Abstract
The developing heart contains an inner tube of specialized endothelium known as endocardium, which performs multiple essential functions. In spite of the essential role of the endocardium in heart development and function, the transcriptional pathways that regulate its development remain largely undefined. GATA4 is a zinc finger transcription factor that is expressed in multiple cardiovascular lineages and is required for endocardial cushion development and embryonic viability, but the transcriptional pathways upstream of Gata4 in the endocardium and its derivatives in the endocardial cushions are unknown. Here, we describe a distal enhancer from the mouse Gata4 gene that is briefly active in multiple cardiac lineages early in cardiac development but restricts to the endocardium where it remains active through cardiogenesis. The activity of this Gata4 cardiac enhancer in transgenic embryos and in cultured aortic endothelial cells is dependent on four ETS sites. To identify which ETS transcription factors might be involved in Gata4 regulation via the ETS sites in the enhancer, we determined the expression profile of 24 distinct ETS factors in embryonic mouse hearts. Among multiple ETS transcripts present, ETS1, FLI1, ETV1, ETV5, ERG, and ETV6 were the most abundant in the early embryonic heart. We found that ETS1, FLI1, and ERG were strongly expressed in the heart at embryonic day 8.5 and that ETS1 and ERG bound to the endogenous Gata4 enhancer in cultured endothelial cells. Thus, these studies define the ETS expression profile in the early embryonic heart and identify an ETS-dependent enhancer from the Gata4 locus.
INTRODUCTION
The embryonic cardiovascular system begins to assemble after gastrulation around embryonic day (E) 7.5 in the mouse. Bilaterally symmetrical mesodermal progenitors in the anterior lateral mesoderm fuse at the midline to fashion a linear heart tube made up of an outer layer of myocardial cells that surround an inner layer of endothelial cells known as the endocardium (Brand, 2003). As the linear heart tube forms, angioblasts in the lateral mesoderm differentiate into endothelial cells and form the first lumenized vessels in the embryo (Flamme et al., 1997). As development proceeds, the heart undergoes extensive morphogenesis, which ultimately separates the right and left sides and inflow and outflow regions (Brand, 2003). In parallel, the vasculature is elaborated, forming a complex network of vessels to deliver blood and nutrients, pumped by the heart, throughout the developing embryo (Drake and Fleming, 2000).
The endocardium is continuous with the vascular system but is composed of specialized endothelial cells (Harris and Black, 2010). Endocardial cells have distinct cytoskeletal and connective characteristics compared to other types of endothelial cells (Andries and Brutsaert, 1993; Brutsaert and Andries, 1992; Melax and Leeson, 1967). During embryonic development, endocardial cells perform unique roles in the developing heart: they induce some myocardial cells to form essential trabeculae, ridges of myocardium within the ventricles (Stankunas et al., 2008; Wagner and Siddiqui, 2007), and a subset of specialized cells from the endocardium undergo endocardial-to-mensenchymal transformation (EMT) to populate the endocardial cushions, which give rise to the cardiac valves and portions of the interventricular and atrial septa (Hutson and Kirby, 2007; Person et al., 2005; Snarr et al., 2008). The unique nature of the endocardium may arise, at least in part, from signals received by the endocardium during cardiogenesis, which are different than signals in other areas of endothelial specification in the embryo (Misfeldt et al., 2009). Importantly, malformations in the endocardium and its derivatives underlie numerous pediatric and adult cardiac diseases (Bruneau, 2008; Markwald et al., 2010). In spite of its role in development and disease, molecular and genetic characteristics that distinguish the endocardium from other endothelial cells remain incompletely defined, and the cell-autonomous transcriptional pathways important for endocardial development are poorly understood (Harris and Black, 2010).
The ETS family of transcription factors is widely appreciated for its role in endothelial cell development and function (De Val and Black, 2009; Hollenhorst et al., 2004; Sato, 2001). The ETS family is composed of thirty factors in mammals. ETS proteins are defined by the presence of a conserved DNA-binding domain and a common consensus target sequence, GGA(A/T) (Hollenhorst et al., 2011). Human endothelial cells express 19 ETS factors (Hollenhorst et al., 2004). Among them, ETS1, ETS2, FLI1, ETV2, and ETV6 are required for normal vascular development in mice (Barton et al., 1998; Lee et al., 2008; Spyropoulos et al., 2000; Wang et al., 1997; Wei et al., 2009). ETS factors regulate vascular development by binding consensus target sequences in many endothelial genes, such as Cdh5, Tie2, and Flk1, which are important in differentiated endothelial cell function (De Val and Black, 2009). In addition, ETS factors directly regulate other transcription factor genes, suggesting ETS factors are important nodes in transcriptional pathways that govern endothelial specification (De Val and Black, 2009; De Val et al., 2008; Pimanda et al., 2008; Pimanda et al., 2007).
The GATA family of transcription factors participates in the transcriptional pathways that regulate the development of several cardiovascular lineages, including the myocardium, epicardium, and endocardial cushions (Lepore et al., 2006; Lugus et al., 2007; Rivera-Feliciano et al., 2006; Rojas et al., 2008; Song et al., 2009; Xin et al., 2006; Zeisberg et al., 2005; Zhao et al., 2008). The GATA family consists of six factors that share a zinc-finger DNA-binding domain that binds to the GATA consensus sequence, (A/T)GATA(A/G), which is found in numerous cardiovascular gene regulatory regions (Charron and Nemer, 1999; De Val and Black, 2009; Molkentin, 2000; Patient and McGhee, 2002). GATA4 is a GATA transcription factor that is required for myocardial and endocardial development (Oka et al., 2006; Rivera-Feliciano et al., 2006; Rojas et al., 2008; Zeisberg et al., 2005). Gata4-null mice have defective endoderm development leading to cardia bifida ((Kuo et al., 1997; Molkentin et al., 1997), and mice lacking GATA4 in the myocardium display defective cardiomyocyte proliferation and embryonic lethality (Rojas et al., 2008; Zeisberg et al., 2005). Mice lacking Gata4 in the endothelium have defective endocardial cushion development and die around E12.5 of apparent heart failure (Rivera-Feliciano et al., 2006). In spite of its importance in heart development, the transcriptional pathways upstream of Gata4 in cardiovascular development are unknown.
In this study, we identify a distal enhancer of the mouse Gata4 gene. This enhancer, referred to as Gata4 G9, is active in the cardiac crescent at E7.5 and in the linear heart at E8.5. As the heart undergoes looping morphogenesis, enhancer activity remains strong in the endocardial layer but rapidly diminishes in the myocardial layer. At E11.5 and later stages, the enhancer is active only in the endocardium and its derivatives in the endocardial cushions. We define a minimal region of the enhancer that is necessary and sufficient for endocardial activity in transgenic mouse embryos and in cultured aortic endothelial cells. We show that the enhancer is dependent on four consensus ETS binding sites, since mutation of the sites completely abrogates enhancer activity in vivo. To determine which ETS factors regulate Gata4 via the ETS sites in the enhancer, we assessed the expression of twenty-four ETS factors in the embryonic heart. Among these factors, transcripts for FLI1, ETV5, ETV6, ETS1 ERG, ELF1, ETV1, and ETS2 were the most enriched, and ETS1 and ERG bound to the endogenous Gata4 G9 enhancer in cultured aortic endothelial cells as determined by chromatin immunoprecipitation. Thus, these studies define the ETS expression profile in the early embryonic heart, and identify a cardiac enhancer from the Gata4 locus that is active predominately in the endocardium and is directly regulated by ETS proteins.
