google Scholar]
- 25.Frisard MI, Broussard A, Davies SS, Roberts LJ, Rood J, et al. Ageing, resting metabolic rate, and oxidative damage: results from the Louisiana Healthy Ageing Study. J Gerontol A Biol Sci Med Sci. 2007;62:752–759. doi: 10.1093/gerona/62.7.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brand MD. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp Gerontol. 2000;35:811–820. doi: 10.1016/s0531-5565(00)00135-2. [DOI] [PubMed] [Google Scholar]
- 27.Wolkow CA, Iser WB. Uncoupling protein homologs may provide a link between mitochondria, metabolism and lifespan. Ageing Res Rev. 2006;5:196–208. doi: 10.1016/j.arr.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mookerjee SA, Divakaruni AS, Jastroch M, Brand MD. Mitochondrial uncoupling and lifespan. Mech Ageing Dev. 2010;131:463–472. doi: 10.1016/j.mad.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harper ME, Bevilacqua L, Hagopian K, Weindruch R, Ramsey JJ. Ageing, oxidative stress, and mitochondrial uncoupling. Acta Physiol Scand. 2004;182:321–331. doi: 10.1111/j.1365-201X.2004.01370.x. [DOI] [PubMed] [Google Scholar]
- 30.Dietrich MO, Horvath TL. The role of mitochondrial uncoupling proteins in lifespan. Eur J Physiol. 2010;459:269–275. doi: 10.1007/s00424-009-0729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fridell YW, Sánchez-Blanco A, Silvia BA, Helfand SL. Functional characterization of a Drosophila mitochondrial uncoupling protein. J Bioenerg Biomembr. 2004;36:219–228. doi: 10.1023/b:jobb.0000031973.20153.c6. [DOI] [PubMed] [Google Scholar]
- 32.Fridell YW, Hoh M, Kréneisz O, Hosier S, Chang C, et al. Increased uncoupling protein (UCP) activity in Drosophila insulin-producing neurons attenuates insulin signaling and extends lifespan. Ageing. 2009;1:699–713. doi: 10.18632/aging.100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Speakman JR, Talbot DA, Selman C, Snart S, McLaren JS, et al. Uncoupled and surviving: individual mice with high metabolism have greater mitochondrial uncoupling and live longer. Ageing Cell. 2004;3:87–95. doi: 10.1111/j.1474-9728.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- 34.Conti B, Sanchez-Alavez M, Winsky-Sommerer R, Morale MC, Lucero J, et al. Transgenic mice with a reduced core body temperature have an increased life span. Science. 2006;314:825–828. doi: 10.1126/science.1132191. [DOI] [PubMed] [Google Scholar]
- 35.Andrews ZB, Horvath TL. Uncoupling protein-2 regulates lifespan in mice. Am J Physiol Endocrinol Metab. 2009;96(E):621–627. doi: 10.1152/ajpendo.90903.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caldeira da Silva CC, Cerqueira FM, Barbosa LF, Medeiros MH, Kowaltowski AJ. Mild mitochondrial uncoupling in mice affects energy metabolism, redox balance and longevity. Ageing Cell. 2008;7:552–560. doi: 10.1111/j.1474-9726.2008.00407.x. [DOI] [PubMed] [Google Scholar]
- 37.Bevilacqua L, Ramsey JJ, Hagopian K, Weindruch R, Harper ME. Long-term caloric restriction increases UCP3 content but decreases proton leak and reactive oxygen species production in rat skeletal muscle mitochondria. Am J Physiol Endocrinol Metab. 2005;289(E):429–438. doi: 10.1152/ajpendo.00435.2004. [DOI] [PubMed] [Google Scholar]
