Abstract
AFNI is an open source software package for the analysis and display of functional MRI data. It originated in 1994 to meet the specific needs of researchers at the Medical College of Wisconsin, in particular the mapping of activation maps to Talairach-Tournoux space, but has been expanded steadily since then into a wide-ranging set of tool for FMRI data analyses. AFNI was the first platform for real-time 3D functional activation and registration calculations. One of AFNI’s main strengths is its flexibility and transparency. In recent years, significant efforts have been made to increase the user-friendliness of AFNI’s FMRI processing stream, with the introduction of “super-scripts” to setup the entire analysis, and graphical front-ends for these managers.
Keywords: Functional data analysis, 3D registration, FMRI group analysis, FMRI connectivity, FMRI software
The Past
Contingency. AFNI is one result of unexpected events. I met a woman and fell in love. She had a tenured faculty position in Milwaukee, so I moved there in 1993, got married, and looked for a job. Meanwhile, FMRI had started up at the Medical College of Wisconsin (MCW) in late 1991, thanks largely to the acumen and efforts of 2 graduate students: Peter Bandettini and Eric Wong (Bandettini et al., 1992). By early 1993, Jim Hyde (chairman of MCW Biophysics) had decided they were “drowning in data” and was casting about for a lifeline. At that moment, I showed up and looked like I knew something about data analysis. I got the job that autumn, and spent much of the next 6 months frantically trying to learn about the fields of MRI, FMRI, neuroscience, and their guardian phalanxes of jargon and tropes. (Plus contribute to grant writing.)
I started developing AFNI (Cox, 1996) in mid-1994 to meet the needs of the MCW researchers using this new FMRI tool to explore brain function. Their most insistently expressed desire was the ability to transform their data to Talairach-Tournoux coordinates—an inherently 3D process. Up until then, all FMRI processing and display at MCW had been in 2D slices. At that time, when the Web was just starting, most research groups processed their data with home-grown software, and good information about the tools used at other sites was hard to glean. After pondering the Talairach-Tournoux atlas (Talairach and Tournoux, 1988) and some FMRI data for a while, I sketched out a plan for a volumetric-based software tool to enable researchers to surf through their datasets in 3D, jumping between multiple datasets, and overlaying functional maps on structural volumes as desired.
My design and implementation from the start was based on a few generic principles:
Let the user stay close to her data and view it in several different ways, providing controls to allow for interactive adjustment of statistical thresholds and colorization. This ability helps the user become familiar with the structure of her data and to understand her results.
Let the user compose and control the data analysis and image processing as needed for her particular research problems and predispositions.
Let the user see the intermediate results of the processing sequence, so that it is easy to backtrack to understand how particular results were obtained. This ability has proved especially important when trying to decipher unexpected results (e.g., activation “blobs” that don’t make sense).
Everything should be open-source and the software should be modular enough that other people could usefully contribute to it.
By September 1994, I had the first version of the AFNI graphical interface ready for MCW faculty to try. Using it, a user could view low-resolution 3D functional activation maps overlaid on sagittal, axial, and/or coronal slices from a high-resolution volume, with the ability to scroll through slices at will in any or all viewer windows, alter the statistical threshold on the activation map, and change the colorization scheme. The first researchers who tried it were ecstatic—they could finally see their brain mapping results they way they wanted, and control how the maps appeared. And they could transform their results to Talairach-Tournoux space, which was why it all had started.
I developed AFNI to run on Unix+X11 workstations with 32+ MB of RAM (which was a lot back in those ancient times). Datasets only had 3 dimensions; it wasn’t practicable to read in and process entire 3D+time collections of images—the basic units of FMRI data. Processing of FMRI data to produce functional activation datasets proceeded slice-by-slice, and only at the end were the 3D functional overlay datasets assembled for visualization in AFNI. By mid-1996, with more memory available, an extensive re-write of AFNI allowed datasets to have a fourth dimension, and all data analyses moved from a 2D basis to 3D. This change dovetailed with the development at MCW of realtime FMRI activation analysis (Cox et al., 1995) and volume registration (Cox and Jesmanowicz, 1999), both of which were incorporated into AFNI soon after they were developed. About the same time, the central FMRI time series analysis methodology in AFNI evolved from the correlation method (Bandettini et al., 1993) to multiple linear regression and hemodynamic impulse response function deconvolution.
