SUMMARY
Identifying the genetic program that leads to formation of functionally and morphologically distinct muscle fibers is one of the major challenges in developmental biology. In Drosophila, the Myocyte Enhancer Factor-2 (MEF2) transcription factor is important for all types of embryonic muscle differentiation. In this study we investigated the role of MEF2 at different stages of adult skeletal muscle formation, where a diverse group of specialized muscles arises. Through stage- and tissue- specific expression of Mef2 RNAi constructs, we demonstrate that MEF2 is critical at the early stages of adult myoblast fusion: mutant myoblasts are attracted normally to their founder cell targets, but are unable to fuse to form myotubes. Interestingly, ablation of Mef2 expression at later stages of development showed MEF2 to be more dispensable for structural gene expression: after myoblast fusion, Mef2 knockdown did not interrupt expression of major structural gene transcripts, and myofibrils were formed. However, the MEF2-depleted fibers showed impaired integrity and a lack of fibrillar organization. When Mef2 RNAi was induced in muscles following eclosion, we found no adverse effects of attenuating Mef2 function. We conclude that in the context of adult myogenesis, MEF2 remains an essential factor, participating in control of myoblast fusion, and myofibrillogenesis in developing myotubes. However, MEF2 does not show a major requirement in the maintenance of muscle structural gene expression. Our findings point to the importance of a diversity of regulatory factors that are required for the formation and function of the distinct muscle fibers found in animals.
Keywords: MEF2, Drosophila, adult myogenesis
INTRODUCTION
Studying myogenesis in model organisms provides insights into the genetic causes of human muscular diseases, as divergent species are thought to utilize similar strategies in muscle development and maintenance. Drosophila melanogaster has long been used as a tool in dissecting genetic and molecular mechanisms of muscle development.
Development of somatic muscles in Drosophila occurs at two stages of the life cycle. First, during the embryonic phase, all muscle types arise from the mesoderm via an intense burst of cell specification and tissue-type differentiation. Fusion of numerous fusion-competent myoblasts to individual founder cells creates multi-nucleate myofibers that elongate and adhere to designated cuticular attachment sites (Baylies and Michelson, 2001; Beckett and Baylies, 2006; Dohrmann et al., 1990). Following fusion, nascent myofibers activate the expression of muscle structural genes, and the larval somatic muscles activate a relatively uniform set of these genes: the muscles invariably express the embryonic muscle actin gene, Act57B (Kelly et al., 2002); as well as the troponin C gene TpnC73F (with other troponin C gene products being non-detectable by hybridization in situ (Herranz et al., 2004)), and other muscle structural genes (Arredondo et al., 2001; Gasch et al., 1988; Zhang and Bernstein, 2001). Hence, at the end of embryonic myogenesis, somatic muscles appear as arrays of individual myofibers, arranged in a largely consistent pattern in each body segment, and sharing a relatively uniform expression of muscle structural genes.
There is compelling evidence that the MADS domain transcription factor Myocyte enhancer factor-2 (MEF2) plays an essential role at the embryonic stage of muscle development. Although specification of muscle precursors proceeds normally in a Mef2 mutant background, these mutants show a profound lack of multinucleate myotubes (Bour et al., 1995; Lilly et al., 1995; Paululat et al., 1999; Ranganayakulu et al., 1995). Consistent with this observation, many structural muscle genes have functional MEF2-binding sites in their enhancers (Kelly et al., 2002; Lin et al., 1996; Sandmann et al., 2006; Tanaka et al., 2008). Drosophila MEF2 is a transcriptional activator, capable of initiating expression of these target genes autonomously, even in foreign environments such as the embryonic ectoderm or S2 cells in tissue culture (Lin et al., 1997a; Tanaka et al., 2008).
At the second phase of Drosophila myogenesis, that occurs during pupal development, pre-existing larval muscles become histolyzed and adult muscles develop de novo. Adult myofibers arise from adult muscle precursor myoblasts, that have been preserved throughout the larval stage as small clusters of cells associated with the nerves and imaginal discs (Dutta et al., 2004; Rivlin et al., 2000). During metamorphosis, adult muscle precursors proliferate and migrate towards fusion sites (Currie and Bate, 1991; Fernandes et al., 1991; Roy and VijayRaghavan, 1997), where myoblast fusion is initiated by sparse founder cells that play essentially the same roles as in embryogenesis (Dutta et al., 2004; Rivlin et al., 2000). In one specific case, being the formation of the adult dorsal longitudinal indirect flight muscles, the role of founder cells is taken by a subset of persistent larval myofibers (Fernandes et al., 1991). Following fusion, newly-developed myofibers enter the hypertrophic phase of growth, where the muscle volume increases due to massive expression of structural genes and assembly of the contractile apparatus.
While superficially the process of adult skeletal myogenesis appears analogous to muscle development in the embryo, muscle formation in adults results in significantly more diverse groups of myofibers, due to the appearance of new specialized muscles. Specialized somatic muscles in adult flies include the indirect flight muscles (IFMs), the tergal depressor of the trochanter (TDT, or jump muscle), direct flight muscles at the base of the wing, as well as head and leg muscles (Bernstein et al., 1993). Muscles in adults that resemble the relatively uniform larval muscle type are restricted to the abdominal body wall (Baker et al., 2005; Currie and Bate, 1991). Clearly, adult myogenesis in Drosophila shows both similarities and differences with embryonic muscle development: specification of founder cells, myoblast fusion, and muscle differentiation are common processes; on the other hand, adult muscles are highly divergent from embryonic muscles in size, arrangement, ultrastructure, and physiology (reviewed in (Bernstein et al., 1993)).
The broad spectrum of specialized adult muscles correlates with the distinct and specific functions that they perform. For example, the most prominent thoracic muscles, the TDTs and IFMs, generate power for jumping and flying, respectively, and their different behavioral functions are correlated with these muscles having different morphological characteristics (see (Peckham et al., 1990)). Based upon their ultrastructure, the IFMs belong to the fibrillar type of muscles, whereas the TDTs are of a tubular type, reflecting the architecture and organization of their myofibrils. These muscles are different at the molecular level, too: IFMs express the muscle-specific actin gene Act88F, and the troponin C gene TpnC41F; while TDTs express Act79B and TpnC41C (Fyrberg et al., 1983; Herranz et al., 2004). The Act57B and TpnC73F genes, expressed in all embryonic and larval skeletal muscles, show strong restriction in expression at the adult stage, to the abdominal body wall muscles (Baker et al., 2005; Herranz et al., 2004). Nevertheless, some muscle-specific genes retain their persistent expression in all adult muscles, including the Myosin heavy chain (Mhc) gene (Hess et al., 2007). Altogether, the understanding of how transcriptional regulation is controlled in somatic muscles during the transition from larval to adult musculature - allowing activation of some new genes and shutting down other ones, while keeping some genes active all along - has become a major research question.
The role of MEF2 in adult myogenesis remains obscure. Our laboratory previously assessed the role of MEF2 in adult myogenesis using Mef2 temperature-sensitive alleles (Baker et al., 2005). The results of our study revealed that Mef2 down-regulation caused relatively mild defects in adult muscle formation, in remarkable contrast to the severe muscle defects observed in embryos under similar experimental conditions. The weak adult phenotype was also demonstrated in earlier studies that employed different Mef2 hypomorphic mutants (Nguyen et al., 2002; Ranganayakulu et al., 1995). Thus, it was suggested that the requirement for MEF2 function in adult myogenesis is somewhat reduced. However, it was still clear that both the hypomorphic and temperature-sensitive mutants retained some small MEF2 function, and we specifically raised the possibility that low levels of residual MEF2 function were still sufficient to support myogenesis. It remained to be determined how a more exacerbated effect upon MEF2 activity might impact adult myogenesis.
In this study, we have taken an RNAi approach to knockdown Mef2 function in the developing adult muscles, at various steps of adult myogenesis. We find that silencing of Mef2 in adult myoblasts leads to a massive loss of somatic muscles, due to inability of the myoblasts to fuse, to activate expression of muscle genes, and to generate myofibers. By contrast, silencing of Mef2 in post-fusion myofibers has a less dramatic effects on fibrillogenesis and expression of structural muscle genes. Finally, silencing of Mef2 in post-eclosion muscles does not produce a detectable deleterious effect on muscular structure and performance during adult life. We conclude that, in adult muscles, MEF2 remains a critical factor for myoblast fusion and initiation of muscle structural gene expression. However, MEF2 becomes less essential for the maintenance of structural gene expression, and its role is taken by other factors. Our study provides new data for understanding the developmental program necessary for formation of divergent, specialized groups of muscles such as those that are found in higher animals.
MATERIALS AND METHODS
Molecular cloning
The cloning of the Mef2 inverted repeat (IR) construct was done following the strategy described before (Bao and Cagan, 2006). An approximately 700-bp region of the Mef2 coding sequence, present in all annotated Mef2 transcripts (Fig. 1A), was amplified using the forward 5′-ACTCTAGACCACCATTTGTCCATTAAGCA and the reverse 5′-GTTCTAGACTGGAGTGGGTGTGATGTGG primers. The underlined regions in the primer sequences are non-genomic sequence, added to achieve cleavage of the PCR product with XbaI. Amplicons digested with XbaI and thereby containing the CTAG overhangs at each end, were cloned into the vector pGEM-WIZ (obtained from the Drosophila Genomic Resource Center (DGRC), Indiana, USA) via the end-compatible AvrII restriction site. The orientation of the insert was verified by analytical restriction digest. In the second round of cloning, the same XbaI-treated amplicons were inserted into the pGEM-WIZ already containing one sense-oriented amplicon, via the XbaI restriction site, and the resulting clones were screened for those bearing tail-to-tail amplicon insertions, separated by a short intron provided by the vector. Selected clones were verified by direct sequencing. Next, the whole construct, containing the two amplicons separated by the intron, was cut out of the pGEM-WIZ with SpeI and inserted into pUAST (Brand and Perrimon, 1993) via compatible ends produced by digesting pUAST with XbaI. The final construct, pUAST-C-Mef2-IR(2), or IR2, was verified by sequencing, and contained the amplified Mef2 part oriented in the sense direction, followed by the intron, and its own antisense-oriented copy.
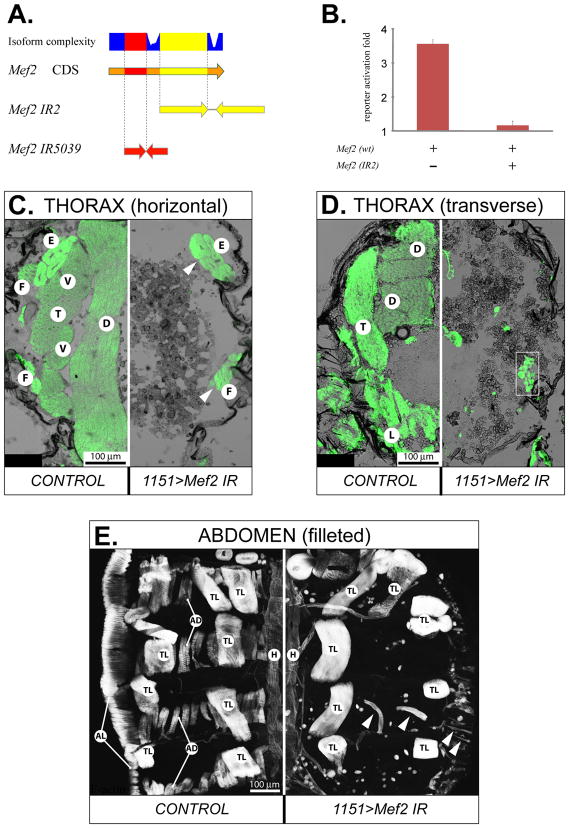
Fig. 1. Structural and functional properties of the inverted repeat (IR) RNAi constructs.
A: Mapping the sequences used in two different IR constructs to the coding DNA sequence (CDS) of Mef2. Isoform complexity depicts the usage of nucleotide sequence in Mef2 annotated transcripts, with the highest plotted value corresponding to usage in all transcripts. B: Mef2 silencing ability of the Mef2 IR2 construct assayed in cell culture co-transfection assays. Reporter activation was determined as fold β-galactosidase accumulation, over the basal β-galactosidase expression level when the activating Mef2-expressing plasmid was omitted. Expression of β-galactosidase from the reporter construct was controlled by a MEF2-resposive enhancer/promoter of the Act57B gene. C, D, E: Comparative analysis of gross muscle morphology between control (on the left of each panel) and RNAi (on the right of each panel) pharate unhatched adults at 90 h APF on indicated planes of thorax sections (C, D) and abdominal fillets (E), stained for F-actin. The RNAi was activated by the 1151-Gal4 driver in adult myoblast precursors. The UAS-Mef2 IR constructs were: IR2 (C) and IR5039 (D, E). Arrowheads point to escaper muscles that did form; white box indicates unusual mini-muscles. E – extracoxal depressor of the trochanter, F – direct flight muscles, T – TDT, D – DLM, V – DVM, L – leg muscles, TL – dorsal internal oblique muscles, which are temporary muscles of larval origin, AD – abdominal dorsal muscles, AL – abdominal lateral muscles, H – cardiac muscles of the dorsal vessel.