- 38.McDonald RB, Walker KM, Warman DB, Griffey SM, Warden CH, et al. Characterization of survival and phenotype throughout the life span in UCP2/UCP3 genetically altered mice. Exp Gerontol. 2008;43:1061–1068. doi: 10.1016/j.exger.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Maedler K, Shu L, Haataja L. UCP-2 and UCP-3 proteins are differentially regulated in pancreatic beta-cells. PLoS One. 2008;3:e1397. doi: 10.1371/journal.pone.0001397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salopuro T, Pulkkinen L, Lindström J, Kolehmainen M, Tolppanen AM, et al. Variation in the UCP2 and UCP3 genes associates with abdominal obesity and serum lipids: the Finnish Diabetes Prevention Study. BMC Med Genet. 2009;10:94. doi: 10.1186/1471-2350-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia JJ, Zhang X, Ge CR, Jois M. The polymorphisms of UCP2 and UCP3 genes associated with fat metabolism, obesity and diabetes. Obes Rev. 2009;10:519–552. doi: 10.1111/j.1467-789X.2009.00569.x. [DOI] [PubMed] [Google Scholar]
- 42.Crocco P, Montesanto A, Passarino G, Rose G. A common polymorphism in the UCP3 promoter influences hand grip strength in elderly people. Biogerontology. 2011;12(3):265–271. doi: 10.1007/s10522-011-9321-z. [DOI] [PubMed] [Google Scholar]
- 43.Rose G, Crocco P, D'Aquila P, Montesanto A, Bellizzi D, et al. Exp Gerontol [Epub ahead of print]; 2011. Two variants located in the upstream enhancer region of human UCP1 gene affect gene expression and are correlated with human longevity. [DOI] [PubMed] [Google Scholar]
- 44.Passarino G, Montesanto A, Dato S, Giordano S, Domma F, et al. Sex and age specificity of susceptibility genes modulating survival at old age. Hum Hered. 2006;62:213–220. doi: 10.1159/000097305. [DOI] [PubMed] [Google Scholar]
- 45.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freidlin B, Zheng G, Li Z, Gastwirth JL. Trend tests for case-control studies of genetic markers: power, sample size and robustness. Hum Hered. 2002;53:146–152. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 47.Zang Y, Fung WK. Robust tests for matched case-control genetic association studies. BMC Genet. 2010;11:91. doi: 10.1186/1471-2156-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zang Y, Fung WK, Zheng G. Simple algorithms to calculate asymptotic null distribution for robust tests in case-control genetic association studies in R. Journal of Statistical Software. 2010;33:1–24. [Google Scholar]
- 49.So HC, Sham PC. Robust Association Tests Under Different Genetic Models, Allowing for Binary or Quantitative Traits and Covariates. Behav Genet. 2011;41:768–75. doi: 10.1007/s10519-011-9450-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 51.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. "Score tests for association of traits with haplotypes when linkage phase is ambiguous. " Amer J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calle ML, Urrea V, Vellalta G, Malats N, Steen KV. Improving strategies for detecting genetic patterns of disease susceptibility in association studies. Stat Med. 2008;27(30):6532–6546. doi: 10.1002/sim.3431. [DOI] [PubMed] [Google Scholar]
- 53.Bouchard C, Perusse L, Chagnon YC, Warden C, Ricquier D. Linkage between markers in the vicinity of the uncoupling protein 2 gene and resting metabolic rate in humans. Hum Mol Genet. 1997;6:1887–1889. doi: 10.1093/hmg/6.11.1887. [DOI] [PubMed] [Google Scholar]