In 2001, the NIH recruited me to establish a group to support the rapidly expanding FMRI research efforts on the Bethesda campus. As this group expanded, AFNI branched out in several directions, most fundamentally to include processing of dataset defined over triangulated cortical surface models, not just 3D regular grids, in the SUMA software (Saad and Reynolds, this issue).
Structure
Originally, AFNI dealt only with 3D (and later, 3D+time) datasets stored in a format of my own devising, where all auxiliary non-image data are stored in a text file of the general form “attribute-name = values”, and the much bulkier image values are in a separate binary file. One advantage of this approach is that arbitrary expansions of the auxiliary data are simple, and at the same time image data I/O is also straightforward. One major disadvantage is that this custom format is not readily useful with other software. In the late 1990s, the NIH became concerned with the interoperability of neuroimaging research software. A working group was formed, chaired by Stephen Strother and including representatives from most major FMRI software developers, which met under the aegis of the NIMH and NINDS extramural programs. The upshot was the new and simple NIfTI-1 format for storing data defined over regular grids (Cox et al., 2004). This format includes the ability to include arbitrary application-specific extensions to the header information, which AFNI uses to store (via XML) various auxiliary information not otherwise defined in the NIfTI-1 specification—for example, the history of the commands that produced a given dataset are saved in its header. AFNI now supports NIfTI-1 for input and output at the same level as the AFNI custom format. All that the user has to do is specify that the output filename end in “.nii” or “.nii.gz” to get a NIfTI-1 output file. We support other formats, such as DICOM, mostly by the use of file converters.
AFNI comprises the interactive visualization program of the same name, a collection of plugins to the graphical interface, plus a large set of batch (Unix command line) data processing programs. Most of these batch program names start with the characters “3d” (e.g., 3dDeconvolve is an FMRI time series deconvolution program)—this naming scheme was chosen to emphasize that they process data stored in 3D image volumes. In the early days of FMRI, most neuroscience researchers still thought in 2D slice terms, not in 3D volumetric terms; I wanted the program names to help change this conceptualization. However, AFNI “3d” programs that operate on a voxel-wise basis are not actually restricted to data defined over 3D grids; for example, it is simple to map a 3D FMRI time series dataset to a cortical surface domain (e.g., via 3dVol2Surf), and then carry out the rest of the analyses on the surface model using the same regression and statistical software as in the volumetric analysis. Only the inter-voxel processing steps, such as spatial smoothing, require special purpose surface-aware software. The recent development of the GIfTI file format for surfaces and surface-based datasets now allows interoperability of surface-analysis software from different packages, much as the NIfTI-1 format does.
A competent C programmer can write an AFNI “3d” batch program without climbing a steep learning curve. For example, the core AFNI library provides utility subroutines to create an output dataset from a programmer-supplied function which takes a collection of numbers from the input dataset at a single voxel and returns another collection of numbers to be stored at that location in the output dataset. For plugin creation, the AFNI graphical user interface has a set of library functions to allow a programmer to create a “fill in the form” style of interface (cf. Figure 1 and the InstaCorr setup control panel). However, my dream of having significant contributions to AFNI from outside developers has only been partially realized. A few such programs have been donated and are distributed with AFNI, but most people (understandably) prefer to work independently. Fortunately, the advent of the NIfTI-1 format makes such third-party efforts readily usable with AFNI batch programs.
Figure 1.