MATERIALS AND METHODS
Bioinformatic analyses, cloning, and mutagenesis
Mouse, human, and cow sequences were compared using BLAST and VISTA (Altschul et al., 1990; Mayor et al., 2000). The 1945 bp Gata4 G9 fragment was generated by PCR from mouse genomic DNA using the following primers: 5’-gctggacctgtctcgagcacacttgttat-3’ and 5’-tctcagaataacccgggatgactattt-3’. The 404-bp Gata4 G9[902–1305] fragment was generated from Gata4 G9 using the following primers: 5’-ggcgtttctcgagtagtccttggatgccagaa-3’ and 5’-ataatcacccgggcctgttgccctccgccct-3’. The deletion construct Gata4 G9 [Δ902–1305] was generated by PCR from Gata4 G9 using the following mutagenic primer and its reverse complement: 5’-gctctgagggacgcaggggatccagtgagc-3’. All Gata4 G9 fragments were cloned into the XhoI-XmaI sites of the transgenic reporter plasmid hsp68-lacZ (Kothary et al., 1989). The 4× ETS mutant was generated by PCR using the following mutagenic primers and their reverse complements:ETS-E mutant, 5’-ttctgataaatgggttgctccacccctccc-3’, ETS-H mutant, 5’-ctcagactgatgggcaataccattgtctg-3’, and ETS-I/J mutant, 5’-ccagggcgacaggcattgcagaggcgtgggg-3’.
Generation of transgenic mice
Transgenic lacZ reporter fragments were generated by gel purifying Xho-SalI fragments from parental hsp68-lacZ plasmids. Pronuclear injection of transgene fragments was performed as described previously (Dodou et al., 2004). The presence of lacZ transgenes in embryo yolks sacs or tail biopsies was detected using PCR and primers specific for the transgene or by Southern blot using a probe specific for lacZ. All experiments using animals were reviewed and approved by the UCSF Institutional Animal Care and Use Committee and complied with all institutional and federal guidelines.
X-gal staining, immunofluorescence, and immunohistochemistry
β-galactosidase activity in transgene-positive embryos was detected by X-gal staining, as described previously (Anderson et al., 2004). Transverse sections from X-gal stained embryos were prepared and counterstained with Neutral Fast Red as described previously (Dodou et al., 2003). For section immunofluorescence and immunohistochemistry, embryos were sectioned, de-waxed, boiled in antigen retrieval solution (Biogenex), and blocked in PBS containing 10% sheep serum and 0.1% Triton X-100. For whole mount immunohistochemistry, embryos were boiled in antigen retrieval solution (Biogenex), treated with 5% H2O2, and blocked in PBS with 5% milk, 5% sheep serum, and 0.1% Triton X-100. The following primary antibodies were used at 1:100 dilutions in blocking serum: rabbit anti-ETS1 (Santa Cruz; sc-350), mouse anti-GABPA (Santa Cruz; sc-28311), rabbit anti-FLI1 (Santa Cruz; sc-356), rabbit anti-ERG1/2/3 (Santa Cruz; sc-353), rabbit anti-ETV6 (Santa Cruz; sc-11382), mouse anti-myosin heavy chain (DHSB; MF20), mouse anti-GATA4 (Santa Cruz; sc-25310), chicken anti-β-galactosidase (Abcam; Ab9361), and rat anti-endomucin (eBioscience; 14-5851-82). The following secondary antibodies were used at 1:300 dilutions in blocking serum: biotinylated goat anti-rabbit IgG (Vector Laboratories; BA-1000), biotinylated goat anti-mouse IgG (Vector Laboratories; BA-9200), Alexa Fluor 594 goat anti-chicken IgG (Invitrogen; A11042), streptavidin Alexa Fluor 488 (Invitrogen; S32354), and Oregon Green anti-mouse IgG (Invitrogen; O-6383). For immunofluorescence, slides were mounted and photographed as described previously (Rojas et al., 2009). For immunohistochemistry, immunoperoxidase staining and photography were performed as described previously (Rojas et al., 2009).
Cell Culture, transfections, and reporter assays
For transfection experiments, full-length Gata4 G9, G9[902–1305], and mutant constructs were subcloned from hsp68-lacZ as KpnI fragments into the KpnI site of a modified pGL2-Basic vector (Promega) that contains the thymidine kinase (TK) minimal promoter. Bovine aortic endothelial cells (BAEC) were purchased from Cambrex Bio and maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 1% L-glutamine and 1% Penicillin/Streptomycin. Transient transfections were performed in 24-well plates using Lipofectamine LTX (Invitrogen) following the manufacturer’s recommendations. Transfection mixtures contained 1 µg of reporter plasmid. Luciferase activity was measured, after normalization to protein concentration, 48 h after transfection, using the Luciferase Assay System (Promega) according to the manufacturer’s protocol.
cDNA preparation and Quantitative Real Time PCR
Embryos were collected between 8.5 and 9.5 days post-coitum and hearts were mechanically dissected and pooled. RNA was extracted using the Qiagen RNeasy Micro Kit, as recommended by the manufacturer. RNA was then used to generate cDNA with the SuperArray RT2 First Strand Kit according to the manufacturer’s directions.
Quantitative real time PCR (qPCR) was performed performed using SYBR Green PCR Master Mix (Applied Biosystems) and primers that have been previously described (Galang et al., 2004). The reaction mixtures were analyzed on an Applied Biosystems 7900HT Fast Real Time PCR System. Analysis of relative gene expression was performed as previously described (Galang et al., 2004). Briefly, for each experiment, a standard curve relating GAPDH cDNA concentration to threshold cycle (Ct) was generated using serial dilutions of cDNA with GAPDH primers. cDNA concentration was calculated according to the standard curve and then normalized to the GAPDH concentration.
Electrophoretic mobility shift assay (EMSA)
DNA binding assays were performed as described previously (Dodou et al., 2003) in modified 1× binding buffer (15mM KCl, 5mM Hepes pH 7.9, 5% glycerol, 0.04 mM EDTA, 0.125 mM DTT, 0.25 mM PMSF). The ETS1 DNA-binding domain (DBD) construct used in EMSA analyses has been described previously (De Val et al., 2004). The ETS1 DBD was transcribed and translated using the TNT Coupled Transcription-Translation system (Promega) according to the manufacturer’s protocol. The ETS1 control (Mef2c–F7 ETS-A ) and mutant control probe sequences have been described previously (De Val et al., 2004) and were: control, 5′-gctcagagaaggaagtggagagt-3′; mutant, 5′-gctcagagaagcttgtgggaggtt-3′. The sequences of the competitor oligonucleotides encompassing the ETS sites from the Gata4 G9 enhancer were: ETS A, 5′- tagtccttggatgccagaaagaaa-3′; ETS B, 5′- agaaagaaaggccaggaattgtggc-3′; ETS C, 5′-ccagctgttagggggaacggggtgaattct-3′; ETS D, 5′-ccagctgttaagggggaacggggtgaattct-3′ ETS E, 5′-ttctgataaatgggttcctccacccctccc-3′; ETS F, 5′-cccggtgcagaaaccacttccaagccaggcatga-3′; ETS G, 5′-aaaaacagcagtggataaggcccgg-3′; ETS H, 5′-ctcagactgatgggaaataccattgtctg-3′, ETS I/J, C 5′-ccagggcgacagggattccagaggcgtgggg-3′. The sequences for the ETS mutant sites were identical to the mutagenic primers described above.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed using the ChIP Assay Kit (Millipore; 17–295). DNA was isolated from confluent BAEC grown on 10 cm plates. Chromatin derived from 1×106 cells was cross-linked, lysed, and sonicated to shear the DNA into 200–500 bp fragments. Samples were pre-cleared and incubated with antibody. Normal rabbit IgG (Santa Cruz; sc-2027), rabbit anti-ETS1 (Santa Cruz; sc-350), and rabbit anti-ERG1/2/3 (Santa Cruz; sc-353) were used at a concentration of 1 µg/ml. After incubation with protein-A-agarose beads, samples were washed and protein-DNA complexes were eluted. The crosslinks were reversed and the DNA was recovered by phenol-chloroform extraction. The primers used to amplify the region of the bovine Gata4 G9 enhancer containing the ETS sites were 5′-gacaatgggtgttagactaatag −3′ and 5′-gcttgtggccgctgggatccctgtcgc −3′. The primers used to amplify a non-conserved sequence of Gata4 outside the G9 enhancer region were 5′-ttagagggctacagtccatacg-3′ and 5′-agattaatcagactcatctggcc-3′.