- 54.Fleury C, Neverova M, Collins S, Raimbault S, Champigny O, et al. Uncoupling protein-2: a novel gene linked to obesity and hyperinsulinemia. Nat Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- 55.Bézaire V, Spriet LL, Campbell S, Sabet N, Gerrits M, et al. Constitutive UCP3 overexpression at physiological levels increases mouse skeletal muscle capacity for fatty acid transport and oxidation. FASEB J. 2005;19:977–979. doi: 10.1096/fj.04-2765fje. [DOI] [PubMed] [Google Scholar]
- 56.Xu K, Zhang M, Cui D, Fu Y, Qian L, et al. Diabetologia; 2011. UCP2 -866G/A and Ala55Val, and UCP3 -55C/T polymorphisms in association with type 2 diabetes susceptibility: a meta-analysis study. DOI 10.1007/s00125-011-2245-y [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 57.Astrup A, Toubro S, Dalgaard LT, Urhammer SA, Sorensen TI, et al. Impact of the v/v 55 polymorphism of the uncoupling protein 2 gene on 24-h energy expenditure and substrate oxidation. Int J Obes Relat Metab Disord. 1999;23:1030–1034. doi: 10.1038/sj.ijo.0801040. [DOI] [PubMed] [Google Scholar]
- 58.Buemann B, Schierning B, Toubro S, Bibby BM, Sorensen T, et al. The association between the Val/Ala-55 polymorphism of the uncoupling protein 2 gene and exercise efficiency. Int J Obes Relat Metab Disord. 2001;25:467–471. doi: 10.1038/sj.ijo.0801564. [DOI] [PubMed] [Google Scholar]
- 59.Walder K, Norman RA, Hanson RL, Schrauwen P, Neverova M, et al. Association between uncoupling protein polymorphisms (UCP2-UCP3) and energy metabolism/obesity in Pima Indians. Hum Mol Genet. 1998;7:1431–1435. doi: 10.1093/hmg/7.9.1431. [DOI] [PubMed] [Google Scholar]
- 60.Yu X, Jacobs DR, Schreiner PJ, Gross MD, Steffes MW, et al. The uncoupling protein 2 Ala55Val polymorphism is associated with diabetes mellitus: the CARDIA Study. Clin Chem. 2005;51:1451–1456. doi: 10.1373/clinchem.2004.044859. [DOI] [PubMed] [Google Scholar]
- 61.Willig AL, Casazza KR, Divers J, Bigham AW, Gower BA, et al. Uncoupling protein 2 Ala55Val polymorphism is associated with a higher acute insulin response to glucose. Metabolism. 2009;58:877–881. doi: 10.1016/j.metabol.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen HH, Lee W, Wang W, Huang MT, Lee YC, et al. Ala55Val polymorphism on UCP2 gene predicts greater weight loss in morbidly obese patients undergoing gastric banding. Obes Surg. 2007;17:926–933. doi: 10.1007/s11695-007-9171-6. [DOI] [PubMed] [Google Scholar]
- 63.Dalgaard LT, Andersen G, Larsen LH, Sorensen TI, Andersen T, et al. Mutational analysis of the UCP2 core promoter and relationships of variants with obesity. Obes Res. 2003;11:1420–1427. doi: 10.1038/oby.2003.191. [DOI] [PubMed] [Google Scholar]
- 64.Wang H, Chu WS, Lu T, Hasstedt SJ, Kern PA, et al. Uncoupling protein-2 polymorphisms in type 2 diabetes, obesity, and insulin secretion. Am J Physiol Endocrinol Metab. 2004;286:E1–E7. doi: 10.1152/ajpendo.00231.2003. [DOI] [PubMed] [Google Scholar]
- 65.Hsu YH, Niu T, Song Y, Tinker L, Kuller LH, et al. Genetic variants in the UCP2- UCP3 gene cluster and risk of diabetes in the women's health initiative observational study. Diabetes. 2008;57:1101–1107. doi: 10.2337/db07-1269. [DOI] [PubMed] [Google Scholar]
- 66.Lee YH, Kim W, Yu BC, Lae Park BL, Kim LH, et al. Association of the Ins/Del polymorphisms of uncoupling protein 2 (UCP2) with BMI in a Korean population. Biochem Biophys Res Commun. 2008;371:767–771. doi: 10.1016/j.bbrc.2008.04.144. [DOI] [PubMed] [Google Scholar]