Fragments of Act79B and Act88F upstream sequences were amplified by PCR and cloned into the pCaSpeR-hs-AUG-βGal (CHAB) vector (Thummel and Pirrotta, 1992). For generation of Gal4 drivers, the enhancer sequences recapitulating the entire expression patterns of Act88F and Act79B genes were first amplified via PCR. The primers used in these reactions were forward 5′-GAAGAGCATTGGCACCAA and reverse 5′-TGACAATAGGCTCTCCGTTT to amplify the Act79B enhancer; and forward 5′-TTGCACTGATAAATGGTCGG and reverse 5′-CGGACCTTAGAAGGACCGA to amplify the Act88F enhancer. Both amplicons were then cloned into the vector pChsGal4 (Apitz, 2002). The orientation and integrity of the inserts were verified by direct sequencing of the final clones.
Cell culture co-transfection assay
In order to test the silencing potency of the IR2 construct, S2 cells were co-transfected with: pPacPl-Gal4, to activate expression from the IR2 construct; pUAST-C-Mef2-IR(2), the Mef2 silencing construct; pPacPl-Mef2(wt) (Tanaka et al 2008), to express MEF2; and the reporter pC9-CHAB, bearing the MEF2-responsive enhancer of Act57B fused to LacZ (Kelly et al., 2002). The base-line expression control contained these constructs, except pPacPl-Mef2(wt); and the positive expression control excluded pUAST-C-Mef2-IR(2). Excluded plasmids in control transfections were replaced by the empty cell culture expression vector pPacPl, to maintain total transfecting pDNA at the same amount. The efficacy of Mef2 silencing was evaluated by measuring β-galactosidase activity in experimental and control wells. Details on cell culture conditions, transfection, and β-galactosidase assays were published previously (Tanaka et al., 2008).
Fly stocks and crosses
All experimental crosses were carried out at 25° C unless otherwise indicated. Transgenic lines for this study were obtained by P-element-mediated transformation (Rubin and Spradling, 1982). Several independent transgenic lines were created and tested for specific transgene expression. Transformation with pUAST-C-Mef-IR(2) resulted in transgenic lines, identified as 40-1 and 15-2, in which the IR2 transgene locations were mapped to the III and X chromosomes, respectively. Both of the IR lines demonstrated similar effects in Mef2 RNAi-mediated knock-down, based on immunological staining for MEF2. In addition, an independent line expressing an alternative Gal4-inducible Mef2 IR construct 5039 (Fig. 1A) was obtained from the Vienna Drosophila RNAi Center (VDRC, Vienna, Austria, http://stockcenter.vdrc.at). This line, designated as 15550, has the Mef2 IR 5039 transgene mapped to the III chromosome. The lines 40-1 and 15550 were additionally combined with the UAS-Dcr2 (II) line (generously provided by Dr Sarah Certel, University of Montana, MT, USA) for enhancement of the RNAi knockdown (Dietzl et al., 2007).
Lines Act79B-Gal4 (79B6) and Act88F-Gal4 (88F2) were obtained from recombinant pChsGal4 plasmids, and the transgenes for each driver have been mapped to the X chromosome. These lines were selected based on the results of a test cross with UAS-LacZ (II) (obtained from the Bloomington Drosophila Stock Center, Bloomington, IN, USA), to confirm the specific and robust β-galactosidase expression in the TDT and IFM for lines that were designated 79B6 and 88F2, respectively.
The fly line 1151-Gal4(X) was a gift from Dr Joyce Fernandes (Miami University of Ohio, OH, USA), and was described earlier (Anant et al., 1998; Roy and VijayRaghavan, 1997). A modification of this line, 1151-Gal4; UAS-mCD8::GFP UAS-H2B::YFP; UAS-mCD8::GFP, that was used for fluorescent tracing of myoblast membranes, was kindly provided by Dr. Alexis Lalouette, Institut Jacques Monod, CNRS-Université Paris Diderot, Paris, France. The Act88F IR strain was obtained from the VDRC (ID#9780). All other fly stocks were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN, USA).
Identification of putative RNAi off-targets
The Mef2 sequence used to generate the IR2 construct was submitted to the on-line tool at the website of the Drosophila RNAi screening center (http://www.flyrnai.org/RNAi_find_frag_free.html), which calculated four putative off-target genes. They are: CG15207, CG9815, CG15186 (for all, molecular function and mutant phenotype are not known), and cwo that is a transcriptional repressor, involved in circadian rhythm regulation. Following the same analysis, the alternative Mef2 IR5039 that was obtained from the VDRC was found to potentially have one off-target gene: Rab-RP4, a GTPase, experimentally shown to be involved in regulation of cell shape. Since the two IR constructs did not have overlapping off-targets, we therefore decided to use them both in experimental crosses, but to consider only common phenotypes in RNAi-induced mutants.
Histology and staining procedures
For paraffin sections, animals removed from the pupal case at indicated stages of development were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS), at 4°C overnight. Sectioning and staining was performed essentially as described earlier (Cripps et al., 1998; Jaramillo et al., 2009). At each indicatedtime-point, at least four specimens (control or RNAi-induced) were analyzed.
Filleting of abdomens of newly eclosed adults was performed as described (Molina and Cripps, 2001). Formaldehyde-fixed abdomen preparations were stained with Alexa488-labeled phalloidin (Molecular Probes) overnight at ambient temperature. Additionally, some abdomens were counterstained with anti-MEF2 antibody, as described below.
For cryosections, adults or pupae that had been removed from pupal cases at indicated developmental times, were mounted in Tissue-Tek freezing media (VWR Scientific Products) and snap-frozen in liquid nitrogen. Sections were cut at approximately 10 μm thickness, using a Minotome Plus cryotome (Triangle Biomedical Sciences, NC, USA). Sections, mounted onto histological slides (VWR Scientific Products), were fixed for 10 min in PBS containing 0.1% (v/v) Triton X-100 (PBSTx), containing 3.7 % formaldehyde, and additionally permeabilized in PBS containing 0.5% (v/v) Triton X-100 for 30 min. Slides with sections were stained overnight at 4°C with rabbit anti-MEF2 serum (a kind gift of Dr Bruce Paterson, NIH), diluted in PBSTx containing 1% (w/v) BSA as a blocking reagent. Antigen-bound primary antibody was visualized with Alexa568-labeled, affinity-purified, goat anti-rabbit Fab immunoglobulin fragments (Molecular Probes). Sections were also counterstained with Alexa488-labeled phalloidin (Molecular Probes), to visualize F-actin, and with the nuclear stain DAPI (Sigma). Analysis of fluorescently labeled sections was performed on a laser confocal microscope, Zeiss 510META in the University of New Mexico & Cancer Center Fluorescence Microscopy Shared Resource. During image acquisition the fluorescent signals were assigned pseudocolors, for better visualization of experimental phenotypes. Microscopic analysis was carried out for at least ten specimens per genotype, per indicated time-point.
Expression of β-galactosidase was detected in situ by histochemistry. For that, cryosections of thoraces were briefly fixed with 3.7 % (v/v) formaldehyde prepared in PBS, washed thoroughly with several changes of PBS, and incubated in a standard X-gal staining solution for 20–30 minutes at ambient temperature. The reaction was terminated by washing slides with sections in PBS.
Expression analyses
Samples for quantitative expression analysis were collected directly from freshly prepared frozen sections of thoraces. For that, 16-μm sections were briefly dried at ambient temperature and scrapes from DLMs and DVMs were collected with a fresh 22 G syringe needle, and deposited directly into the RLT buffer provided by the RNeasy mini-spin kit (Qiagen). The process of scraping was tightly controlled under a dissection microscope; a special precaution was made not to collect material from the TDT and other non-IFM tissues. Each comparative pair of similarly staged samples contained material collected from one control (Act88F-Gal4 × w1118) and one RNAi-induced pupa (Act88F-Gal4 xUAS-Dcr2; UAS-Mef2 IR15550). Total RNA from each sample was extracted using the RNeasy mini-spin kit (Qiagen), following the manufacturer’s protocol. Subsequently, cDNA synthesis was performed from 55–80 ng/sample of freshly isolated RNA with the use of random hexamer primers (Roche) and the SuperScript II kit (Invitrogen). Diluted cDNA was used as template for real time qPCR amplification. The primers used to detect experimental Act88F and Mhc, as well as reference 18S and 28S targets are listed in Table 1. The amplification efficiencies for these primers were 80–85%, as determined experimentally. Amplification reactions were set up in triplicates using SYBR Green PCR Master kit (Applied Biosystems) and run in an ABI Prism 7000 machine (Applied Biosystems); background and threshold cycles (Ct) were determined in the automated mode by the ABI Prism 7000 SDS Software (Applied Biosystems). For each sample, qPCR data were calculated from an average difference between Ct of gene of interest and Ct of the two reference ribosomal RNA genes, 18S and 28S (Schmittgen and Livak, 2008). ΔCt values pooled from 3–4 independent biological repeats of similarly staged control and RNAi samples were subjected to a paired, two-tail Student’s t-test. Expression between control and RNAi samples was considered statistically non-significant for p-value > 0.05.
Table 1.
Primer sequences used in this study for analytical PCR analysis.
| TARGET | FORWARD PRIMER | REVERSE PRIMER |
|---|---|---|
| Act88F | 5′-CTTCATGGCCATTTCATCG | 5′-TACTCGACATGGAGCACAGC |
| Mhc | 5′-TTGATGACCACTCTGCGTTC | 5′-TTCAAGCACACCGTTACAGG |
| 28S | 5′-CAGGTTGAAGTCAGGGGAAA | 5′-ATGGTTCGATTGGTCTTTCG |
| 18S | 5′-TTCATGCTTGGGATTGTGAA | 5′-GGGACGTAATCAATGCGAGT |
| sing | 5′-TTCGGCAGTCACTATTCGAG | 5′-GAACTGCGGATGTAGCCATAC |
| lmd | 5′-CAGACAACTGCCGGAAATG | 5′-TCTCTGGTGATTTGCTGTCG |
| RpL30 | 5′-TGAAGTCCAGCACTACAGCG | 5′-ATGATGTCCGAATCTCCAGG |
| blow | 5′-GAGGCAAGCATCTCCAGATAAC | 5′-TGGTCGACTGTGTCGATAGC |
| mbc | 5′-CATCGACATGATTGGCTGTC | 5′-CGCACATGTTCAAAGACCTG |
| Act5C | 5′-AAGGATCGCTTGTCTGGG | 5′-GTATATCATATCATATCTCATGTGG |
In the analysis of transcripts recovered from fusing myoblasts, real time PCR was not practical due to the low RNA yield in dissected samples, and the subsequent low abundance of the target templates. However, significant differences in the expression levels for some genes between control and RNAi samples allowed them to be visualized by the means of end-point PCR. For that, samples of the DLM fusion templates and surrounding myoblasts were collected from serial frozen sections of four control (w1118) and Mef2-silenced (UAS-Dcr2; UAS-Mef2 IR15550 × 1151-Gal4; Twi-LacZ) male pupae, staged at 25° C for 24 h APF. Samples were scraped from the appropriate region of a cryosection using a pulled glass capillary needle and a hydraulic manipulator, visualized under an inverted microscope. RNA was purified from these samples as described above. The primers used to amplify selected targets from the extracted DLM template/myoblast RNA are summarized in Table 1. All PCRs were run simultaneously, under identical conditions. The resulting products were loaded equally and resolved in a 2% agarose gel for image analysis.