RESULTS
Identification of a Gata4 enhancer with activity in the developing heart
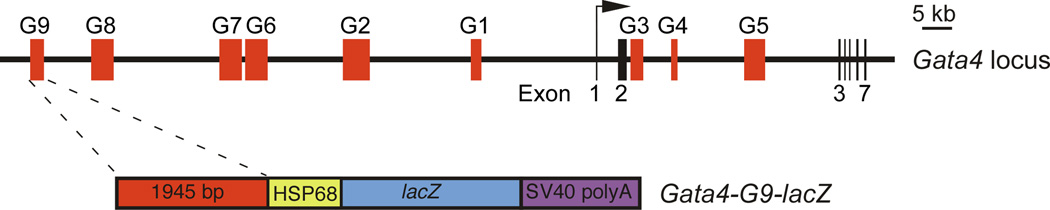
To identify enhancers of Gata4 expression, we screened nine regions of conserved noncoding sequence from the Gata4 locus for enhancer activity in transgenic mice (Fig. 1). Multiple enhancers were identified. Two enhancers (G4 and G8) have early- and late-acting endoderm specificities (Rojas et al., 2009; Rojas et al., 2010). A third enhancer (G2) directs activity to the lateral mesoderm and its derivatives in the liver mesenchyme (Rojas et al., 2005). The most distal region that we tested spans 1945 bp and is located approximately 93 kb away from the Gata4 transcriptional start site (Fig. 1). We cloned this region, referred to as Gata4 G9, into the transgenic reporter plasmid Hsp68-lacZ (Kothary et al., 1989) and assayed β-galactosidase reporter activity by X-gal staining of transgenic embryos (Fig. 2). At E7.5, enhancer activity was observed in the embryo in a crescent-shaped region of splanchnic mesoderm corresponding to the developing heart field (Fig. 2A). The expression pattern of GATA4 protein at the same embryonic stage was similar because it included the crescent-shaped region of precardiac mesoderm marked by the Gata4-G9-lacZ transgene but was also broader because GATA4 protein was detected the lateral mesoderm (LM) (Fig. 2C). This was expected since endogenous GATA4 is expressed in multiple lineages in addition to the heart (Arceci et al., 1993; Molkentin, 2000).
Fig. 1. Schematic representation of the mouse Gata4 locus and the Gata4-G9-LacZ transgene.
The black horizontal line represents the mouse Gata4 locus and the black vertical bars represent exons in the locus. The red boxes represent non-coding regions of deep conservation between mouse, human, and opossum. A 1945-bp region, encompassing G9, was cloned into the hsp68-lacZ transgenic reporter (lower schematic).
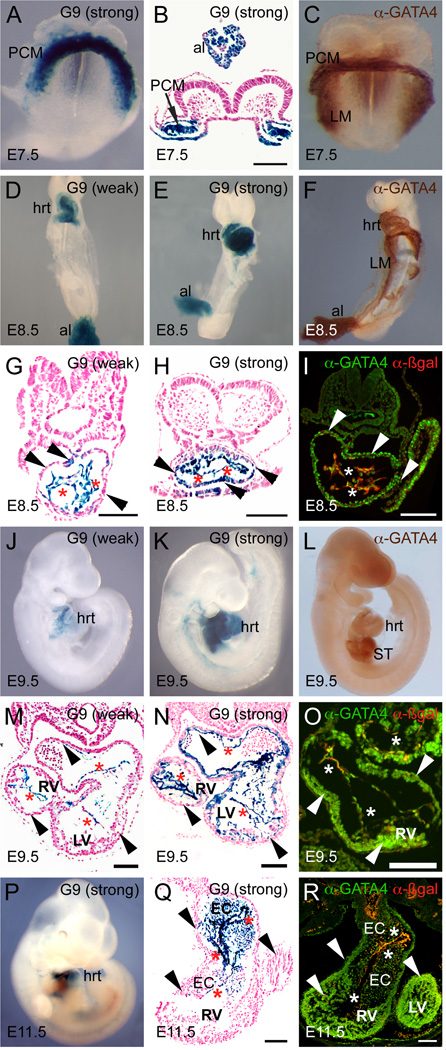
Fig. 2. TheGata4G9 enhancer is active in cardiac progenitors in the cardiac crescent and the linear heart tube and restricts to endocardium during mouse cardiogenesis.
X-gal stained embryos photographed as whole-mounts (A, D, E, J, K, P) or following transverse sectioning of stained embryos (B, G, H, M, N, Q) are shown. For comparison, wild type embryos stained as whole mounts with anti-GATA4 antibody are shown (C, F, L). In addition, transverse frozen sections stained for immunofluorescence with anti-p-galactosidase and anti-GATA4 antibodies are also shown (I, O, R). Strong Gata4-G9-lacZ transgenic lines are pictured in B, E, H, I, K, N, and O; weak Gata4-G9-lacZ lines are pictured in D, G, J, M. Asterisks mark endocardium and arrowheads mark myocardium. al, allantois; EC, endocardial cushions; hrt, heart; LM, lateral mesoderm; LV, left ventricle; PCM, precardiac mesoderm; RV, right ventricle; ST, septum transversum. Bars in all panels = 100 µM.
Detailed analyses of transverse sections showed X-gal staining in the precardiac mesoderm and allantois at E7.5 (Fig. 2B). At E8.5, the Gata4 G9 enhancer was active in the linear heart tube and allantois (Fig. 2D, E) in a pattern similar to endogenous GATA4 expression (Fig. 2F). Transverse sections through the hearts of transgenic lines with enhancer activity revealed that the majority (9/12) of Gata4-G9-lacZ lines had activity restricted to the endocardium at E8.5 with at most, a few cells stained in the myocardium (Fig. 2G). However, a minority of the lines (3/12) with strong expression of β-galactosidase in the endocardium also exhibited X-gal activity in the myocardium at E8.5 (Fig. 2H). Importantly, the expression of β-galactosidase from weak and strong lines overlapped with a subset of the endogenous GATA4 expression pattern in the heart, as determined by anti-β-galactosidase and anti-GATA4 immunofluorescence (Fig. 2I). Three transgenic lines displayed no activity in the heart or elsewhere at this stage, probably due to insertion into inactive regions in chromatin (data not shown).
Because X-gal staining in strong transgenic lines appeared to be present in both the endocardial and myocardial layers of the heart at E8.5 (Fig. 2E,H), we compared the expression of β-galactosidase from Gata4-G9-lacZ transgenic embryos to endothelial and myocardial markers at this stage (Fig. 3). β-galactosidase expression completely overlapped the expression of the endothelial marker endomucin within the endocardium at E8.5 (Fig. 3A). As noted above, in the majority of transgenic lines examined at E8.5, transgene activity was mostly restricted to the endocardium (Fig. 2D,G). However, in lines with the strongest X-gal staining, the expression of β-galactosidase also overlapped with the expression of myosin heavy chain in some, but not all, cells of the myocardium (Fig. 3B). Thus, the Gata4-G9-lacZ is active in presumptive endocardial and myocardial progenitors in precardiac mesoderm at E7.5 and is active throughout the endocardium at E8.5. The enhancer also has weak and incomplete activity in the myocardium, which is only apparent in the strongest transgenic lines.