- 67.Paradis E, Clavel S, Bouillaud F, Ricquier D, Richard D. Uncoupling protein 2: A novel player in neuroprotection. Trends Mol Med. 2003;9:522–525. doi: 10.1016/j.molmed.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 68.Horvath TL, Diano S, Barnstable C. Mitochondrial uncoupling protein 2 in the central nervous system: Neuromodulator and neuroprotector. Biochem Pharmacol. 2003;65:1917–1921. doi: 10.1016/s0006-2952(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 69.Cortez-Pinto H, Yang SQ, Lin HZ, Costa S, Hwang CS, et al. Bacterial lipopolysaccharide induces uncoupling protein-2 expression in hepatocytes by a tumor necrosis factor-alpha-dependent mechanism. Biochem Biophys Res Commun. 1998;251:313–319. doi: 10.1006/bbrc.1998.9473. [DOI] [PubMed] [Google Scholar]
- 70.Murray AJ, Anderson RE, Watson GC, Radda GK, Clarke K. Uncoupling proteins in human heart. Lancet. 2004;364:1786–1788. doi: 10.1016/S0140-6736(04)17402-3. [DOI] [PubMed] [Google Scholar]
- 71.Kopecky J, Rossmeisl M, Flachs P, Brauner P, Sponarova J, et al. Energy metabolism of adipose tissue--physiological aspets and target in obesity treatment. Physiol Res. 2004;53:S225–S232. [PubMed] [Google Scholar]
- 72.Samec S, Seydoux J, Dulloo AG. nSkeletal muscle UCP3 and UCP2 gene expression in response to inhibition of free fatty acid flux through mitochondrial beta-oxidation. Pflugers Arch. 1999;438:452–457. doi: 10.1007/s004249900080. [DOI] [PubMed] [Google Scholar]
- 73.Saleh MC, Wheeler MB, Chan CB. Uncoupling protein-2: Evidence for its function as a metabolic regulator. Diabetologia. 2002;45:74–187. doi: 10.1007/s00125-001-0737-x. [DOI] [PubMed] [Google Scholar]
- 74.Diao J, Allister EM, Koshkin V, Lee SC, Bhattacharjee A, et al. UCP2 is highly expressed in pancreatic alpha-cells and influences secretion and survival. PNAS. 2008;105:12057–1262. doi: 10.1073/pnas.0710434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andrews ZB. Uncoupling protein-2 and the potential link between metabolism and longevity. Curr Aging Sci. 2010;3:102–112. doi: 10.2174/1874609811003020102. [DOI] [PubMed] [Google Scholar]
- 76.Barbieri M, Boccardi V, Esposito A, Papa M, Vestini F, et al. Age; 2011. A/ASP/VAL allele combination of IGF1R, IRS2, and UCP2 genes is associated with better metabolic profile, preserved energy expenditure parameters, and low mortality rate in longevity. DOI: 10.1007/s11357-011-9210-z [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manini TM. Energy expenditure and aging. Ageing Res Rev. 2010;9:1–11. doi: 10.1016/j.arr.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barbieri M, Gambardella A, Paolisso G, Varricchio M. Metabolic aspects of the extreme longevity. Exp Gerontol. 2008;43:74–78. doi: 10.1016/j.exger.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 79.Rizzo MR, Mari D, Barbieri M, Ragno E, Grella R, et al. Resting Metabolic Rate and Respiratory Quotient in Human Longevity. J Clin Endocrinol Metab. 2005;90:409–413. doi: 10.1210/jc.2004-0390. [DOI] [PubMed] [Google Scholar]
- 80.Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- 81.Schrauwen P, Xia J, Walder K, Snitker S, Ravussin E. A novel polymorphism in the proximal UCP3 promoter region: effect on skeletal muscle UCP3 mRNA expression and obesity in male non-diabetic Pima Indians. Int J Obes Relat Metab Disord; 1999;23:1242–1245. doi: 10.1038/sj.ijo.0801057. [DOI] [PubMed] [Google Scholar]
- 82.Schrauwen P, Xia J, Bogardus C, Pratley R, Ravussin E. Skeletal muscle UCP3 expression is a determinant of energy expenditure in Pima Indians. Diabetes. 1999;48:146–149. doi: 10.2337/diabetes.48.1.146. [DOI] [PubMed] [Google Scholar]