RESULTS
Generation and verification of a Mef2 RNAi line
To circumvent the absolute requirement of MEF2 for embryonic myogenesis, we applied an inducible tissue-specific RNAi technique to knock down Mef2 expression at later developmental stages. We created an inverted repeat construct, termed IR2, that produced double-stranded RNA corresponding to an approximately 700-bp region of the Mef2 coding sequence, to initiate RNAi against native Mef2 transcripts (Fig. 1A). This construct was first tested in Drosophila cultured S2 cells: when co-transfected with a Mef2 expression plasmid and a MEF2-dependent reporter, the dsRNA expression from IR2 completely abrogated reporter gene activation, which demonstrated strong silencing potency provided by IR2 to its target (Fig. 1B). This finding enabled us to use an RNAi approach to attenuate Mef2 expression, using this construct in transgenic animals. For comparison and confirmation, we also obtained a Mef2 RNAi transgenic line from the Vienna Drosophila RNAi Center (Dietzl et al., 2007). The Mef2 IR construct 5039 (IR5039), expressed by this line, was designed to target a different portion of the Mef2 coding region (Fig. 1A, and the VDRC web resource).
Silencing of Mef2 in adult myoblast progenitors results in the loss of adult somatic muscles
Experimental induction of Mef2 RNAi was carried out in transgenic flies with the use of the Gal4/UAS binary inducible expression system (Brand and Perrimon, 1993). First, we evaluated the progeny of the cross between Mef2 IR2-carrying flies (UAS-Mef2 IR2) and the adult myoblast-specific driver line 1151-Gal4 (Anant et al., 1998). Since both of the transgenes in the parental lines were located on the X chromosome, only female progeny were expected to have RNAi activation and Mef2 silencing in developing adult myoblasts. Therefore, male offspring served in this cross as the control. Consistently, no adult female progeny were recovered from this cross (UAS-Mef2 IR2 × 1151-Gal4, Table 2). In contrast, the abundance of female progeny from a control cross was normal (w1118 × 1151-Gal4, Table 2). Preliminary examination showed that 1151>Mef2 IR2 females developed normally, but remained in their pupal cases and eventually died, unable to eclose. Subsequent histological analyses revealed that these animals lacked essentially all major somatic muscles in their bodies, including IFMs, TDTs, leg muscles and abdominal body wall muscles (Fig. 1C-E). The only muscles remaining in the thorax of the RNAi flies were a few escaper muscles (Fig. 1C) and some miniature muscles (Fig. 1D, boxed region). The thorax cavity in 1151>Mef2 IR2 pharate adults contained no obvious structures or tissues in place of the muscles. In the abdomen of these flies, virtually all of the transverse and longitudinal muscles were absent (Fig. 1E). There were also a few escaper muscles observed. In addition, other muscle types, including the temporary oblique dorsal muscles of larval origin, were unaffected. These muscles were positive for MEF2 expression (not shown). Cardiac and visceral musculature in the RNAi animals remained intact and continued expressing Mef2 (Fig. 1E and data not shown), presumably due to the inactivity of the 1151-Gal4 driver in these muscles.
Table 2.
The viability of Mef2 RNAi and control genotypes.
| Cross (♀ × ♂) | ♂♂ # eclosed |
♀♀ # eclosed |
♀♀ eclosed, % |
|---|---|---|---|
| [control] w1118 × 1151-GAL4 (X) | 112 | 124 | 53% |
|
|
|||
| UAS-Mef2 IR2 (X) × 1151-GAL4 (X) | 204 | 0 | 0% |
|
|
|||
| UAS-Mef2 IR5039 (III) × 1151-GAL4 (X) | 170 | 0 | 0% |
To exclude the possibility that the observed phenotype might arise from silencing of possible “off-target” genes by the RNAi ((Ma et al., 2006), see Materials and Methods), we also tested another RNAi transgene, Mef2 IR5039 (Dietzl et al., 2007). When UAS-Mef2 IR5039 females were crossed with males of the 1151-Gal4 line, their female progeny essentially recapitulated all of the described phenotypes observed for crosses with UAS-Mef2 IR2 flies (Table 2 and data not shown). Thus, these studies have validated two Mef2 RNAi constructs as having similar effects. More importantly, our data expand upon our earlier studies (Baker et al., 2005) by showing a more important requirement for MEF2 function in the formation of the adult skeletal muscles in Drosophila.
Silencing of Mef2 at early stages of adult myogenesis impairs myoblast fusion and muscle structural protein gene expression
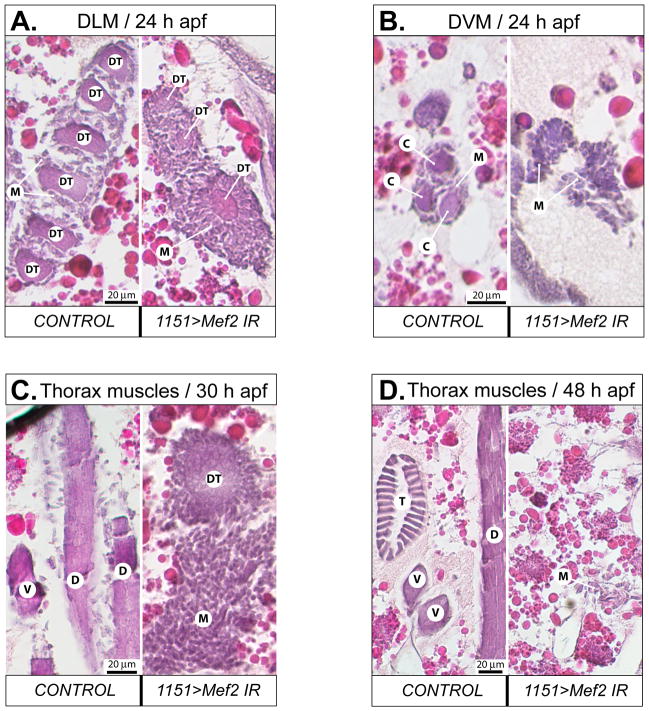
To investigate what problems in muscle development arise upon Mef2 silencing in adult myoblasts, samples from a series of developmentaltime-points were subjected to comparative analysis between control and RNAi pupae, in which Mef2 silencing was induced by the 1151-Gal4 driver. This driver is active in adult myoblasts at the larval stage and extending into the pupal stage (Anant et al., 1998; Roy and VijayRaghavan, 1997), thus bracketing the earliest expression of Mef2 in all developing adult muscles. During this study we focused on the developing IFMs: these muscles are the largest in the fly body, and therefore easily identified at all stages of development. Paraffin sections of thoraces were obtained for times corresponding to the active phase (24 h after puparium formation (APF)) and the finishing phase (30 h APF) of hyperplastic muscle growth (i.e. by myoblast fusion), as well as the phase of hypertrophic (i.e. by size increase) muscle growth (48 h APF). All sections were stained with hematoxylin and eosin (H&E) to reveal general tissue morphology.
In developing pupae, adult muscles begin to form via myoblast fusion. Among the IFMs, the dorsal longitudinal muscles (DLMs) have pre-established fusion templates, that are formed on the basis of the larval oblique muscles (LOMs). By contrast, the dorso-ventral muscles (DVMs) are formed de novo, fusion being initiated by individual founder cells (Atreya and Fernandes, 2008; Fernandes et al., 1991). In each hemisegment, the original three LOMs undergo splitting to give rise to six DLM templates. On transverse sections of 24 h APF control pupae, all six developing DLM fibers could be identified, surrounded by numerous swarming myoblasts participating in fusion (Fig. 2A, left panel). In similarly staged Mef2-knockdown pupae, template splitting did not proceed, and the number of DLM templates remained at three. These templates were smaller than their control counterparts, whereas the layer of swarming myoblasts surrounding them was significantly thicker than in control (Fig. 2A, right panel). For the DVMs, each wild-type DVM fiber was detected at 24 h APF as a central core element, being the product of myoblast fusion to founder cells, and surrounded by swarming myoblasts (Fig. 2B, left panel). Identification of nascent DVMs in Mef2- knockdown pupae was problematic due to the lack of myoblast fusion products. Groups of myoblasts could be found at the putative sites of DVM formation, but they never were accompanied by a fusion core (Fig. 2B, right panel).
Fig. 2. Induction of RNAi in adult muscle precursors severely affects myogenesis by preventing myoblasts from fusing into myofiber syncytia.
A: Transverse, hematoxylin/eosin stained section of the thorax of control and RNAi pupae at 24 h APF, showing DLM templates (DT) surrounded with swarming myoblasts (M). Note that the number of DLM templates in RNAi-induced animal is not doubled, and remains at three. B: Transversely cut developing DVM fibers on horizontal sections of the thorax of control and RNAi pupae at 24 h APF. Control has a myofiber core for each fiber (C), which is absent in the RNAi animal. C, D: horizontal sections of the thorax of control and RNAi pupae at 30 h (C) and 48 h (D) APF, respectively. In control animals, diverse types of muscles have been formed, while in RNAi animals there are only myoblasts which become sparse at 48 h APF. The RNAi was induced with the 1151-Gal4 driver activating UAS-Mef2 IR5039. DT – DLM template, M – myoblasts, C – fusion core, D – DLM, V – DVM, T – TDT.
At 30 h APF, the fusion of IFMs is largely complete in wild-type Drosophila, and these muscles become morphologically distinct. At this time-point, freshly formed myofibers were apparent in control animals as intensely stained structures, associating with sparse myoblasts (Fig. 2C, left panel). Fused myofibers were not found in Mef2-knockdown pupae, but instead the presence of large numbers of unfused myoblasts was apparent (Fig. 2C, right panel). These myoblasts concentrated into irregularly shaped swarms at the dorsal side of the thorax, and on transverse sections the myoblast swarms had wide and long multicellular extensions protruding ventrally (not shown). Anchorless, rounded-up DLM fusion templates were often found localizing sideways to the myoblast swarms, and were surrounded by somewhat less myoblasts than seen around them at 24 h APF (Fig. 2C, right panel).
At 48 h APF, control IFM myofibers demonstrated adult-like appearance in their shape, fiber number, and position (Fig. 2D, left panel). In contrast, RNAi pupae at 48 h APF continued to show a lack of fused myotubes. Moreover, the number of myoblasts in these pupae decreased significantly, and they could be detected only as small groups of loosely aggregated cells (Fig. 2D, right panel). We hypothesize that by this stage most of the unfused myoblasts have undergone cell death, and that the residual thoracic mini-muscles that we see in adult RNAi mutants (Fig. 1D, frame) arise from the myoblasts remaining at 48 h APF.
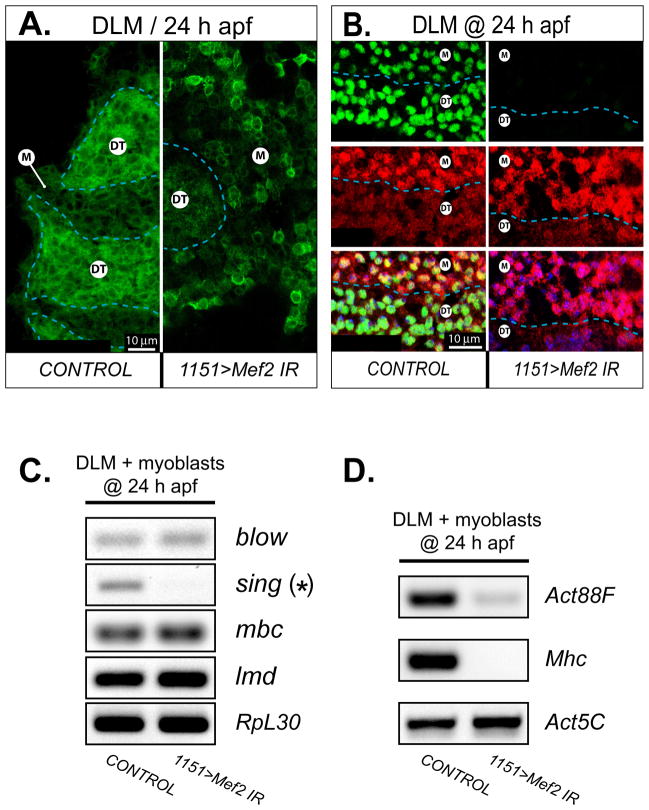
We additionally traced myoblasts with a membrane-associated GFP marker, mCD8-GFP, that was expressed under the control of the 1151-Gal4 driver. At 24 h APF, wild-type myoblasts concentrating around the DLM templates appeared as a compact mass of cells with their membranes in close contact with each other (Fig. 3A, left panel). Induction of Mef2 RNAi by the 1151-Gal4 driver resulted in a significant decrease of the MEF2 protein level in DLM fusion templates and free swarming myoblasts, as determined by immunofluorescence at 24 h APF (Fig. 3B). Mef2-knockdown myoblasts were not closely compacted around their fusion templates. Rather, these myoblasts localized loosely, with noticeable space between individual cells (Fig. 3A, right panel). Both H&E and fluorescent preparations strongly suggested that Mef2-silenced myoblasts could not complete cell fusion to form multinucleate myotubes. To test whether the knock-down of Mef2 impairs expression of genes essential for myoblast fusion, we carried out end-point RT-PCR analysis on RNA samples obtained from myoblasts at 24 h APF (the method used to collect pure muscle/myoblast samples is detailed in Materials and Methods). Our molecular data indicated that expression of at least one fusion-critical gene, singles bar (sing), was clearly down-regulated in Mef2- knockdown myoblasts (Fig. 3C). The sing gene encodes a membrane protein that is critical for myoblast fusion during embryonic myogenesis, being expressed in both fusion-competent myoblasts and founder cells (Estrada et al., 2007). Therefore, the loss of sing expression in adult myoblasts can certainly account for the fusion failure that we have documented. In contrast to sing, expression of other well-known fusion genes, blow and mbc, were unaltered in Mef2-knockdown myoblasts (Fig. 3C). Our data provide a mechanistic explanation for the failure of fusion in the Mef2-knockdown myoblasts, although it is interesting to note that MEF2 does not control expression of the whole repertoire of fusion genes.