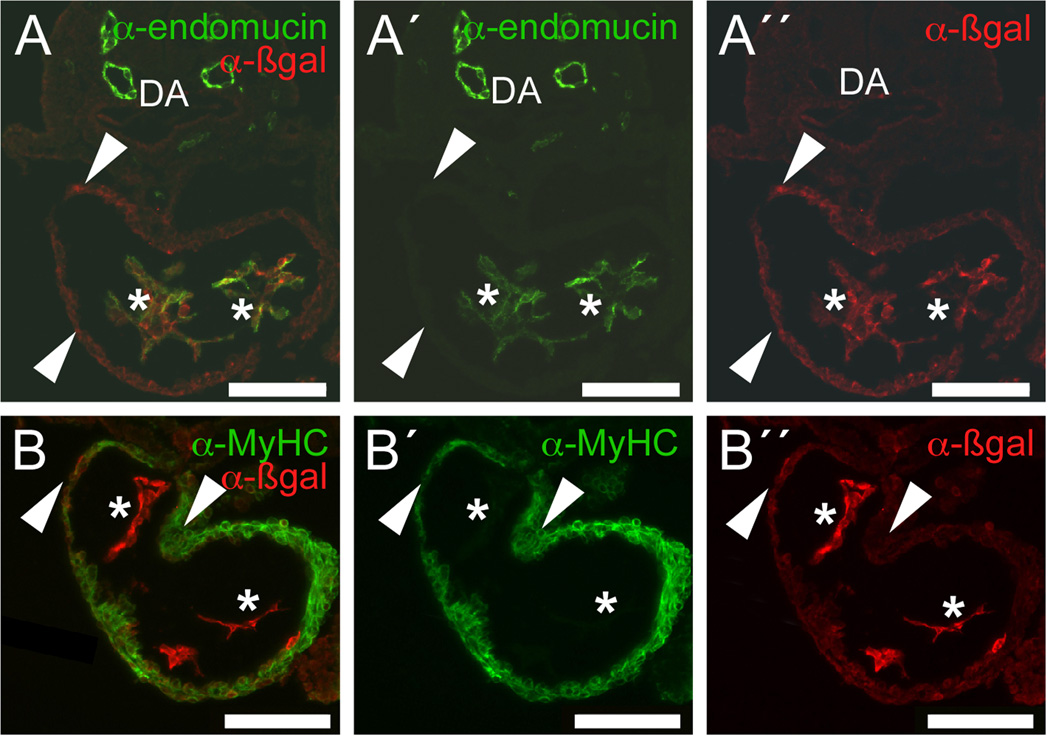
Fig. 3. TheGata4G9 enhancer is active in the endocardium and myocardium of the linear heart.
Sections from a strong transgenic Gata4-G9-lacZ embryo at E8.5 were stained with anti-β-galactosidase (A, A″, B, B″), anti-endomucin (A, A′), and anti-myosin heavy chain (MyHC) (B, B′) antibodies. The Gata4 G9 enhancer directed β-galactosidase expression throughout the endocardium (asterisks). A minority of cells in the myocardial layer also showed anti-β-galactosidase staining (arrowheads). Merged images show endomucin-β-galactosidase-double-positive orange cells (A) and MyHC-β-galactosidase-double-positive orange cells (B). Note that there are no β-galactosidase-negative endocardial cells at this stage (A), that there are endomucin-positive but β-galactosidase-negative endothelial cells in the dorsal aorta (DA), and that the majority of the MyHC-positive cells are β-galactosidase-negative (B″). Bars in all panels = 100 µM.
At E9.5, Gata4-G9-lacZ transgene expression was restricted to the endocardium of the looping heart and to the endothelium of the proximal outflow tract in the majority of the independent transgenic lines with enhancer activity (7/11) (Fig. 2J,M). However, as was the case at E8.5, X-gal staining in myocardial cells could be detected in the strongest lines at E9.5 (4/11) (Fig. 2K,N). In these strong lines, X-gal activity was more restricted within the myocardium compared to E8.5, suggesting further restriction to the endocardium (Fig. 2, compare panels H and N). The pattern of transgene activity in the heart, even in the strongest lines, was more restricted than endogenous GATA4 (Fig. 2J,L,O). Endogenous GATA4 protein was detectable in the septum transversum and developing gut at this stage (2L), but neither the weak nor strong lines displayed X-gal staining in either of those tissues (2J,K). Thus, Gata4 G9 enhancer activity marks a restricted subset of the broader endogenous GATA4 expression pattern.
By E11.5, Gata4-G9-lacZ enhancer activity was restricted to the endocardial cell layer of the developing heart in all lines examined and activity was never observed in the myocardium (Fig. 2P,Q). We also observed robust enhancer activity in the mesenchymal cells populating the proximal outflow tract and atrioventricular endocardial cushions (Fig. 2Q). The staining of these cells likely reflects their endocardial origin since they are derived primarily from endothelial cells overlying the cushions that have undergone endothelial-to-mesenchymal transition (Combs and Yutzey, 2009). We cannot rule out the possibility that the enhancer may have activity in mesenchymal cells of neural crest origin, but we consider this unlikely since enhancer activity was never observed in the cardiac neural crest itself. As in earlier stages, β-galactosidase-positive cells overlapped with a subset of endogenous GATA4 (Fig. 2R). At later stages of embryonic development and in neonatal mice, Gata4-G9-lacZ transgene expression was detected in the outflow tract endocardium and in the aortic endothelium in some lines (data not shown). Thus, while the enhancer is strongly active in endocardial cells and weakly in myocardial cells when the linear heart first appears, its activity remains strong only in the endocardium and its derivatives in the endocardial cushions at subsequent stages. Importantly, this dynamic enhancer activity occurs within the broader endogenous GATA4 expression pattern. These results, and the location of the enhancer proximal to the Gata4 locus, suggest the Gata4 G9 enhancer is indeed a bona fide enhancer of mouse Gata4 transcription.
A 404-bp fragment of Gata4 G9 is necessary and sufficient for enhancer activity in vivo
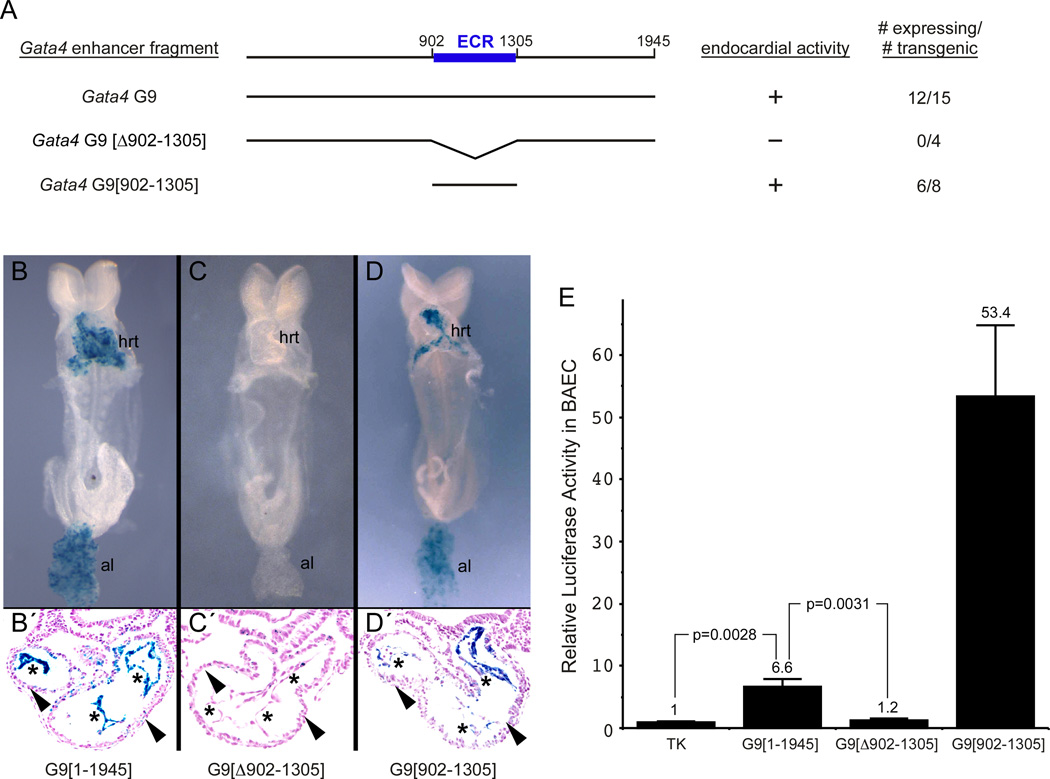
To delimit the essential transcription factor binding elements within the Gata4 G9 fragment, we wanted to define a minimal region necessary and sufficient for cardiac activity in embryos. Comparison of mouse, human, and bovine Gata4 sequence revealed one 404-bp evolutionarily-conserved region (ECR), spanning base pairs 902–1305, within the 1945-bp Gata4 G9 enhancer (Fig 4A). To determine whether this region was necessary and sufficient for enhancer activity in vivo, we tested deletion constructs of Gata4-G9-lacZ for activity in transgenic mouse embryos (Fig 4A–D). The evolutionarily conserved 404 bp region directed activity to the heart at E8.5 in a pattern similar to the full-length 1945-bp enhancer (Fig 4, compare panels B and D), indicating that the ECR was sufficient for Gata4 G9 enhancer activity in the heart and allantois. As with the full-length construct, a majority of the lines in which the transgene was active exhibited X-gal activity mainly in the endocardium. The ECR was also required for enhancer activity in vivo, as evidenced by the inactivity of the Gata4 G9 enhancer transgene in which the conserved region was deleted (Fig. 4C).