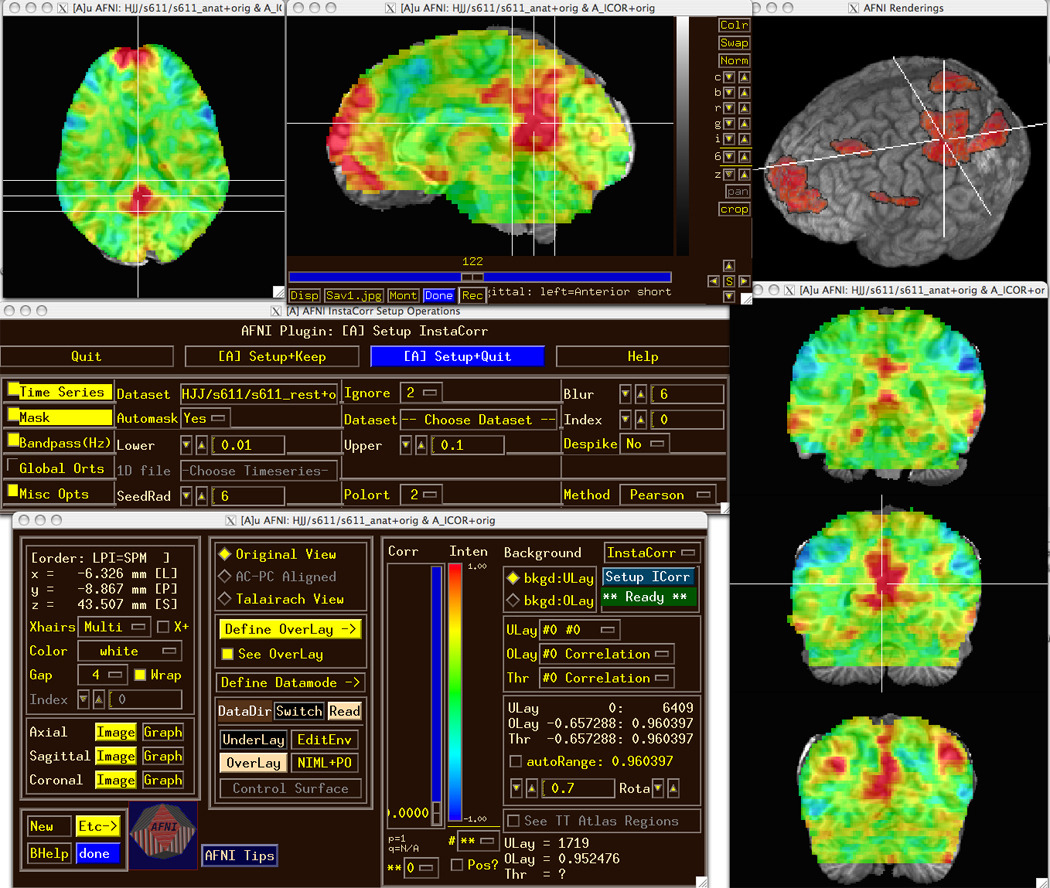
The AFNI graphical user interface (on OS X), with the individual subject InstaCorr module setup window shown, and the InstaCorr results from clicking in the posterior cingulate region (at the crosshair location) overlaid on the subject’s SPGR volume—un-thresholded in slice views and thresholded at r > 0.6 in a 3D see-thru volume rendered view. The sagittal slice viewer shows the AFNI controls which have been turned off (for de-cluttering) in the other image windows.
With the introduction of SUMA, Ziad Saad and I had to design a “live” interchange protocol for external programs to talk to AFNI, exchanging data and commands with the independently running program. At the NIfTI working group meetings, it was clear that no other FMRI software group was interested in such an effort, so we developed our own XML table-based format for this purpose. An external program can attach to AFNI (or SUMA) via a TCP/IP socket—if the programs are on the same computer, the communication channel can be switched to shared memory, which is faster for exchanging large amounts of data. Once this I/O channel is established, the external program can send commands to AFNI to open windows for image or time series graph viewing, and send new 3D datasets for display. In this way, a user can script the viewing of large numbers of datasets—we have found this ability to be very useful when processing large volumes of data, such as the FCON1000 collection (Biswal et al., 2010). When computing with a lot of datasets, some algorithm (e.g., registration) always seems to do badly on a few cases, and being able to scan systematically through the entire collection to find and fix such problems is important. The external driving program need not be in C; we also provide sample programs in Matlab and Python to demonstrate this functionality. The realtime FMRI acquisition system at the NIH FMRI facility uses a set of Perl scripts to control AFNI. SUMA uses this capability to be closely linked to AFNI, exchanging crosshair locations, datasets, and colorized overlays between the folded 2D surface domains and the 3D grid domains.