Fig. 3. Mef2 knockdown in adult myoblasts compromises myoblast behavior at fusion centers and prevents expression of fusion and muscle structural genes.
A: Transverse sections across developing DLM fibers at 24 h APF are shown for control and RNAi (1151>Mef2 IR5039) conditions. The signal is recorded from membrane-associated mCD8:GFP protein driven by the same 1151-Gal4 driver as used for RNAi induction. Individual myoblasts make tight formations around the outlined DLM fusion templates (DT) in the control animal, but only loosely aggregate around DT in the RNAi animal. Note, that mCD8 also labels the nuclear envelopes within the fusion templates. B: A boundary area between a DT and unfused myoblasts (M) at 24 h APF, demonstrating a substantial decrease in MEF2 immunofluorescence (green) in both myoblasts and fusion templates, upon induction of Mef2 RNAi with the 1151-Gal4 driver. Myoblasts are traced with the twi-LacZ transgene whose expression is detected by immunofluorescent staining for β-galactosidase (red). The twi-lacZ activity is subdued in DT, identifying its border with unfused myoblasts (dashed line). The overlaid images at the bottom of the panel are additionally counter-stained for nuclei (blue). C: Expression analysis of several fusion-critical genes in samples obtained at the sites of developing DLMs at 24 h APF, from control and RNAi (1151>Mef2 IR5039) animals. Fusion genes are indicated with red font. The asterisk marks the sing gene, that shows a significantly reduced expression in response to Mef2 knockdown. The transcription factor lmd is used as a genetic marker of myoblasts; and the ribosomal protein gene, RpL30, is used as a general marker for RT-PCR loading control. D: Activation of muscle structural genes using RT-PCR and detecting expression of the pan-muscular Mhc, and the IFM-specific Act88F structural genes in the same samples as in C. The housekeeping actin, Act5C, is used as loading control.
We also investigated how the initiation of muscle structural gene expression was affected in Mef2-knockdown myoblasts and fusion templates at 24 h APF, using identical sampling technique, as described above. We chose Act88F and Mhc as representatives of two distinct classes of adult muscle genes, whose expression is either restricted to only a subset of adult muscles (Act88F) or present in all adult muscles (Mhc). In our analysis, the control sample demonstrated robust presence of Act88F and Mhc transcripts, while the RNAi sample failed to demonstrate any significant expression of both of the genes (Fig. 3D). We conclude that in nascent IFMs, as exemplified by DLM templates at 24 h APF, Mef2 knockdown with the 1151-Gal4 driver largely abolished the initiation of expression for both Act88F and Mhc.
These findings expand upon our earlier studies (Baker et al., 2005), by demonstrating a more crucial role for MEF2 function in the formation of the adult muscles, where Mef2-knockdown myoblasts fail to fuse to form myotubes and mature muscles, and this occurs at least in part through a requirement of MEF2 for the expression of the fusion gene, sing. Interestingly, myoblast migration was not suppressed by Mef2 knockdown, as judged by the ability of Mef2-knockdown myoblasts to migrate away from developing imaginal disks and to accumulate in the thorax near their muscle templates. We further demonstrate that MEF2 function in adults is critical for the initiation of structural gene expression.
Generation of Gal4 drivers active in post-fusion adult muscles
Immunofluorescent analysis of the very few mini-muscles that formed in the 1151>Mef2 IR pharate adults (Fig. 1D, right panel) revealed that the myofibers contained severely reduced amounts of MEF2 (not shown). However, these mini-muscles could complete their differentiation, as confirmed by the presence of detectable F-actin. Hence, it was possible that MEF2 could be less strictly required for steps in muscle development following myoblast fusion.
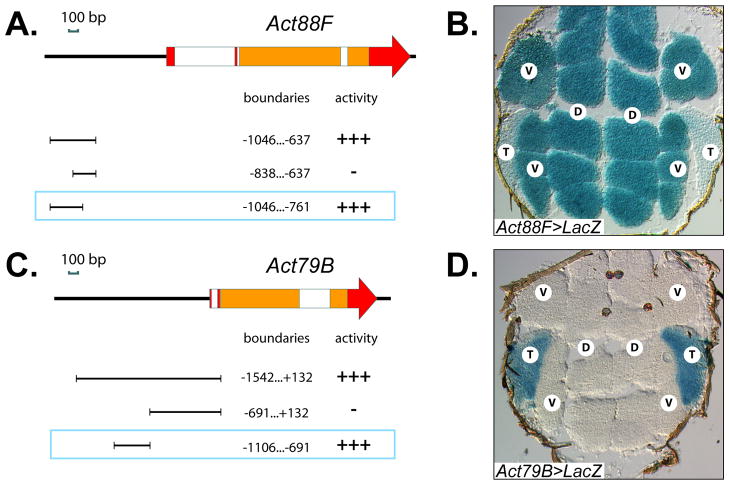
To test this possibility, Mef2 had to be silenced later in adult myogenesis, so that its requirement for myoblast fusion could be circumvented. Thus, a Gal4 driver should initiate expression of Mef2 IR constructs in myotubes immediately after fusion. Assuming that enhancers for adult muscle-specific structural genes would be good candidates for this task, we generated two such drivers, by identifying and cloning the enhancer regions for the IFM-specific Act88F gene, and for the TDT-specific Act79B gene (Fig. 4A,C). For Act88F, a potent enhancer for expression in IFMs, ~300-bp in length, lies approximately 1kb upstream of the transcription start site; and for Act79B, a ~400-bp genomic region with TDT-specific enhancer activity, lies in a similar position relative to the transcription start site of its gene. When fused to Gal4, proper stage- and tissue-specific activities of the cloned enhancers were confirmed by crossing transformant lines with UAS-LacZ flies, and detecting β-galactosidase activity in sections. As expected, the Act88F driver was active specifically in IFMs and not the TDTs (Fig. 4B), while the Act79B driver was active in a complimentary pattern, in TDTs but not IFMs (Fig. 4D). Importantly, neither driver was active in unfused myoblasts (data not shown). In a separate experiment, we subjected Gal4-expressing flies to microscopic analysis and verified that expression of these Gal4 transgenes alone did not cause muscle structural defects (not shown).
Fig. 4. Characterization of the enhancer regions used in post-fusion-activated Gal4 drivers.
A, C: The structure of Act88F (A) and Act79B (C) genes. Red and orange rectangles indicate non-coding and coding exons, respectively; white boxes represent introns. The cloned fragments used for enhancer activity analysis, their precise boundaries related to the transcriptional initiation site, and in vivo activities are indicated below each gene scheme. All drawings are to scale and oriented to the respective gene schemes. The selected enhancers, used in Gal4 drivers, are framed. B, D: Expression of Act88F-Gal4 (B) and Act79B-Gal4 (D) drivers, as revealed by activation of a UAS-LacZ construct. β-galactosidase enzymatic activity was detected in situ on horizontal thorax sections using X-gal. T – TDT, D – DLM, V – DVM.
Silencing of Mef2 in indirect flight muscles perturbs myofibril organization, but does not affect maintenance of two major structural genes
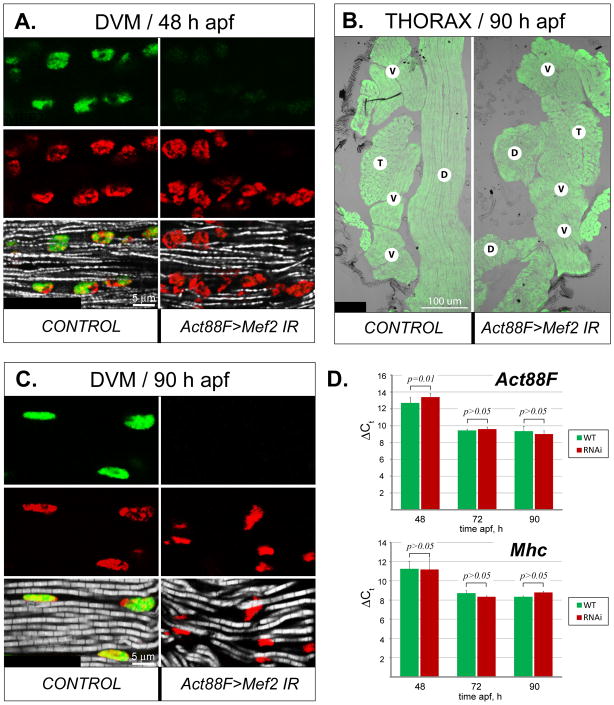
We first analyzed the effects of Mef2 silencing in the post-fusion IFMs, using the Act88F-Gal4 driver. In this cross, IFM myofibrillogenesis following post-fusion Mef2-knockdown did not stall, and myofibrils were observed developing at 48 h APF (Fig. 5A). By the end of the pupal stage, Mef2 knockdown induced by the Act88F driver had a specific impact on the integrity of a subset of IFMs: in most cases, Mef2-knockdown DLMs often collapsed to the back of the thorax (Fig. 5B). Nevertheless, RNAi DVMs developed normal-looking, myofibrils by the end of the pupal stage, although they were somewhat wavy in their organization (Fig. 5C).
Fig. 5. Morphological and molecular effects of Mef2 silencing in post-fused IFM fibers.
A: Expression of MEF2 (green) in the nuclei (red), as well as nascent myofibrils (gray), in longitudinally sectioned DVM fibers shortly after completion of myoblast fusion in control and Act88F-Gal4-induced RNAi (Act88F>Mef2 IR5039) pupae at 48 h APF. B: General muscle localization in the thorax of control and RNAi pharate adults at 90 h APF, visualized by phalloidin staining (green). The DLM fibers (D) in RNAi animals (Act88F>Mef2 IR5039) are collapsed. The positions of other major thoracic muscles, DVMs (V) and TDT (T), remain unchanged. C: Longitudinally sectioned DVMs, stained for MEF2 (green), nuclei (red), and myofibrils (gray) in control and RNAi pharate adults (Act88F>Mef2 IR5039) at 90 h APF. Lack of MEF2 results in a wavy arrangement of myofibrils, although their structure is similar to control. D: Comparative analysis of expression of Act88F (upper panel) and Mhc (lower panel) between control (green bars, WT) and Act88F>Mef2 IR5039 (red bars, RNAi) samples, obtained from IFMs (combined DVM and DLM fibers) at 48 h, 72 h, and 90 h APF. Realtime qRT-PCR data are shown as the lag in detection (ΔCt) between transcripts of interest, and 18S and 28S rRNA, used as internal references (details are in Material and Methods) ± s.d. Student’s t-test p value > 0.05 indicates statistically non-significant differences. Note that the expression of Act88F lags slightly in the RNAi samples at the 48 h APF time-point, but not at any other stages. Mhc transcript levels are not significantly different between control and knockdown samples.
To assess how Mef2 knockdown can influence expression of myofiber-specific and pan-muscular-specific structural genes, we examined by quantitative RT-PCR the levels of expression of Act88F and Mhc at different developmental time-points in wild-type and Act88F-directed Mef2 knockdown pupae. We found that, at three time-points tested, the levels of Act88F and Mhc transcripts did not show any statistically significant changes in knockdown versus control muscle fibers (Fig. 5D). The only exception to this result was for Act88F transcripts at 48 h APF, where transcripts in knockdown muscles were significantly reduced compared to controls. This effect upon Act88F transcripts was not observed at later pupal time-points.