Fig. 4. The Gata4 G9 enhancer contains a deeply conserved region that is necessary and sufficient for activityin vivo and in cultured aortic endothelial cells.
(A) The black horizontal lines represent the Gata4 G9 enhancer and deletion mutants. The blue box between positions 902 and 1305 represents the evolutionary conserved region (ECR). The column to the right indicates the number of independent transgenic lines that expressed β-galactosidase in the endocardium at E8.5 over the total number of transgene-positive embryos. (B–D) Representative transgenic embryos collected at 9.5 days post-coitum from the three constructs tested. Whole mount X-gal-stained embryos are shown in B, C, D. Transverse sections through the heart from three distinct embryos from those shown in B, C, D are shown in B′, C′, D′. Asterisks mark the endocardium. Arrowheads mark the myocardium. al, allantois; hrt, heart. (E) BAEC were transfected with TK-luciferase, full-length Gata4-G9[1–1945]-TK-luciferase, Gata4-G9[∆902-1305]-TK-luciferase, or Gata4-G9[902–1305]-TK-luciferase reporters. The activity of the Gata4 G9[1–1945] enhancer was approximately 7-fold higher than the parental TK vector while the Gata4 G9[902–1305] construct was inactive (lanes 1–3). The construct containing Gata4 G9[902–1305] showed 53-fold more activity compared to the parent vector. Data are shown as the mean fold activation over TK-luciferase. Error bars represent SEM for 8 independent transfections and analyses. Indicated p values were calculated by two-tailed, unpaired t test.
We also tested the activity of the full-length Gata4 G9 enhancer, the 902–1305 sufficiency region, and the Δ902–1305 fragment in bovine aortic endothelial cells (BAEC) (Fig. 4E). Bovine aortic endothelial cells (BAEC) are a well-established endothelial system (Gory et al., 1998; Ronicke et al., 1996; Schlaeger et al., 1997), and we reasoned that because the Gata4 G9 enhancer was active in endocardial endothelium and in the proximal aortic and outflow tract endothelium in transgenic mice, BAEC would serve as a relevant cell culture system to further characterize enhancer function, particularly since there are no established endocardial cell lines. The full-length and minimal 404 bp Gata4 G9 enhancer fragments directed robust and significant reporter activity in BAEC compared to the parental TK vector (Fig. 4E). Interestingly, the ECR was more active than the full-length construct, suggesting elements outside of the minimal region might repress activity in cultured cells. However, in the developing embryo, these putative repressor elements appear to play no role, as the minimal region did not direct activity outside of the tissues in which the full-length enhancer was active (4B,D). Alternatively, the smaller 404-bp fragment might position critical regulatory elements closer to the promoter than the full-length, 1945-bp fragment, allowing for more potent activation in cultured cells. Importantly, the mutated enhancer (Δ902–1305), in which the ECR was deleted, displayed only background activation in BAEC (Fig. 4E). These cell culture experiments were consistent with the in vivo data in transgenic mouse embryos and establish that Gata4 G9[902–1305] contains elements that are necessary and sufficient for enhancer activity in the heart, including the endocardium, and aortic endothelium in vivo and in cultured aortic endothelial cells.
The Gata4 G9 enhancer contains four ETS sites essential for enhancer function
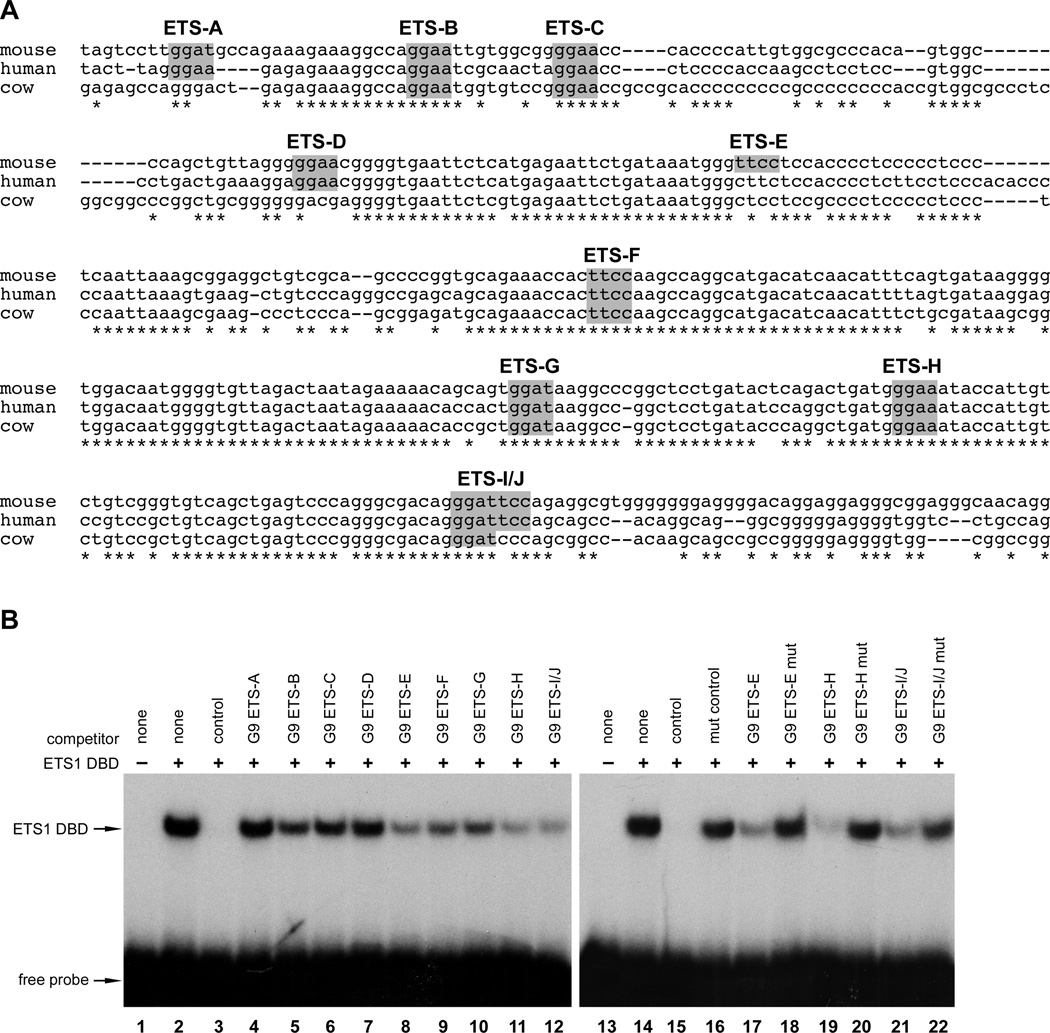
Among multiple potential candidate sites within the 404-bp conserved region of Gata4 G9, we identified ten consensus ETS binding sites in the mouse sequence (Fig. 5A; ETS sites A-J). The presence of multiple ETS sites was intriguing since several ETS transcription factor family members are well-established essential regulators of endothelial development (De Val and Black, 2009; Graves and Petersen, 1998). To determine which of the consensus ETS elements within the 404-bp region of the Gata4 G9 enhancer function as bona fide ETS transcription factor binding sites, we designed probes encompassing each of the ten consensus ETS elements and tested their ability to compete for the binding of the ETS1 DNA-binding domain (DBD) to a bona fide control ETS site. We used the ETS1 DBD as a general ETS domain because ETS1 binding has been well characterized and because all ETS transcription factors share this highly conserved motif (Graves and Petersen, 1998; Hollenhorst et al., 2011). As expected, ETS1 efficiently bound to a radiolabeled probed encompassing the bona fide control ETS site (Fig. 5B, Lane 2), and excess unlabeled control probe competed for binding (Fig. 5B, Lane 3). Each of the Gata4 G9 probes containing ETS consensus elements competed for ETS1 DBD binding to the control probe to some extent, although most competed only weakly (Fig. 5B, lanes 4–12). Among the ten Gata4 G9 ETS site probes, ETS-E, ETS-H, and ETS-I/J competed most effectively (Fig. 5B, Lanes 8, 11, 12). The binding of the ETS1 DBD to the ETS-E, ETS-H, and ETS-I/J sites was specific since competitor probes with mutations in each of those ETS sites abolished competition for binding to the control probe (Fig. 5B, lanes 17–22). These results demonstrate that the Gata4 G9 ECR contains several ETS sites capable of ETS protein binding and that ETS-E, ETS-H, and ETS-I/J are the sites with the highest affinity for ETS1 in vitro.