The Present
AFNI has had the reputation for being hard to use. In the past, users were required to create their own processing scripts to chain together the various “3d” programs to achieve the goals they wanted, using our sample scripts as starting points. Physicists and statisticians like this style of operation, since they generally want complete transparency of and control over every detail of the analysis process. It is common in neuroscience, however, that a researcher wants first to see some activation results, and possibly later go back to refine the analysis—such a user wants an easy-to-use standard and reliable analysis path to follow at the start.
We have worked hard to make it simple to run the basic and common analysis pathways through the AFNI suite of programs. Our first big step was the creation of the afni_proc.py super-script, which is a (Python) command line program that takes as input the list of MRI 3D and FMRI 3D+time dataset files for a single subject, the stimulus timing files, and a few more pieces of information, and produces a script that can process all the data from that subject to give the statistical maps ready for group analysis. Different classes of stimuli are given user-chosen labels, and contrasts are specified symbolically (e.g., “+Faces –Houses”). All the intermediate datasets are saved, making it easy to check the analysis if the final output seems unusual. The generated processing script itself serves as a record of what was done, and can be manually edited if non-standard steps need to be injected into the calculations. We strongly recommend that all but the most experienced AFNI users start with or switch to using afni_proc.py to generate their FMRI analysis pipeline.
Our next step was the creation of a graphical interface, uber_subject.py, to make it easy to run afni_proc.py by button-clicking and menu-selection. This GUI is now available with AFNI, but is still a work in progress, growing in flexibility at a brisk pace. We also have a command line script, align_epi_anat.py, which is the recommended tool for 3D registration processing in AFNI—despite its name, suggesting it is only for aligning anatomical and EPI volumes, it is capable of other tasks, including mapping 3D and 3D+time datasets to template spaces. We are currently developing a graphical interface for these alignment tasks, which are often needed in non-FMRI applications. A usable super-script for setting up group (inter-subject) analyses has also been written and is being refined.
It is the flexibility of AFNI programs that is one of our software’s greatest strengths. Most of our analysis codes have dozen of options—one source of the obsolescent “AFNI is hard to use” meme, from the time when it was necessary for each user to create her own processing script ex nihilo. Each option is there to meet some particular need. For example, the 3dDeconvolve linear regression program has many different built-in hemodynamic response models, including amplitude- and duration-modulated stimuli, fixed-shape and deconvolution hemodynamic response models, because FMRI-based investigations are so mutable and the questions asked about the response amplitude and shape can vary so much. Response amplitudes can be calculated for each stimulus separately, to be merged at a later step, or pooled across stimuli that are classed together—the most common form of analysis in FMRI. For more specialized purposes, other time series regression programs are available, including nonlinear voxel-wise regression (e.g., for fitting pharmaco-kinetic and -dynamic models) and a code for linear regression with voxel-dependent models, including parameter sign constraints and L1, L2, and LASSO fitting options. In addition, over the years we have implemented a large number of utility programs for various statistical and 3D image processing purposes, available to users who have particular processing needs. Of these, perhaps the most useful is 3dcalc, a general purpose voxel-wise calculator program—the Swiss Army knife of AFNI, not well-adapted for any single purpose, but very helpful for getting some computation done quickly.
At the group analysis level, AFNI has options ranging from simple t-tests to a sophisticated mixed-effects meta-analysis tool (3dMEMA) that incorporates voxel-wise estimates of the per-subject variance of the FMRI response amplitude and can model outliers. The linear mixed effects code (3dLME) can model and fit correlations between data points when multiple parameter estimates are carried to the group level from each subject simultaneously (e.g., when comparing and contrasting the shape of the hemodynamic response among subjects, not just its amplitude). Subject-wise covariates are easily incorporated into these analyses by including a file containing a table of these values, and the t-test program also allows for voxel-wise covariates for each subject (e.g., gray matter volume fraction). AFNI also includes several flavors of task-based and resting-state connectivity and network analyses.