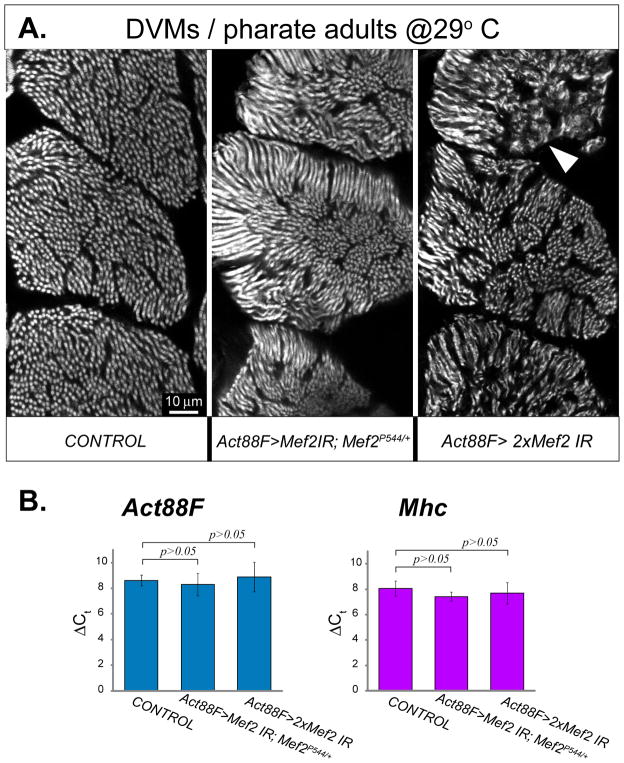
To determine if the observed minimal effect upon structural gene expression and myofibril assembly in the Act88F>Mef2 IR cross was due to residual MEF2 activity in the knockdown muscles, we carried out two additional experiments: we performed the RNAi experiment in a Mef2P544/+ genetic background, generating haploinsufficiency for the endogenous Mef2 gene on top of the knockdown; and we combined the two RNAi lines to maximize the effects of the RNAi. In addition, to maximize the transcriptional activity of the GAL4 driver, and thereby to further increase the effectiveness of RNAi knockdown, pupae were incubated at 29°C. Interestingly, there was worsening of the myofibrillar phenotype in these “enhanced” knockdown experiments with DVM myofibrils appearing more disoriented, slightly thinner and with less distinct Z-lines and M-lines (Fig. 6A). Nevertheless, we also saw no reduction in Act88F nor Mhc transcription levels at the end of pupal development in any of the enhanced crosses (Fig. 6B).
Fig. 6. Enhanced knockdown of Mef2 in post-fused DVMs worsens myofibril morphology, but does not affect expression of two major structural genes.
A: Phalloidin-stained transverse sections through posterior DVM fibers, from pharate adults in which Mef2 RNAi was either not induced (CONTROL), induced with a single IR5039 construct in an heterozygous Mef2P544/+ null allele background (Act88F>Mef2 IR; Mef2P544/+), or induced with both IR2 and IR5039 constructs (Act88F>2xMef2 IR). RNAi was initiated and maintained via the IFM-specific Act88F-Gal4 driver. To reach maximum RNAi effectiveness, pupae were allowed to develop at 29°C. Note that under these exacerbated conditions, myofibril structure is significantly perturbed, showing misaligned, thinned myofibrils, occasionally lacking any structure (arrowhead). The phenotype conferred by double IRs was most severe. B: Expression analysis of Act88F and Mhc genes in combined samples of DLM and DVM fibers collected from the pharate adults described above. The qRT-PCR data, shown as detection delay (ΔCt) from internal reference rRNA transcripts, do not show statistically significant differences between all samples (t-test p value > 0.05), indicating that expression of both Act88F and Mhc does not readily respond to Mef2 knockdown, even under enhanced conditions.
From these observations, we make two important conclusions: firstly, since haploinsufficiency for Mef2 enhanced the mutant phenotype, this suggests that there is yet some residual MEF2 activity in the knockdown muscles. On the other hand, since the levels of two key myofibrillar protein genes was not attenuated in the haploinsufficient background nor in the double-RNAi background, we conclude that the much of adult myofibrillogenesis proceeds independently of MEF2. This will be discussed at greater length in the Discussion.
Silencing of Mef2 in post-fusion TDT myotubes does not prevent fibrillogenesis, but results in myofibrillar disorganization
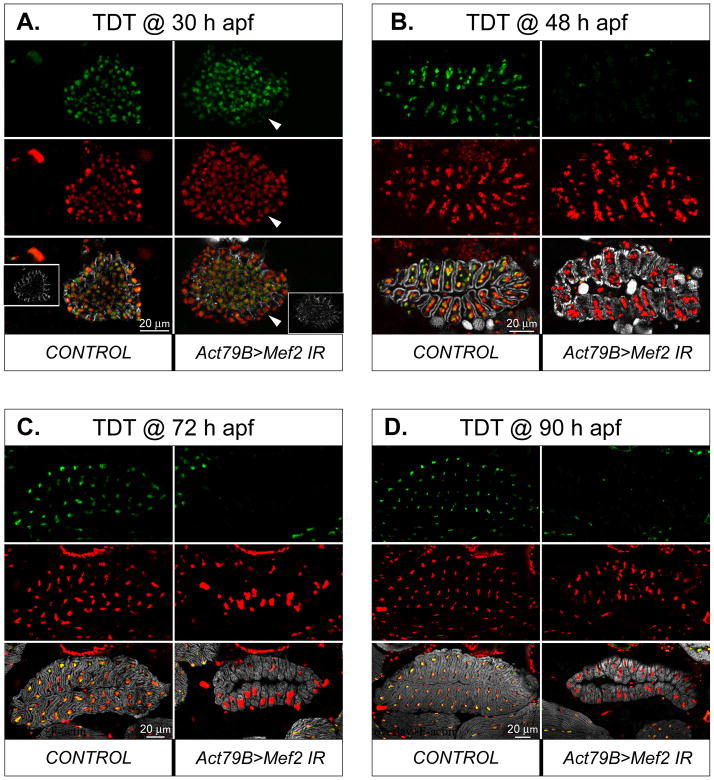
We next analyzed the effects of Mef2 reduction in the post-fusion TDT. To understand the kinetics of Mef2 silencing in this muscle, we analyzed a time series of samples collected at 30 h APF (the active phase of TDT myoblast fusion in wild-type), 48 h APF (when TDT myoblast fusion is complete), 72 h APF (an intermediate point of hyperplastic muscular growth), and past 90 h APF (the end of pupal myogenesis). The pupae in which Mef2 RNAi was activated by the Act79B-Gal4 driver were compared with similarly staged control animals in which the driver was not present. All samples were collected as cryosections transverse to the TDT; these sections were then analyzed by confocal microscopy. Moreover, both RNAi lines were used for this analysis, and we observed identical phenotypes in each experimental case.
In the TDT, myofibers form via fusion that occurs at the outer periphery of the myoblast swarm (Jaramillo et al., 2009; Rivlin et al., 2000). Therefore, at 30 h APF, the newly forming TDTs had a core that contained tightly packed individual myoblasts expressing Mef2, while the peripheral ring was a product of myoblast fusion, in which expression of structural genes was activated (Fig. 7A, left panel, inset). Induction of Mef2 RNAi via the Act79B-gal4 driver resulted at 30 h APF in clearly diminished immunofluorescence of MEF2 in the fused myotubes at the periphery, but not in the centrally-located myoblasts (Fig. 7A, right panel, arrowheads).
Fig. 7. Mef2 knockdown in developing TDTs results in moderate, but persistent, changes in muscle morphology.
A-D: Transverse section through developing TDT fibers at 30 h (A), 48 h (B), 72 h (C), and 90 h [pharate adult] (D) APF in control and (1151>Mef2 IR5039) RNAi pupae. Samples are immunofluorescently stained for MEF2 (green), DNA (red), and F-actin (gray). The Mef2 silencing was induced with the Act79B-Gal4 driver and was evident by reduced MEF2 immunofluorescence, readily noticeable in the nuclei of fused fibers at 24 h APF (arrowhead). Insets in panel A show the appearance of developing muscle fibers at the periphery of the myoblast swarm, that contain increasing amount of polymerized actin; the MEF2 positive nuclei in the center belong to unfused myoblasts. Note that, at later stages, MEF2 immunofluorescence is either severely reduced (B) or not detected (C, D) in TDT, but stay at high levels in unaffected neighboring muscles. Anterior is to the left.
At 48 h APF, when the fusion process was complete, control TDT muscles adopted their typical oval cross-sectional shape, comprising 24–28 individual myofibers linked together (Fig. 7B; (O’Donnell et al., 1989; Peckham et al., 1990)). By this time-point, MEF2 immunofluorescence in RNAi TDTs was very close to the background level, whereas MEF2 immunofluorescence in the adjacent IFMs was normal.
At 72 h and 90 h APF, control TDT myofibers demonstrated expanded amounts of F-actin that filled out most of the myofiber volume, tightly surrounding the nuclei (Fig. 7C-D, left panels). At these time-points MEF2 protein could not be immunologically detected in Mef2-silenced TDTs. Nevertheless, such RNAi-induced TDT myofibers contained densely packed myofibrils (Fig. 7C-D, right panels), indicating that accumulation of muscle structural genes was not stalled in the absence of MEF2. The other persistent features of Mef2- knockdown TDTs were: the presence of a central cavity within the center of TDT myofiber rosette, that is usually minimized in mature wild-type TDT; wider gaps between individual TDT myofibers; mis-localized and aggregated nuclei; and reduced overall size of the muscle (Fig. 7C-D, right panels). By counting the numbers of TDT fibers in RNAi samples at 48, 72, and 96 h APF time points, the reduction in TDT size was not due to a loss of fibers, and therefore arose from reduced size of the individual fibers.
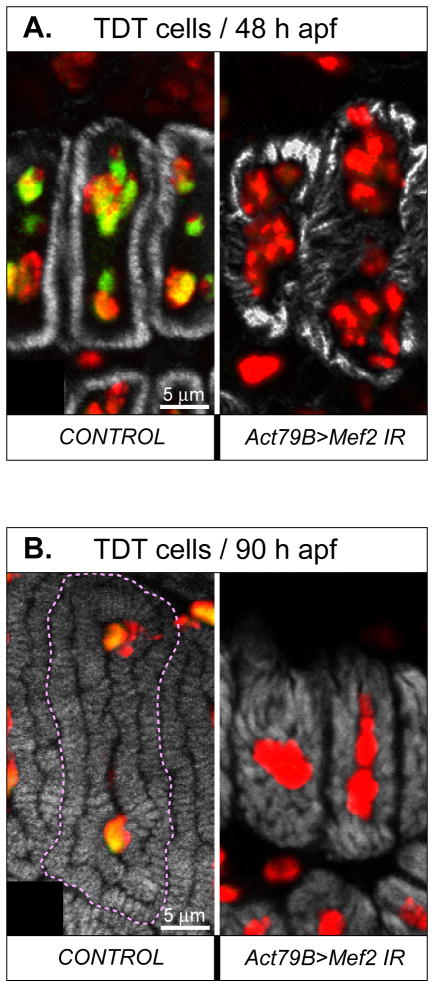
We also studied fibrillar organization within individual myofibers, in control and Mef2- knockdown TDT muscles. Each mature TDT fiber contains a double-ring of tightly-packed parallel myofibrils surrounding centrally-located nuclei (see Fig. 7D, left panel). At higher magnification of pupal samples, it was apparent that the outer ring of myofibrils was laid down first during development (Fig. 8A, left panel), and by the pharate adult stage individual myofibrils could be discerned in cross section as small oblongs, numbering 150–200 per TDT large muscle fiber (Fig. 8B, left panel (O’Donnell et al., 1989; Peckham et al., 1990)). In Mef2- knockdown TDT at 48 h APF, there was also evidence of F-actin accumulation, although the intensity of staining was highly irregular across individual myofibers (Fig. 8A, right panel). By 90 h APF, the F-actin accumulation in RNAi TDT myofibers was even more significant (Fig. 8B, right panel). However, at bothtime-points in RNAi TDT samples, it was difficult to discern individual myofibrils in cross-section, arguing that the myofibrils were not arranged parallel to the axis of the muscle, but were irregular in their organization.
Fig. 8. Mef2 knockdown in the TDT does not prevent myofibrillogenesis, but affects organization of myofibril arrays and general myofiber morphology.
A, B: Transversely cut individual TDT muscle fibers at 48 h (A) and 90 h (B) APF, in control and RNAi (1151>Mef2 IR5039) pupae. The superimposed images of three fluorescent channels, corresponding to MEF2 (green), DNA (red), and F-acting (grey) are shown. The yellow color indicates an overlap between the green and the red signals. Note that nuclei of RNAi fibers contain only the red signal and are significantly disorganized. Fibrillar organization is disturbed in RNAi fibers at both time points, but the sizes of individual myofibrils have increased between 48 h and 90 h APF.