Fig. 5. TheGata4G9 enhancer contains ETS binding sites.
(A) ClustalW analysis of the 404-bp Gata4 G9 ECR, comparing mouse, human, and bovine sequences. The region contains 10 ETS consensus sequences (gray boxes). (B) Recombinant ETS1 DNA-binding domain (DBD) was used in EMSA with radiolabeled probe encompassing a bona fide ETS-binding site from the Mef2c locus. Lanes 1 and 13 contain unprogrammed rabbit reticulocyte lysate. ETS1 DBD bound to the bona fide ETS-binding site in the control probe (lanes 2, and 14) and this specific binding was competed by unlabeled control probe (lane 3, 15). Unlabeled probes corresponding to each of the ETS sites in the Gata4 G9 ECR were used as competitors for ETS1 DBD binding to the control probe (lanes 4–12). The Gata4 G9 ETS sites competed to varying degrees. Probes encompassing ETS-E, -H, and -I/J competed most efficiently (lanes 8, 11, and 12), and were further examined for competition with the control probe using wild type and mutant versions of each (lanes 17–22). Mutation of the ETS-binding consensus sequences in ETS-E, H, and I/J reduced competition compared to wild type sequences (compare lanes 17–18, 19‒20, and 21–22).
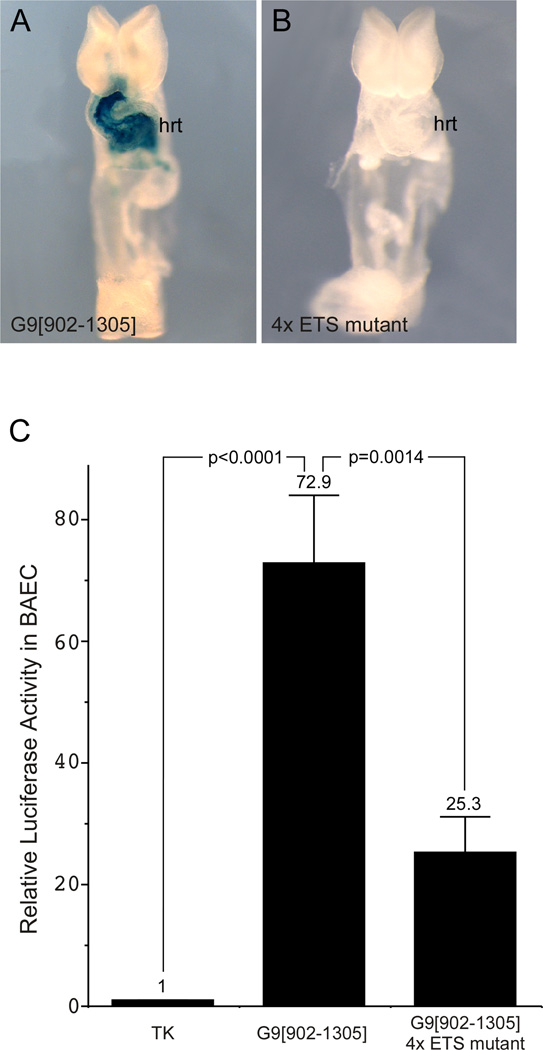
To examine the Gata4 G9 ETS sites for functionality in vivo, we generated transgenic mice bearing the minimal Gata4-G9[902–1305]-lacZ transgene in which ETS-E, ETS-H, and ETS-I/J sites were mutated and compared transgene activity of the mutant construct to a wild type Gata4-G9[902–1305]-lacZ transgene (Fig. 6A,B). As expected, Gata4-G9[902–1305] directed β-galactosidase activity to the developing heart at E8.5 (Fig. 6A). In contrast, mutation of the four ETS binding sites completely abolished enhancer activity (Fig. 6B), indicating that the four most robust in vitro-binding ETS sites are required for Gata4 G9 enhancer function in vivo. Consistent with this observation, mutation of the same four ETS sites resulted in a significant reduction of enhancer activity in BAEC compared to the wild type construct (Fig. 6C). Thus, these results establish that Gata4 G9 cardiac enhancer is a direct transcriptional target of ETS factors in vivo and in cultured endothelial cells.
Fig. 6. Gata4 G9 enhancer activity is dependent on four ETS sites.
(A, B) Representative transgenic embryos harboring Gata4-G9[902–1305]-lacZ and Gata4-G9[902–1305] 4× ETS mutant-lacZ transgenes are shown. 6 of 8 independently generated Gata4-G9[902–1305]-lacZ transgenic lines showed the pattern shown in (A). No activity of the 4× ETS mutant transgene was observed in any of the 6 independent lines examined. (C) BAEC were transfected with TK-luciferase, Gata4-G9[902–1305]-TK-luciferase, or G9[902–1305] 4× ETS mutant-TK-luciferase, and activity was determined. The activity of Gata4-G9[902–1305] was approximately 73-fold higher than the parent TK reporter (lanes 1, 2). Mutation of the 4 ETS sites (ETS-E, ETS-H, and ETS-I/J) reduced activity by approximately 65% compared to Gata4-G9[902–1305]-TK-luciferase. Data are shown as the mean fold activation over TK-luciferase control. Error bars represent SEM for 13 independent transfections and analyses. Indicated p values were calculated by two-tailed, unpaired t test.