The AFNI graphical interface contains several tools for quick interactive data analyses. The InstaCorr module allows the user to load a resting-state FMRI dataset and pre-process it— masking, despiking, blurring, bandpassing—then, when the user clicks on a voxel location, the correlation map of that voxel’s time series with all other voxels’ data is computed and displayed within milliseconds. The group version of InstaCorr also performs a 1- or 2-sample t-test (optionally allowing for subject-wise covariates) across the individual subject tanh−1 transformed correlation maps; on my desktop, 100 rs-FMRI datasets from the FCON1000 collection (≈69K voxels and 170 TRs per dataset: 1.2 Gbytes total) can be correlated, t-tested, and transmitted to AFNI for display in about 0.4 seconds per seed voxel click. (CPU-intensive AFNI programs, such as group InstaCorr, are generally parallelized using OpenMP.) My purpose in developing these interactive computational tools is to allow researchers to explore the structure of their data, to aid in forming hypotheses, and to help identify problems such as scanner artifacts (Jo et al., 2010). Figure 1 shows a screen snapshot of the individual subject InstaCorr setup and results windows.
We provide support for non-NIH AFNI users via an active forum at our Web site http://afni.nimh.nih.gov, including the ability for users to upload datasets if needed to help us understand their issues. (Such datasets go to a directory that is not accessible except to the AFNI group, and are deleted when no longer needed.) We have no particular release schedule or version numbering for AFNI. When we add new features to the software, an updated package is put up on our Web site. No significant software is bug-free, but we pride ourselves on fixing reported problems with AFNI programs rapidly—within a day or two if at all possible. Bugs that might have widespread impact are further highlighted with a popup message-of-the-day (dynamically fetched from the AFNI server), explaining the issues and our recommendations.
Our close interaction with users, particularly at the NIH, has been the source of many developments. For example, our EPI-structural registration algorithm (Saad et al., 2009) was developed when a few discerning users complained that mutual information did not do a good job for this type of image alignment. At first we were skeptical, and suggested using the correlation ratio cost function instead; however, when these users uploaded some sample datasets, we played with them and discovered that datasets with high visual quality can still be very hard to register accurately—the ventricles did not match no matter what cost function or software package we used. Not liking to admit defeat so easily, I examined the joint histogram of the datasets carefully and then developed a cost function specialized for this particular application. This example illustrates the importance of one of my generic rules: Always look at your data and intermediate results. Complicated algorithms and software can fail without giving any obvious indication of error. Only by staying close to the data and the processing stream can a researcher expect to get quality results.
The Future
One of the most exciting things we are working on now is a set of tools for “atlasing”—to make it easy to incorporate brain atlas datasets into AFNI, to serve as guides for the user, to use in ROI generation, and in more advanced forms of group analysis now under development. AFNI already has a number of atlases built-in, including the San Antonio Talairach Daemon (Lancaster et al., 2000) and the Eickhoff-Zilles probabilistic atlas (Eickhoff et al., 2005), but each of these was incorporated by custom coding and manipulations. With our new software, we aim to make it easy to build atlas datasets and then transform individual subject coordinates to these spaces automatically and on-the-fly. In our current software, a user can click a button and pop up a report on the overlap of each activation blob with the regions defined in a set of atlases. We are also developing software and protocols to allow AFNI to query remote atlases (e.g., from commercial entities such as Elsevier). Our efforts are not limited to human brain mapping, since there are many non-human FMRI studies carried out at the NIH and other sites. We are enhancing our nonlinear 3D registration capabilities to go along with this atlasing suite, and are also developing improved methods for tissue segmentation to be used for this and other purposes (Vovk et al., 2011).