In summary, depletion of MEF2 from TDT muscles shortly after fusion did not prevent muscle structural gene expression. Nevertheless, similar to the case for the IFMs, MEF2 function was required in developing TDTs for proper arrangement of the muscle fibers, and correct arrangement of myofibrils within muscle fibers.
Altogether, our combined morphological and molecular data indicate that knockdown of Mef2 in post-fused adult muscles does not abolish myofibrillogenesis, which is in contrast to the extensive requirement for MEF2 in myoblast fusion and in the initiation of muscle structural gene expression during early (24 h APF) myogenesis. However, the myofibril disorganization and DLM collapse that are associated with post-fusion Mef2 knockdown in IFMs and TDTs, indicate that some aspects of muscle development remain sensitive to the presence of MEF2.
Silencing of Mef2 in established adult muscles does not produce a deteriorative effect
Protein synthesis significantly slows down in adult flies (Clarke and Smith, 1966; Smith et al., 1970), although Mef2 expression in muscles continues throughout their whole life. We therefore sought to define a role for MEF2 in adult animals, a question that had yet to be addressed in detail.
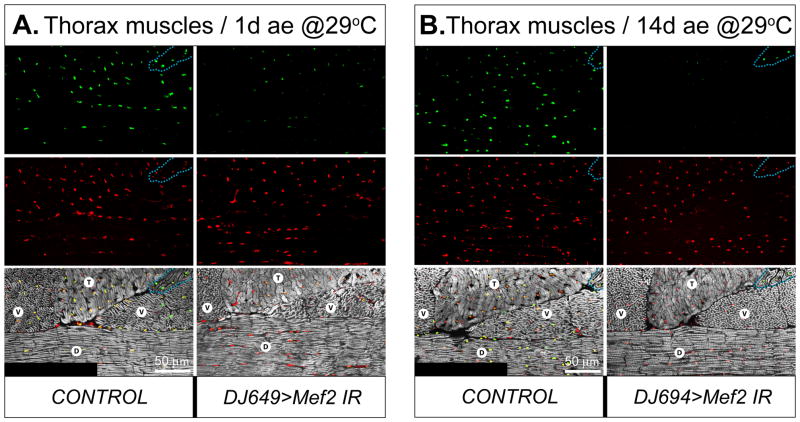
For induction of Mef2 RNAi in fully grown adult muscles, the DJ694-Gal4 driver was employed, which shows a dramatic induction of Gal4 expression immediately following eclosion. To ensure effective and quick removal of the MEF2 protein from the muscles of freshly eclosed adults, all staging times were conducted at 29°C, which enhances the effectiveness of the Gal4 system (Michelson, 1994). The DJ694-Gal4 driver becomes initiated in adults, especially in IFMs, upon eclosion. This driver remains active during the whole adult life span (Seroude et al., 2002). Indeed, the progeny of the cross between DJ694-Gal4 flies and Mef2 IR lines developed normally at 29°C and showed intact, Mef2-expressing thoracic muscles within the first day after eclosion (ae) (Fig. 9A). However, within the first two days ae MEF2 protein levels in IFMs decreased dramatically, and by the third day ae MEF2 could not be detected by means of immunofluorescence in the nuclei of IFMs, while the TDT had significantly reduced levels of MEF2 immunofluorescence. In contrast to IFMs and TDTs, MEF2 levels remained unchanged in direct flight muscles (see Fig. 9B, for example), and other types of adult muscles (not shown).
Fig. 9. Mef2 knockdown in post-developed IFMs does not compromise muscle morphology.
A, B: Major thoracic muscles of control and RNAi (DJ694>Mef2 IR2) males at 1 day ae (ae) (A), or 14 days ae (B), stained for MEF2 (red), DNA (green), and F-actin (grey). The flies were raised at 29°C. Note that MEF2 immunofluorescence in DLMs (D) and DVMs (V) is reduced at 1day ae, and not detected at 14 days ae in RNAi flies, while staying unchanged in control and RNAi non-affected direct flight muscles (outlined with dashed line). MEF2 immunofluorescence in the TDT (T) of RNAi flies is also reduced, but to a lesser extent than in DVMs and DLMs. Anterior is to the left.
Despite the efficient knockdown of Mef2 in IFMs shortly after eclosion, the RNAi-induced flies did not show any gross aberrations in their locomotion until day 6 ae. At that time, the flies in which the RNAi was mediated by the Mef2 IR5039 construct demonstrated a significant decrease in responses to external stimuli and then rapidly died within the following day. In contrast, the flies in which the DJ694-induced RNAi was mediated by the Mef2 IR2 construct, targeting a different region in Mef2 (Fig. 1A), did not show enhanced death for more than 3 weeks of monitored observation at 29°C. Mef2 silencing directed by the IR2 construct was complete. The knockdown also effectively targeted the IFMs and, to a lesser degree, the TDT (Fig. 9B). Since both of the RNAi constructs, IR5039 and IR2, can potentially mediate RNAi to a few off-target genes, and since their off-targets are not overlapping (see Materials and Methods), we reason that the effects on the life span rendered by the IR5039 is not specific to MEF2 depletion, and that the phenotypes arising from expression of the IR2 construct represents the most relevant effect of Mef2 knockdown.
The loss of MEF2 protein in IFMs of the DJ694>Mef2 IR2 flies was confirmed at day 3 ae (not shown) and persisted until at least day 14 ae (Fig. 9B). MEF2-depleted IFMs did not bear detectable morphological abnormalities in myofibril structure (Fig. 9B, right panels). Concomitantly, the flies with silenced Mef2 at this age did not show any significant reduction in their flight ability (not shown).
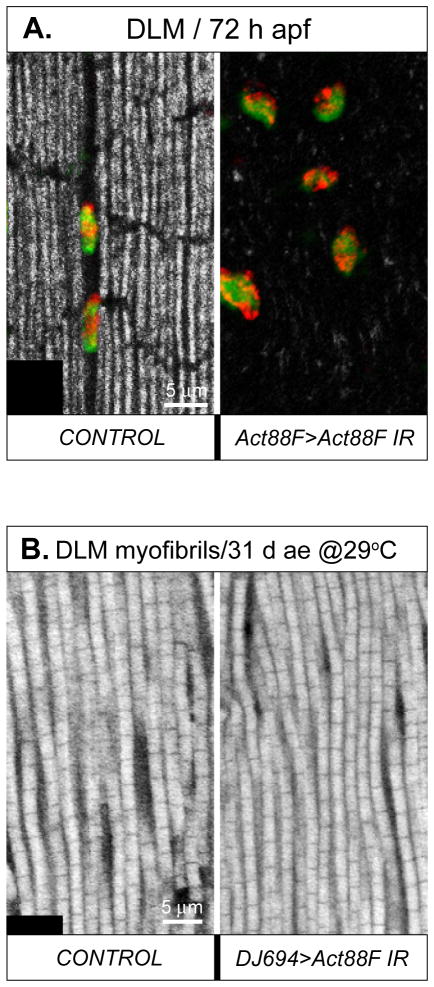
Since the turnover of structural muscle proteins in adult flies was reported to stay at very low levels (Clarke and Smith, 1966; Smith et al., 1970), we decided to test the effect of direct silencing of one of the major IFM structural components, Act88F. First, the effectiveness of the Act88F IR construct mediating RNAi was tested in post-fused IFMs, using the Act88F-Gal4 driver. In RNAi pupae at 72 h APF, IFMs did not show mature myofibrils. This result confirmed that Act88F depletion was functioning efficiently, and justified the silencing potency of the Act88F IR construct (Fig. 10A). Next, we conducted Act88F RNAi in adult flies using the DJ694 driver essentially in the same way as in the Mef2 silencing experiments. Male flies, aged with activated Act88F RNAi up to 31 day at 29°C, did not show compromised flight ability nor structural defects to IFM myofibrils (Fig. 10B).
Fig. 10. RNAi knockdown of the actin gene Act88F in post-developed muscles does not alter myofibril morphology.
A: Overlaid images of an enlarged area of the DLM in control and RNAi pupae at 72 h APF, stained for MEF2 (green), DNA (red), and F-actin (grey); overlaid red and green signals produce yellow. The RNAi was induced by Act88F-Gal4 driving the Act88F IR construct. In RNAi-induced DLM myofibrils are missing, demonstrating the effectiveness of the Act88F IR construct. B: Enlarged area of the DLM stained for F-actin in control and RNAi flies staged for 31 days at 29°C. The adult-specific RNAi was induced by the DJ694-Gal4 driver and the same Act88F IR construct. Unlike the situation in the developing muscles, post-developed muscles in adults do not produce a fibrillar phenotype in response to actin silencing, even after a prolonged exposure to RNAi targeting Act88F.
We conclude that, in mature adult muscles, MEF2 is dispensable for the maintenance of normal muscle ultrastructure. It seems likely that this result is due to the very low turnover of structural proteins in adult flies, as demonstrated by our Act88F experiment and in agreement with classical studies. We nevertheless acknowledge that MEF2 may play a role in other muscles not impacted by the DJ694 driver, and it may play a roles in muscles that is not assessed by our existing methods.
To summarize our findings, we demonstrate that there is an essential early function for MEF2 in promoting myoblast fusion and the onset of muscle structural gene expression in the developing adult muscles. By contrast, depletion of MEF2 from nascent muscle fibers later in development resulted in morphological defects in the muscles, but did not completely block myogenesis. Depletion of MEF2 from skeletal muscles after eclosion of the adults did not result in significant nor severe phenotypes. These data were collected using a series of stage- and tissue-specific Gal4 drivers, and two different RNAi lines, targeting different portions of the Mef2 transcript. Similar phenotypic effects were seen for each of the IR constructs when expressed during the pupal stage. These findings, and the similarity in effects of the two RNAi lines, are documented in Table 3.
Table 3.
Summary of phenotypes obtained with Mef2 IR constructs.
| Mef2 IR construct | Observed phenotype with 1151 driver | |||
|---|---|---|---|---|
| Stalled myoblast fusion | Absent Muscles | Escaping mini- muscles | Stalled activation of Act88F and Mhc | |
| IR2 | Yes | Yes | Yes | Not Tested |
|
| ||||
| IR5039 | Yes | Yes | Yes | Yes |
|
| ||||
| Observed phenotype with Act79B driver | ||||
| Reduced TDT size | Disorganized myofibrils | Mis-shapen fibers | Perturbed nuclear localization | |
|
| ||||
| IR2 | Yes | Yes | Yes | Yes |
|
| ||||
| IR5039 | Yes | Yes | Yes | Yes |
|
| ||||
| Observed phenotype with Act88F driver | ||||
| Collapsed DLMs | Disorganized myofibrils | Mis-shapen nuclei | Continued expression of Act88F and Mhc | |
|
| ||||
| IR2 | Yes | Yes | Yes | Not Tested |
|
| ||||
| IR5039 | Yes | Yes | Yes | Yes |
|
| ||||
| IR2+IR5039 | Yes | Yes | Yes | Yes |
| Observed phenotype with DJ694 driver | ||||
| Shortened lifespan | Weak flier | |||
|
| ||||
| IR2 | No | No | ||
|
| ||||
| IR5039 | Yes | Not Tested | ||
DISCUSSION
The phenotypes of Mef2 RNAi versus Mef2 mutants
In this manuscript, we demonstrate an important requirement for MEF2 in the formation of the adult muscles, where early reduction in MEF2 levels blocks myoblast fusion, and obliterates formation of the adult somatic muscles. These findings are more severe than we had previously reported for a Mef2 temperature-sensitive combination (Baker et al., 2005); why should this be the case? The most reasonable explanation for this discrepancy is that the temperature-sensitive mutants, while severely impacting MEF2 function, nevertheless still showed some low level of MEF2 activity and myogenic potential, and that this low level of MEF2 function was sufficient to support adult myogenesis. An additional factor to consider is the period of time that it takes for the first steps of myogenesis to occur, at the adult stage versus the embryonic stage. In embryos, the fusion process is completed within a time window of just a few hours (Bate, 1990); whereas in adult myogenesis, fusion proceeds over the course of at least 12 hours of pupal development (Fernandes et al., 1991). It is feasible then, that low levels of sustained MEF2 function over an extended period of time are sufficient to support the earliest steps of adult muscle development, while such an effect is not possible in embryonic myogenesis, due to its strictly limited timing.