Expression of ETS genes in the early mouse heart
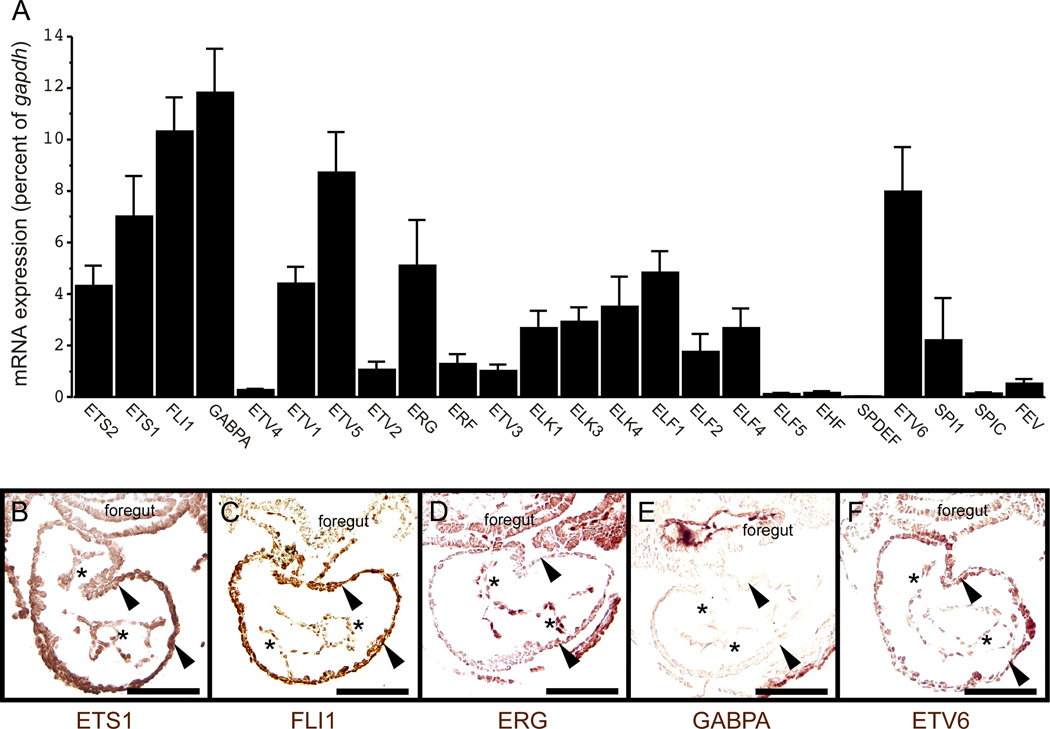
The ETS family contains more than thirty members, all of which contain a DNA-binding domain that specifically binds to a GGA(A/T) motif in target enhancers, such as those identified in the Gata4 G9 enhancer. Expression analyses have shown 19 ETS factors are expressed in endothelial cells and 20 ETS factors are expressed in the adult heart (Hollenhorst et al., 2004). Thus, a large number of ETS factors could potentially bind the candidate ETS sites in the Gata4 G9 enhancer. As a first step to determine the ETS factors that are the most highly expressed in the developing heart when the Gata4 G9 enhancer is active, we performed qPCR using validated primers designed to examine mRNA expression of 24 unique ETS genes in dissected hearts and associated tissues at E8.5 (Fig. 7A). Transcripts for multiple ETS factors were detected by qPCR, which showed that the most abundant ETS transcripts in the early embryonic mouse heart were GABPA, FLI1, ETV5, ETV6, ETS1, ERG, ELF1, ETV1, and ETS2 (Fig. 7A).
Fig. 7. ETS factor expression profile in embryonic mouse hearts.
(A) Quantification of ETS transcript levels in embryonic hearts at E8.5 by qPCR showed that GABPA, FLI1, ETV5, ETV6, ETS1, ERG, ELF1, ETV1, and ETS2 were the most abundant. Data are expressed as the mean transcript abundance relative to GAPDH from 5 independent tissue isolations and qPCR analyses. Error bars represent SEM. (B–F) Immunohistochemical analyses of wild type E8.5 mouse hearts with antibodies to ETS1, FLI1, ERG, GABPA, and ETV6. FLI1, ETS1, and ERG proteins were expressed in locations with Gata4 G9 enhancer activity (compare panels B–D with Fig. 2G–I). ETS1 and FLI1 proteins were expressed in the endocardium (asterisks in B, C) and myocardium (arrowheads in B, C). ERG expression appeared to be largely restricted to the endocardium (asterisks in D). GABPA protein was strongly expressed in the foregut and largely absent from the embryonic heart (E). ETV6 protein was present in the myocardium (arrowheads in F) with little or no expression detected in the endocardium (asterisks in F). Bars in all panels = 100 µM.
Among those ETS genes expressed in the early heart, several are known to be required for embryonic development and endothelial gene expression. Gabpa, Ets1, Ets2, Fli1, Etv6, and Erg are essential for embryonic development and regulate endothelial gene expression (De Val and Black, 2009; Loughran et al., 2008; Ristevski et al., 2004; Spyropoulos et al., 2000; Wang et al., 1997; Wei et al., 2009). By contrast, Etv5-, Etv1-, and Elf1-null mice have apparently normal endothelial development and are viable (Arber et al., 2000; Garrett-Sinha et al., 2001; Hippenmeyer et al., 2007). Because Gata4 is also required in the endothelium for normal endocardial development and embryonic viability (Rivera-Feliciano et al., 2006), we further analyzed by immunohistochemistry the expression patterns of those ETS proteins that were abundantly expressed in the early heart and that also play essential roles in development (Fig. 7B–F).
We analyzed the expression patterns of ETS1, FLI1, ERG, GABPA, and ETV6 by immunohistochemistry to determine whether the expression of those ETS proteins overlapped with Gata4 G9 enhancer activity at E8.5. ETS1, FLI1, and ERG were expressed in the endocardium at E8.5 (Fig. 7B–D). ETS1 and FLI1 were also strongly expressed in the myocardium, whereas ERG was not readily detected in the myocardium at this stage (Fig. 7B–D). GABPA and ETV6 were not strongly expressed in the endocardium (Fig. 7E,F). The apparent abundance of GABPA as determined by qPCR might have been the result of strong expression in the neighboring foregut (Fig. 7E), which may have been inadvertently included when embryonic hearts were isolated. Alternatively, this may reflect a discrepancy between GABPA RNA and protein expression. The abundance of ETV6 in the early embryonic heart by PCR was likely the result of strong expression in the myocardium rather than expression in the endocardium, which was undetectable by immunohistochemistry at E8.5 (Fig 7F). Because the Gata4 G9 enhancer is active primarily in the endocardium at E8.5 (Fig. 2, Fig. 3), we reasoned that ETS factors present throughout the endocardium could be potential regulators of the enhancer. The data in Fig. 7 show that ETS1, FLI1, and ERG protein expression correlated with Gata4 G9 enhancer activity in the endocardium, therefore, we pursued these factors as potential direct regulators of the Gata4 G9 enhancer.
ETS1 and ERG occupy the endogenous Gata4 G9 enhancer
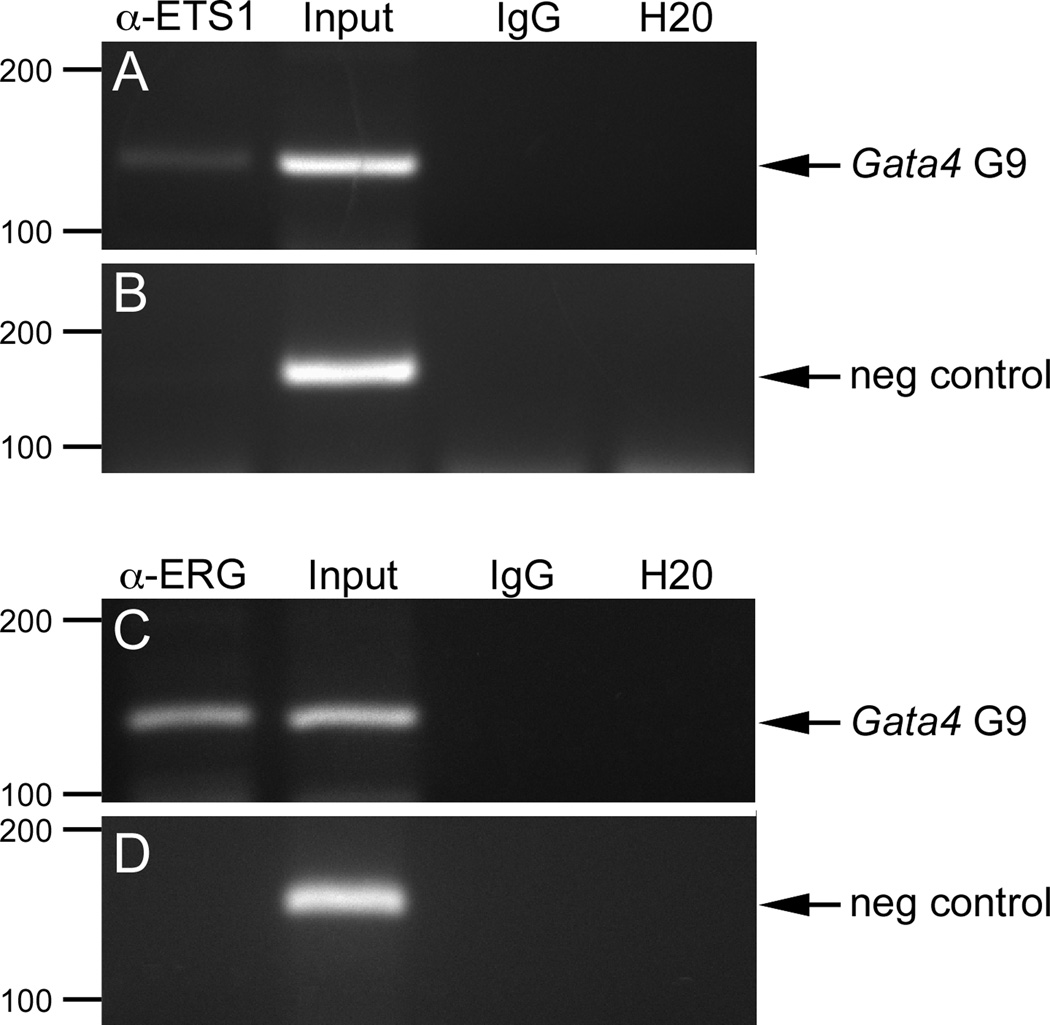
To determine if ETS1, FLI1, and ERG occupied the ETS sites in the endogenous Gata4 G9 enhancer in endothelial cells, we performed ChIP assays in BAEC (Fig. 8). Binding of FLI1 to the endogenous enhancer was not reproducibly detected (data not shown). However, anti-ETS1 and anti-ERG antibodies consistently immunoprecipitated genomic DNA fragments encompassing the region containing ETS binding sites in the endogenous Gata4 G9 enhancer, as indicated by amplification with primers specific for the region (Fig. 8A,C, a-ETS1 and α-ERG). A nonconserved region 5 kb away from the ETS binding sites was not amplified under conditions in which the region containing ETS binding sites in the endogenous Gata4 G9 enhancer was efficiently amplified (Fig. 8B,D a-ETS1 and α-ERG), showing that anti-ETS1 and anti-ERG antibodies were binding specifically to the ECR in Gata4 G9 and not to adjacent non-conserved regions. Importantly, neither the ECR containing the ETS binding sites nor the nonconserved region 5 kb away were immunoprecipitated by non-specific isotype control antibody (Fig. 8A–D, IgG). Thus, these results establish that ETS1 and ERG bind directly to the Gata4 G9 enhancer in the ECR containing multiple bona fide essential ETS sites.