- 83.Nabben M, Hoeks J, Briede JJ, Glatz JF, Moonen-Kornips E, et al. The effect of UCP3 overexpression on mitochondrial ROS production in skeletal muscle of young versus aged mice. FEBS Lett. 2008;582:4147–4152. doi: 10.1016/j.febslet.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 84.Karakelides H, Nair KS. Sarcopenia of ageing and its metabolic impact. Curr Top Dev Biol. 2005;68:123–148. doi: 10.1016/S0070-2153(05)68005-2. [DOI] [PubMed] [Google Scholar]
- 85.Rossi P, Marzani B, Giardina S, Negro M, Marzatico F. Human skeletal muscle ageing and the oxidative system: cellular events. Curr Ageing Sci. 2008;1:182–191. doi: 10.2174/1874609810801030182. [DOI] [PubMed] [Google Scholar]
- 86.Amara CE, Shankland EG, Jubrias SA, Marcinek DJ, Kushmerick MJ, et al. Mild mitochondrial uncoupling impacts cellular aging in human muscles in vivo. PNAS. 2007;104:1057–62. doi: 10.1073/pnas.0610131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dalgaard LT. Genetic Variance in Uncoupling Protein 2 in Relation to Obesity, Type 2 Diabetes, and Related Metabolic Traits: Focus on the Functional -866G>A Promoter Variant (rs659366). J Obes. 2011 doi: 10.1155/2011/340241. doi: 10.1155/2011/340241 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andrews ZB, Diano S, Horvath TL. Mitochondrial uncoupling proteins in the CNS: in support of function and survival. Nat Rev Neurosci. 2005;6:829–840. doi: 10.1038/nrn1767. [DOI] [PubMed] [Google Scholar]
- 89.Liu D, Chan SL, De Souza-Pinto NC, Slevin JR, Wersto RP, et al. Mitochondrial UCP4 mediates an adaptive shift in energy metabolism and increases the resistance of neurons to metabolic and oxidative stress. Neuromolecular Med. 2006;8:389–414. doi: 10.1385/NMM:8:3:389. [DOI] [PubMed] [Google Scholar]
- 90.Wei Z, Chigurupati S, Bagsiyao P, Henriquez A, Chan SL. The brain uncoupling protein UCP4 attenuates mitochondrial toxin-induced cell death: role of extracellular signal-regulated kinases in bioenergetics adaptation and cell survival. Neurotox Res. 2009;16:14–29. doi: 10.1007/s12640-009-9039-8. [DOI] [PubMed] [Google Scholar]
- 91.de la Monte SM, Wands JR. Molecular indices of oxidative stress and mitochondrial dysfunction occur early and often progress with severity of Alzheimer's disease. J Alzheimers Dis. 2006;9:167–181. doi: 10.3233/jad-2006-9209. [DOI] [PubMed] [Google Scholar]
- 92.Szolnoki Z, Kondacs A, Mandi Y, Bodor A, Somogyvari F. A homozygous genetic variant of mitochondrial uncoupling protein 4 exerts protection against the occurrence of multiple sclerosis. Neuromolecular Med. 2009;11:101–105. doi: 10.1007/s12017-009-8071-4. [DOI] [PubMed] [Google Scholar]
- 93.Szolnoki Z, Kondacs A, Mandi Y, Bodor A, Somogyvari F. A homozygous genetic variant of mitochondrial uncoupling protein 4 affects the occurrence of leukoaraiosis. Acta Neurol Scand. 2011;123:352–357. doi: 10.1111/j.1600-0404.2010.01391.x. [DOI] [PubMed] [Google Scholar]
- 94.Girousse A, Tavernier G, Tiraby C, Lichtenstein L, Iacovoni JS, et al. Transcription of the human uncoupling protein 3 gene is governed by a complex interplay between the promoter and intronic sequences. Diabetologia. 2009;52:1638–1646. doi: 10.1007/s00125-009-1385-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotypic and allelic frequencies in the analyzed sample by group.
(PDF)