We present data indicating that there is yet some residual MEF2 accumulation in the skeletal muscles of some of the RNAi lines that we have characterized, since haploinsufficiency for Mef2, or use of a double-RNAi, exacerbated the myofibrillar defects in post-fusion Mef2 knockdowns. Nevertheless, these levels must be extremely low, given our immunofluorescent data indicating that indeed levels of MEF2 are reduced to background detection levels from as early as 24 h APF time-points. More importantly, neither haploinsufficiency for Mef2 nor the double-RNAi background reduced the levels of Act88F and Mhc expression. We therefore conclude that there are clear stage-specific requirements for this important transcription factor, where high levels of MEF2 are required for early myogenesis, but later myogenesis appears to proceed with only minimal levels (if any) of MEF2 function required.
Regulation of adult myoblast fusion by MEF2
Fusion defects occur in null Mef2 mutants during embryogenesis (Bour et al., 1995; Lilly et al., 1995), however such a phenotype has not yet been reported for adult myogenesis. Our study, demonstrating a requirement for MEF2 in adult myoblast fusion, therefore underlines a similarity between these two distinct phases of Drosophila muscle development. Our data suggest that the abolition of myoblast fusion is at least partly due to the significant down-regulation of sing transcripts, in samples of purified myoblasts plus myotubes isolated from cryosections. sing expression was also decreased in Mef2 mutant embryos (Sandmann et al., 2006), conferring another level of similarity between the molecular mechanisms regulating myoblast fusion in embryonic and adult myogenesis. Importantly, the expression levels of two other tested fusion genes, blow and mbc, did not show any detectable alterations in expression in response to Mef2 knockdown. We conclude that MEF2 is not a key regulator for multiple fusion genes, although it certainly is essential for fusion via activation of sing transcription. We note that mbc is not exclusively expressed in developing muscles, and is in fact more broadly-distributed (Erickson et al., 1997), reducing the likelihood that its expression solely responds to MEF2 activity. Whether there are more MEF2-dependent fusion genes, and whether MEF2 is a direct transcriptional activator of such genes, is yet to be fully established at either the embryonic or the adult stages.
Initiation of myofibrillar protein gene expression depends upon MEF2
Our findings also demonstrate that the transcriptional initiation of genes encoding adult myofibrillar protein genes depends upon MEF2 being present early in muscle formation. Whereas a sample from control muscle fusion templates plus myoblasts showed robust expression of both Act88F and Mhc at 30 h APF, Mef2-knockdown samples showed sparse activation of these muscle structural genes (Fig 3D). Our findings are consistent with the role of Mef2 in embryonic myogenesis, where the majority of muscle-specific genes fail to become expressed in the absence of MEF2 function (Bour et al., 1995; Lilly et al., 1995). Clearly, a role for MEF2 in the early stages of muscle formation appears to be a commonality between embryonic and adult early myogenesis. Nevertheless, some intermediate factors must also be important in this initiation process, since we have yet to demonstrate that adult actin gene expression is directly responsive to MEF2 (see discussion below).
A limited role for MEF2 in differentiation of post-fused myofibers
In agreement with our earlier data, there is clearly a more moderate requirement for MEF2 function in formation of the adult muscles following myoblast fusion. Despite MEF2 being immunologically undetectable shortly after fusion in RNAi animals, significant myogenesis occurs, and this process results in the formation of muscles in pharate adults. This finding is in contrast to Mef2 knockdown during the early stages of adult myogenesis, where it was absolutely required for activation of fibrillogenesis.
Importantly, adult-specific actin gene expression does not become stalled or aborted in the Mef2 knockdown animals, as we can judge based on three major observations: 1) F-actin clearly accumulates in Mef2 knockdown myofibers; 2) the tissue-specific drivers, derived from adult muscle actin genes, function effectively to suppress MEF2 accumulation until the end of myogenesis, and therefore are not dependent on MEF2 activity; and 3) quantitative RT-PCR analysis did not reveal statistically significant differences in Act88F expression levels between experimental and control samples at late time-points of myogenesis (72 h and 90 h APF), even in a Mef2 haploinsufficient background and in a double-RNAi background. These data provide further support for the notion that MEF2 is at most minimally required for the sustained expression of some adult muscle genes. Moreover, neither the Act88F nor the Act79B enhancers contain canonical MEF2 binding sites (Andres et al., 1995), and neither enhancer is activated by MEF2 in transient co-transfection assays (data not show).
In the light of the demonstrated independence of adult actin gene transcription from Mef2 expression, one must ask how known MEF2-dependent genes, with general muscle expression, respond to Mef2 silencing during adult myogenesis? In this study we used Mhc as an example of such a pan-muscle-specific gene, since in the embryo initiation of Mhc expression depends on Mef2 activity (Bour et al., 1995; Lilly et al., 1995). In addition, the cloned minimal Mhc promoter/enhancer contains three functional MEF2 sites (Hess et al., 2007; Tanaka et al., 2008). Our data, however, indicate that MEF2 depletion does not alter Mhc expression at statistically significant levels, even under exacerbated conditions. We conclude that while MEF2 is required to initiate expression of Mhc, MEF2 is likely to be just one of the factors participating in the maintenance of Mhc expression.
Our results are supported by the studies of others (Gajewski and Schulz, 2010; Hess et al., 2007), in which the authors show that the Mhc gene receives regulatory input from numerous evolutionarily conserved cis-regulatory elements scattered throughout its complex regulatory region, expanding from as far as 10-kb upstream of the transcription start site and into the first intron. Moreover, some of the cloned Mhc enhancer elements, lacking MEF2 sites, directed myofiber-specific reporter gene expression in adult flies. Apparently, maintenance of Mhc expression in adults depends on multiple transcription factors.
Besides Mhc, other pan-specifically expressed muscle genes have been investigated for transcriptional regulatory elements. Tropomyosin-1 and -2, Troponin T, and Paramyosin/mini-Paramyosin also possess complex enhancers comprising multiple regulatory elements that can be activated in a myofibril-specific fashion, often independently of MEF2 sites (Arredondo et al., 2001; Garcia-Zaragoza et al., 2008; Lin and Storti, 1997; Marco-Ferreres et al., 2005). These, and other observations (such as (Marin et al., 2004)), suggest that MEF2, while being important for activation of muscle structural genes, becomes less critical in their maintenance, which concurs with our observations. The latter postulate can explain why enhancers containing generic MEF2 sites, being functionally active in cell culture co-transfection assays, fail to support muscle-specific expression of the LacZ reporter gene in transgenic animals (data not shown); and why a single MEF2 site with flanking sequences, taken out of the context of a functional embryonic Act57B enhancer, is not capable of driving reporter gene expression in embryonic muscles (Kelly et al., 2002).
Nevertheless, MEF2 remains a key regulator of early steps of myogenesis in Drosophila. According to our experimental data, MEF2 functions in early stages of adult myogenesis to support myoblast fusion and we postulate that at this time MEF2 also begins to activate additional muscle-specific regulatory factors that then go on to activate muscle structural genes during the remainder of adult myogenesis.
So far, we are aware of very few factors, other than MEF2, that directly regulate muscle structural genes in Drosophila. Chorion Factor-2 (CF2) is an important MEF2 co-factor for muscle gene expression in the embryo/larva (Tanaka et al., 2008), and it does play a role in adult myogenesis as well (Gajewski and Schulz, 2010; Garcia-Zaragoza et al., 2008). In addition, PAR-domain protein 1 (PDP1), with broad expression in a range of tissues, was shown to specifically bind and activate the enhancers of Tropomyosin-1 and mini-Paramyosin genes (Lin et al., 1997b; Marco-Ferreres et al., 2005) cooperatively with MEF2 (Lin et al., 1996; Lin et al., 1997b). The Cf2 gene is genetically downstream of Mef2 (Bagni et al., 2002), while Pdp1 regulation is complicated due to the presence of multiple alternative promoters, but late accumulation of products of this gene in skeletal muscles during embryogenesis may suggest that it is also MEF2-dependent (Reddy et al., 2000). Thus, MEF2 can be an initiator for expression of additional myogenic regulators, and these factors can later acquire their own positive auto-regulatory loop. In adults, the transcriptional co-activator vestigial, participating in IFM development, is activated by MEF2 in adult-specific myoblasts and then participates in self-activation (Bernard et al., 2009). It is plausible that other transcriptional regulators participating in myofiber-specific myogenesis first receive activation of their expression from MEF2 – to be later maintained by different means.
Further, there is clearly a subset of muscle protein genes that are activated after myoblast fusion and that must nevertheless be significantly affected by loss of MEF2 function. Evidence for this comes from the pathology that does occur in Mef2- knockdown myofibers, which includes loss of proper myofibril organization, detachment of muscle fibers from the attachment sites, and separation of neighboring myofibers. Some of the Mef2- knockdown phenotypes were clearly visible even after 18 hours of Mef2 RNAi initiation (Fig. 6B), indicating that there must be targets that are heavily dependent on MEF2 activity. Identification of such target genes should become a focus of future research efforts.
The role of MEF2 in post-eclosion adult muscles
In the animals in which Mef2 knockdown was induced post eclosion, we do not consider the severe changes in lifespan conferred by the IR 5039 as being a true phenotype of MEF2 loss. Most likely, this phenotype is a reflection of RNAi off-target activity. Thus, post-eclosion, there appears to be relatively little requirement for MEF2, at least over the time period and under the environmental conditions that we have tested. This finding is consistent with the classical observations of Smith et al (1970), who demonstrated that there is extremely low turnover of thoracic proteins following eclosion, and we additionally demonstrate that sustained expression of Act88F is not required following the first day of adult life. We cannot rule out that MEF2 may be participating in other aspects of muscle physiology, besides structural protein turn-over. Hence, MEF2 might be involved in expression of muscle-specific enzymes, controlling muscle metabolism (Deak, 1977). Another intriguing possibility could be a potential participation of MEF2 in muscle repair after an extensive muscle exercise such as long-term flying, or pathology. The sustained presence of MEF2 in mature adult muscles argues strongly for a role in these cells, which additional studies should be designed to uncover.
Drosophila as a model for mammalian myogenesis
Vertebrate slow/fast twitch muscles consist of fibers with different functional, metabolic, and molecular properties (Zierath and Hawley, 2004), just as Drosophila adult muscles are fine-tuned in order to carry out their unique physiological functions. In both cases, it is still unclear how initial fiber types are specified in the nascent muscle. In mammals, there is evidence that fiber-type switching relies on MEF2 working in combination with other transcription factors to selectively activate muscle structural genes (Lin et al., 2002). Given the broad conservation in the basic mechanisms of myogenesis, as exemplified by the role of MEF2 in muscle differentiation from flies to vertebrates, we anticipate that studying adult myogenesis in Drosophila, which had been hindered by the lack of appropriate molecular tools, now can start providing new genetic data on myofiber-specific development.
Research highlights.