Does it make sense for there to continue to be a number of highly overlapping open-source software packages for analysis of FMRI datasets? Each software package for FMRI is not just a set of tools, but is also a social ecosystem, with its own style of support and types of users. When I started at the NIH in 2001, the NIMH Scientific Director at the time (Bob Desimone) was enthusiastic about the idea of a central software platform to which all developers would “plug in”. A decade later, this vision has not been realized, or even seriously attempted. More recently, there have been efforts to develop and popularize FMRI pipeline toolkits which can incorporate standalone programs from a variety of sources. We have found it difficult to incorporate AFNI programs, which have many user-adjustable parameters, into these pipelines; as a result, we have chosen to develop our own customized pipelines, such as afni_proc.py and align_epi_anat.py.
At present, there is little evidence of convergence on any higher level of interoperability than the NIfTI-1 and GIfTI file formats. From the viewpoint of developers, this situation makes sense, since agreeing and adapting to some centralized system would be a lot of effort with no self-evident payoff. From the viewpoint of advanced users, cleaner interoperability is desirable; however, the science that we subserve should not come to depend on a single software source—failures (gross or subtle) in such a system could easily contaminate the entire FMRI-based literature for a long time. Multiple packages can avoid this problem, and also provide a competitive prod for the developers to do better. For these reasons, I believe that instead of trying to unify the major FMRI software packages, a useful intermediate step would be a common input format for specifying a task-based FMRI analysis; this format would make it simpler to run the same data through several toolkits and determine the sensitivity of the results to the myriad assumptions underlying the packages’ algorithms. A similar idea is the definition of a similar platform-independent way to specify a network analysis on pre-specified regions, either of task-based FMRI or rs-FMRI datasets.
Going forward, what is the place of AFNI in the functional neuroimaging world? AFNI is best for researchers who want software that preserves the intermediate results and is transparent about its processing steps, who want the ability to easily craft their own analysis streams, who want flexibility in the display and thresholding of their results, and who want the rapid responses that our team endeavors to give to feature requests, to bug reports, and to calls for help on our forum. In turn, we are ready to cooperate and collaborate with such users to extend and expand the capabilities of AFNI. The AFNI core team at the NIH comprises people who have worked with FMRI data, methods, statistics, and software development for years, and who are ready to continue innovating and helping.
Acknowledgments
There is no space to acknowledge the many many people who have encouraged and inspired the creation and development of AFNI since 1994. Here, I want to call attention to those who have contributed directly and significantly over the years to this continuing work:
Gang Chen (NIH), who has developed a veritable host of ideas, methods, and software tools for FMRI group and network/connectivity analyses, and whose statistical expertise has been critically important.
Daniel Glen (NIH), who has created the AFNI infrastructure for storing and querying atlas datasets, developed the align_epi_anat.py script for managing multiple types of 3D registration tasks, and implemented the nonlinear diffusion tensor estimation software in AFNI.
Rich Hammett (formerly NIMH), who implemented a number of important library functions and statistical tools during his too-brief tenure in the AFNI group.
Andrzej Jesmanowicz (MCW), whose 2D FMRI FD program was a major source of ideas in the early days of AFNI’s creation—I was planning to call AFNI “FD3” for a few days, until I came to my senses.
Stephen LaConte (Virginia Tech), who created and contributed the 3dsvm software for multi-voxel pattern analysis (Laconte et al., 2005).
Rick Reynolds (NIH), who has done truly heroic work in making AFNI analyses user-friendly, in implementing the NIfTI-1 and GIfTI C libraries, in developing behind-the-scenes software for the meshing of AFNI and SUMA analyses, and in improving the AFNI realtime image data acquisition and analysis plugin.
Tom Ross (NIDA), who contributed several important tools for pre- and post-processing FMRI datasets.
Ziad Saad (NIH), who—besides creating SUMA ab ova—has contributed a multitude of invaluable new ideas and concepts for improving FMRI data analyses and AFNI; it is now impossible for me to imagine what AFNI would be like without his influence.
Doug Ward (MCW), who wrote the first edition of 3dDeconvolve, the suite of 1–3 way 3dANOVA programs, and who first brought FDR statistics to AFNI.