Bryanstev et al
Differential requirements for Myocyte Enhancer Factor-2 during adult myogenesis in Drosophila
Drosophila adults lacking MEF2 during muscle development were generated
MEF2 is required for myoblast fusion in adult myogenesis
MEF2 is dispensable for actin gene expression during adult myogenesis
MEF2 shows a partial, but not complete, requirement for adult muscle differentiation following myoblast fusion
Acknowledgments
We thank Drs. Bruce Paterson, Alexis Lalouette, Joyce Fernandes, and Sarah Certel for generously providing reagents. This work was supported by GM061738 from the NIH awarded to RMC, and by a Research Grant from the Muscular Dystrophy Association to RMC. Confocal images in this paper were generated in the University of New Mexico & Cancer Center Fluorescence Microscopy Shared Resource, funded as detailed at: http://hsc.unm.edu/crtc/microscopy/Facility.html. We acknowledge technical support from the Department of Biology’s Molecular Biology Facility, supported by NIH grant number 1P20RR18754 from the Institute Development Award (IDeA) Program of the National Center for Research Resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Anant S, Roy S, VijayRaghavan K. Twist and Notch negatively regulate adult muscle differentiation in Drosophila. Development. 1998;125:1361–9. doi: 10.1242/dev.125.8.1361. [DOI] [PubMed] [Google Scholar]
- Andres V, Cervera M, Mahdavi V. Determination of the consensus binding site for MEF2 expressed in muscle and brain reveals tissue-specific sequence constraints. J Biol Chem. 1995;270:23246–9. doi: 10.1074/jbc.270.40.23246. [DOI] [PubMed] [Google Scholar]
- Apitz H. pChs-Gal4, a vector for the generation of Drosophila Gal4 lines driven by identified enhancer elements. Dros Inf Serv. 2002;85:118–120. [Google Scholar]
- Arredondo JJ, Ferreres RM, Maroto M, Cripps RM, Marco R, Bernstein SI, Cervera M. Control of Drosophila paramyosin/miniparamyosin gene expression. Differential regulatory mechanisms for muscle-specific transcription. J Biol Chem. 2001;276:8278–87. doi: 10.1074/jbc.M009302200. [DOI] [PubMed] [Google Scholar]
- Atreya KB, Fernandes JJ. Founder cells regulate fiber number but not fiber formation during adult myogenesis in Drosophila. Dev Biol. 2008;321:123–40. doi: 10.1016/j.ydbio.2008.06.023. [DOI] [PubMed] [Google Scholar]
- Bagni C, Bray S, Gogos JA, Kafatos FC, Hsu T. The Drosophila zinc finger transcription factor CF2 is a myogenic marker downstream of MEF2 during muscle development. Mech Dev. 2002;117:265–8. doi: 10.1016/s0925-4773(02)00176-4. [DOI] [PubMed] [Google Scholar]
- Baker PW, Tanaka KK, Klitgord N, Cripps RM. Adult myogenesis in Drosophila melanogaster can proceed independently of myocyte enhancer factor-2. Genetics. 2005;170:1747–59. doi: 10.1534/genetics.105.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Cagan R. Fast cloning inverted repeats for RNA interference. RNA. 2006;12:2020–4. doi: 10.1261/rna.258406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Michelson AM. Invertebrate myogenesis: looking back to the future of muscle development. Curr Opin Genet Dev. 2001;11:431–9. doi: 10.1016/s0959-437x(00)00214-8. [DOI] [PubMed] [Google Scholar]
- Beckett K, Baylies MK. The development of the Drosophila larval body wall muscles. Int Rev Neurobiol. 2006;75:55–70. doi: 10.1016/S0074-7742(06)75003-6. [DOI] [PubMed] [Google Scholar]
- Bernard F, Kasherov P, Grenetier S, Dutriaux A, Zider A, Silber J, Lalouette A. Integration of differentiation signals during indirect flight muscle formation by a novel enhancer of Drosophila vestigial gene. Dev Biol. 2009;332:258–72. doi: 10.1016/j.ydbio.2009.05.573. [DOI] [PubMed] [Google Scholar]
- Bernstein SI, O’Donnell PT, Cripps RM. Molecular genetic analysis of muscle development, structure, and function in Drosophila. Int Rev Cytol. 1993;143:63–152. doi: 10.1016/s0074-7696(08)61874-4. [DOI] [PubMed] [Google Scholar]
- Bour BA, O’Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–41. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Clarke JM, Smith JM. Increase in the rate of protein synthesis with age in Drosophila subobscura. Nature. 1966;209:627–9. doi: 10.1038/209627a0. [DOI] [PubMed] [Google Scholar]
- Cripps RM, Black BL, Zhao B, Lien CL, Schulz RA, Olson EN. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev. 1998;12:422–34. doi: 10.1101/gad.12.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie DA, Bate M. The development of adult abdominal muscles in Drosophila: myoblasts express twist and are associated with nerves. Development. 1991;113:91–102. doi: 10.1242/dev.113.1.91. [DOI] [PubMed] [Google Scholar]
- Deak II. A histochemical study of the muscles of Drosophila melanogaster. J Morphol. 1977;153:307–16. doi: 10.1002/jmor.1051530209. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–6. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dohrmann C, Azpiazu N, Frasch M. A new Drosophila homeo box gene is expressed in mesodermal precursor cells of distinct muscles during embryogenesis. Genes Dev. 1990;4:2098–111. doi: 10.1101/gad.4.12a.2098. [DOI] [PubMed] [Google Scholar]
- Dutta D, Anant S, Ruiz-Gomez M, Bate M, VijayRaghavan K. Founder myoblasts and fibre number during adult myogenesis in Drosophila. Development. 2004;131:3761–72. doi: 10.1242/dev.01249. [DOI] [PubMed] [Google Scholar]
- Erickson MR, Galletta BJ, Abmayr SM. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. J Cell Biol. 1997;138:589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Maeland AD, Gisselbrecht SS, Bloor JW, Brown NH, Michelson AM. The MARVEL domain protein, Singles Bar, is required for progression past the pre-fusion complex stage of myoblast fusion. Dev Biol. 2007;307:328–39. doi: 10.1016/j.ydbio.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Bate M, Vijayraghavan K. Development of the indirect flight muscles of Drosophila. Development. 1991;113:67–77. doi: 10.1242/dev.113.1.67. [DOI] [PubMed] [Google Scholar]
- Fyrberg EA, Mahaffey JW, Bond BJ, Davidson N. Transcripts of the six Drosophila actin genes accumulate in a stage- and tissue-specific manner. Cell. 1983;33:115–23. doi: 10.1016/0092-8674(83)90340-9. [DOI] [PubMed] [Google Scholar]
- Gajewski KM, Schulz RA. CF2 represses Actin 88F gene expression and maintains filament balance during indirect flight muscle development in Drosophila. PLoS One. 2010;5:e10713. doi: 10.1371/journal.pone.0010713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Zaragoza E, Mas JA, Vivar J, Arredondo JJ, Cervera M. CF2 activity and enhancer integration are required for proper muscle gene expression in Drosophila. Mech Dev. 2008;125:617–30. doi: 10.1016/j.mod.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Gasch A, Hinz U, Leiss D, Renkawitz-Pohl R. The expression of beta 1 and beta 3 tubulin genes of Drosophila melanogaster is spatially regulated during embryogenesis. Mol Gen Genet. 1988;211:8–16. doi: 10.1007/BF00338387. [DOI] [PubMed] [Google Scholar]
- Herranz R, Diaz-Castillo C, Nguyen TP, Lovato TL, Cripps RM, Marco R. Expression patterns of the whole troponin C gene repertoire during Drosophila development. Gene Expr Patterns. 2004;4:183–90. doi: 10.1016/j.modgep.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Hess NK, Singer PA, Trinh K, Nikkhoy M, Bernstein SI. Transcriptional regulation of the Drosophila melanogaster muscle myosin heavy-chain gene. Gene Expr Patterns. 2007;7:413–22. doi: 10.1016/j.modgep.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo MS, Lovato CV, Baca EM, Cripps RM. Crossveinless and the TGFbeta pathway regulate fiber number in the Drosophila adult jump muscle. Development. 2009;136:1105–13. doi: 10.1242/dev.031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly KK, Meadows SM, Cripps RM. Drosophila MEF2 is a direct regulator of Actin57B transcription in cardiac, skeletal, and visceral muscle lineages. Mech Dev. 2002;110:39–50. doi: 10.1016/s0925-4773(01)00586-x. [DOI] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–93. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Lin MH, Bour BA, Abmayr SM, Storti RV. Ectopic expression of MEF2 in the epidermis induces epidermal expression of muscle genes and abnormal muscle development in Drosophila. Dev Biol. 1997a;182:240–55. doi: 10.1006/dbio.1996.8484. [DOI] [PubMed] [Google Scholar]
- Lin MH, Nguyen HT, Dybala C, Storti RV. Myocyte-specific 2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosin gene muscle expression. Proc Natl Acad Sci U S A. 1996;93 I: 10:4623–8. doi: 10.1073/pnas.93.10.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lin MH, Horvath P, Reddy KL, Storti RV. PDP1, a novel Drosophila PAR domain bZIP transcription factor expressed in developing mesoderm, endoderm and ectoderm, is a transcriptional regulator of somatic muscle genes. Development. 1997b;124:4685–96. doi: 10.1242/dev.124.22.4685. [DOI] [PubMed] [Google Scholar]
- Lin SC, Storti RV. Developmental regulation of the Drosophila Tropomyosin I (TmI) gene is controlled by a muscle activator enhancer region that contains multiple cis-elements and binding sites for multiple proteins. Dev Genet. 1997;20:297–306. doi: 10.1002/(SICI)1520-6408(1997)20:4<297::AID-DVG1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–63. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- Marco-Ferreres R, Vivar J, Arredondo J, Portillo F, Cervera M. Co-operation between enhancers modulates quantitative expression from the Drosophila Paramyosin/miniparamyosin gene in different muscle types. Mech Dev. 2005;122 I: 5:681–94. doi: 10.1016/j.mod.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Marin MC, Rodriguez JR, Ferrus A. Transcription of Drosophila troponin I gene is regulated by two conserved, functionally identical, synergistic elements. Mol Biol Cell. 2004;15:1185–96. doi: 10.1091/mbc.E03-09-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson AM. Muscle pattern diversification in Drosophila is determined by the autonomous function of homeotic genes in the embryonic mesoderm. Development. 1994;120:755–68. doi: 10.1242/dev.120.4.755. [DOI] [PubMed] [Google Scholar]
- Molina MR, Cripps RM. Ostia, the inflow tracts of the Drosophila heart, develop from a genetically distinct subset of cardial cells. Mech Dev. 2001;109:51–9. doi: 10.1016/s0925-4773(01)00509-3. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Wang J, Schulz RA. Mutations within the conserved MADS box of the D-MEF2 muscle differentiation factor result in a loss of DNA binding ability and lethality in Drosophila. Differentiation. 2002;70:438–46. doi: 10.1046/j.1432-0436.2002.700806.x. [DOI] [PubMed] [Google Scholar]
- O’Donnell PT, Collier VL, Mogami K, Bernstein SI. Ultrastructural and molecular analyses of homozygous-viable Drosophila melanogaster muscle mutants indicate there is a complex pattern of myosin heavy-chain isoform distribution. Genes Dev. 1989;3:1233–46. doi: 10.1101/gad.3.8.1233. [DOI] [PubMed] [Google Scholar]
- Paululat A, Holz A, Renkawitz-Pohl R. Essential genes for myoblast fusion in Drosophila embryogenesis. Mech Dev. 1999;83:17–26. doi: 10.1016/s0925-4773(99)00029-5. [DOI] [PubMed] [Google Scholar]
- Peckham M, Molloy JE, Sparrow JC, White DC. Physiological properties of the dorsal longitudinal flight muscle and the tergal depressor of the trochanter muscle of Drosophila melanogaster. J Muscle Res Cell Motil. 1990;11:203–15. doi: 10.1007/BF01843574. [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev Biol. 1995;171:169–81. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Wohlwill A, Dzitoeva S, Lin MH, Holbrook S, Storti RV. The Drosophila PAR domain protein 1 (Pdp1) gene encodes multiple differentially expressed mRNAs and proteins through the use of multiple enhancers and promoters. Dev Biol. 2000;224:401–14. doi: 10.1006/dbio.2000.9797. [DOI] [PubMed] [Google Scholar]
- Rivlin PK, Schneiderman AM, Booker R. Imaginal pioneers prefigure the formation of adult thoracic muscles in Drosophila melanogaster. Dev Biol. 2000;222:450–9. doi: 10.1006/dbio.2000.9676. [DOI] [PubMed] [Google Scholar]
- Roy S, VijayRaghavan K. Homeotic genes and the regulation of myoblast migration, fusion, and fibre-specific gene expression during adult myogenesis in Drosophila. Development. 1997;124:3333–41. doi: 10.1242/dev.124.17.3333. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–53. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Seroude L, Brummel T, Kapahi P, Benzer S. Spatio-temporal analysis of gene expression during aging in Drosophila melanogaster. Aging Cell. 2002;1:47–56. doi: 10.1046/j.1474-9728.2002.00007.x. [DOI] [PubMed] [Google Scholar]
- Smith JM, Bozcuk AN, Tebbutt S. Protein turnover in adult Drosophila. J Insect Physiol. 1970;16:601–13. doi: 10.1016/0022-1910(70)90094-6. [DOI] [PubMed] [Google Scholar]
- Tanaka KK, Bryantsev AL, Cripps RM. Myocyte enhancer factor 2 and chorion factor 2 collaborate in activation of the myogenic program in Drosophila. Mol Cell Biol. 2008;28:1616–29. doi: 10.1128/MCB.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CS, Pirrotta V. New pCaSpeR P element vectors. Dros Inform Serv. 1992;71:150. [Google Scholar]
- Zhang S, Bernstein SI. Spatially and temporally regulated expression of myosin heavy chain alternative exons during Drosophila embryogenesis. Mech Dev. 2001;101:35–45. doi: 10.1016/s0925-4773(00)00549-9. [DOI] [PubMed] [Google Scholar]
- Zierath JR, Hawley JA. Skeletal muscle fiber type: influence on contractile and metabolic properties. PLoS Biol. 2004;2:e348. doi: 10.1371/journal.pbio.0020348. [DOI] [PMC free article] [PubMed] [Google Scholar]