Without the efforts of these people (and others), AFNI would not have “grown up” as well has it has—thank you all. The development of AFNI has been supported over the years by the American people: when I was at MCW, via grants from the NIMH and NINDS extramural programs, and currently via the NIMH and NINDS intramural programs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bandettini PA, Wong EC, Hinks RS, Tikofsky RS, Hyde JS. Time course EPI of human brain function during task activation. Magn. Reson. Med. 1992;25:390–397. doi: 10.1002/mrm.1910250220. [DOI] [PubMed] [Google Scholar]
- Bandettini PA, Jesmanowicz A, Wong EC, Hyde JS. Processing strategies for time-course data sets in functional MRI of the human brain. Magn. Reson. Med. 1993;30:161–173. doi: 10.1002/mrm.1910300204. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, Dogonowski AM, Ernst M, Fair D, Hampson M, Hoptman MJ, Hyde JS, Kiviniemi VJ, Kotter R, Li SJ, Lin CP, Lowe MJ, Mackay C, Madden DJ, Madsen KH, Margulies DS, Mayberg HS, McMahon K, Monk CS, Mostofsky SH, Nagel BJ, Pekar JJ, Peltier SJ, Petersen SE, Riedl V, Rombouts SA, Rypma B, Schlaggar BL, Schmidt S, Seidler RD, Siegle GJ, Sorg C, Teng GJ, Veijola J, Villringer A, Walter M, Wang L, Weng XC, Whitfield-Gabrieli S, Williamson P, Windischberger C, Zang YF, Zhang HY, Castellanos FX, Milham MP. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW, Jesmanowicz A, Hyde JS. Real-time functional magnetic resonance imaging. Magn. Reson. Med. 1995;33:230–236. doi: 10.1002/mrm.1910330213. [DOI] [PubMed] [Google Scholar]
- Cox RW. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Computers and Biomedical Research. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Cox RW, Jesmanowicz A. Real-time 3D image registration for functional MRI. Magn. Reson. Med. 1999;42:1014–1018. doi: 10.1002/(sici)1522-2594(199912)42:6<1014::aid-mrm4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Cox RW, Ashburner J, Breman H, Fissell K, Haselgrove C, Holmes CJ, Lancaster JL, Rex DE, Smith SM, Woodward JB, Strother SC. A (sort of) new image data format standard: NiFTI-1,”. 10th Annual Meeting of the Organization for Human Brain Mapping (OHBM 2004); Budapest. 2004. http://nifti.nimh.nih.gov/nifti-1/documentation/hbm_nifti_2004.pdf. [Google Scholar]
- Eickhoff SB, Stephan KE, Mohlberg H, Grefkes C, Fink GR, Amunts K, Zilles K. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. NeuroImage. 2005;25:1325–1335. doi: 10.1016/j.neuroimage.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Jo HJ, Saad ZS, Simmons WK, Milbury LA, Cox RW. Mapping sources of correlation in resting state FMRI, with artifact detection and removal. NeuroImage. 2010;52:571–582. doi: 10.1016/j.neuroimage.2010.04.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaConte S, Strother S, Cherkassky V, Anderson J, Hu X. Support vector machines for temporal classification of block design FMRI data. NeuroImage. 2005;26:317–329. doi: 10.1016/j.neuroimage.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, Rainey L, Kochunov PV, Nickerson D, Mikiten SA, Fox PT. Automated Talairach atlas labels for functional brain mapping. Hum. Brain. Map. 2000;10:120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad ZS, Glen DR, Chen G, Beauchamp MS, Desai R, Cox RW. A new method for improving functional-to-structural MRI alignment using local Pearson correlation. NeuroImage. 2009;44:839–848. doi: 10.1016/j.neuroimage.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad ZS, Reynolds RC. SUMA. NeuroImage. 2012 doi: 10.1016/j.neuroimage.2011.09.016. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. In: Co-planar stereotaxic atlas of the human brain: 3-dimensional proportional system: an approach to cerebral imaging. Rayport M, translator. New York: Thieme; 1988. [Google Scholar]
- Vovk A, Cox RW, Stare J, Suput D, Saad ZS. Segmentation priors from local image properties: Without using bias field correction, location-based templates, or registration. NeuroImage. 2011;55:142–152. doi: 10.1016/j.neuroimage.2010.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]