Fig. 8. ETS1 and ERG bind to the endogenousGata4 G9 enhancer in aortic endothelial cells.
BAEC were used as a source of genomic material in ChIP to examine ETS1 and ERG binding to the Gata4 G9 ECR, which contains the essential ETS sites. Input material not subjected to ChIP was used a positive PCR control. ChIP with isotype-matched IgG was used as negative control. Isolated genomic material was subjected to PCR, and the resulting products were analyzed by agarose gel electrophoresis. (A, C) PCR with primers specific to the Gata4 G9 ECR demonstrated that the region of the endogenous bovine Gata4 G9 enhancer containing the ETS sites was bound by ETS1 (A) and ERG (C) in vivo. (B, D) PCR with primers designed to detect a region of the bovine Gata4 locus 5 kb from the G9 enhancer shows that neither ETS1 (B) nor ERG (D) bound to this region distal to G9 under identical conditions in which G9 was bound in (A, C). Sizes in bp are indicated at the left of each panel. H2O (water) indicates a negative PCR control with no added template.
DISCUSSION
Regulation of Gata4 expression in the heart
GATA4 is an important transcriptional regulator of myocardial and endocardial development, and numerous studies have defined its requirement and downstream gene targets in those lineages (Garg et al., 2003; Oka et al., 2006; Rivera-Feliciano et al., 2006; Rojas et al., 2008; Zeisberg et al., 2005). However, prior to the present study, direct transcriptional regulation of the Gata4 gene itself had not been defined in any cardiovascular lineage. The Gata4 G9 enhancer described here overlaps almost perfectly with GATA4 expression in the endocardium but, unlike endogenous GATA4 protein, it is largely absent from the the myocardium after E8.5 (Fig. 2 and Fig. 3; Oka et al., 2006; Zeisberg et al., 2005), suggesting that other regulatory modules must control Gata4 expression in myocardial cells. To date, we have not identified any myocardial enhancers based solely on sequence conservation, suggesting the Gata4 locus includes non-conserved enhancer elements. Consistent with this notion, recent work identifying enhancers by combining ChIP and deep-sequencing demonstrated that many functional enhancers in numerous cardiac genes exist outside of regions of sequence conservation (Blow et al., 2010; Visel et al., 2009).
A recent genome-wide ChIP-seq analysis using biotinylated cardiac transcription factors expressed in the cardiomyocyte cell line HL1 identified the Gata4 G9 region as a bound target, suggesting myocardial occupancy of the enhancer by GATA4 and myocardial-specific activation (He et al., 2011). By contrast, our detailed analyses in transgenic mice demonstrate that the element is predominantly endocardial, although we observed incomplete, weak myocardial activity in a minority of the lines at linear and looping heart stages (Fig. 2, Fig. 3). Furthermore, mutation of GATA sites in the ECR, which were bound by GATA4 in EMSA, did not negatively affect enhancer activity in the hearts of transgenic embryos at any stage examined (data not shown). Thus, it is clear that additional approaches will be needed to fully describe the regulation of Gata4 in the myocardium.
Regulation of endocardial gene expression by ETS transcription factors
ETS factors are essential regulators of endothelial-restricted gene expression (De Val and Black, 2009), and a regulatory role in the endocardium further reinforces the notion that endocardium is a specialized endothelium (Harris and Black, 2010). Here, we analyzed the ETS transcript expression profile in the early embryonic heart and found that transcripts for FLI1, ETV5, ETV6, ETS1 ERG, ELF1, ETV1, and ETS2 were the most highly expressed in the early embryonic heart (Fig. 7). Based on our data, the ETS expression profile of the embryonic heart shares more similarities with the HUVEC profile than the adult heart profile (Hollenhorst et al., 2004), suggesting distinct embryonic and adult ETS profiles for the heart and that the expression profiles of ETS factors in the embryonic heart are dominated by expression in the endocardium.
Our data support a model in which Gata4 is directly regulated by ETS1, ERG, or both factors in the developing heart because enhancer activity requires ETS-binding sites and ETS1 and ERG occupy the enhancer in BAEC. However, it is important to note that our studies do not exclude other ETS factors as potential transcriptional regulators nor do they prove that ETS1 and ERG regulate Gata4 in the embryonic heart. ETS1 and ERG each play important roles in endothelial gene regulation and function (Gao et al., 2010; Hashiya et al., 2004; Loughran et al., 2008; Nikolova-Krstevski et al., 2009; Pham et al., 2007; Taoudi et al., 2011; Wei et al., 2009; Ye et al., 2010), and ETS1 and ERG are expressed broadly throughout the endothelium, including the endocardium (Fig. 7; Ye et al., 2010). The Gata4 G9 enhancer is active only in a subset of the ETS1 and ERG expression domains, so it is unclear how Gata4 G9 enhancer activity becomes restricted within these domains. There are several possible explanations. It is possible that the Gata4 G9 enhancer responds only to high ETS protein levels, as a recent study that analyzed ERG expression in different vascular beds of the heart found that ERG was enriched in endocardial endothelial cells compared to venule, artery, and capillary endothelial cells (Yuan et al.). A second possibility is that additional activators expressed exclusively in the Gata4-G9-lacZ expression domain may be required in combination with ETS factors for enhancer activation to occur. A third possibility is that the Gata4 G9 enhancer may contain binding sites for a repressor that restricts its activity in areas of endothelium other than endocardium. Identification of the mechanisms that restrict expression should help explain the long-standing observation that endocardium is a molecularly and functionally unique endothelium, and future studies will address the molecular mechanisms controlling this restriction.
Origins of the endocardium
The endocardium shares many properties with other endothelial populations, but mounting evidence suggests the endocardium is more than just a spatially restricted subpopulation of endothelium. Cardiovascular lineages, including endocardium, arise from multipotent FLK1+ cells that are present during gastrulation (Fehling et al., 2003; Kattman et al., 2006), and cardiogenic mesoderm leaves the primitive streak after mesoderm fated to become other endothelial populations has already exited (Kouskoff et al., 2005; Misfeldt et al., 2009). Interestingly, GATA4 protein was detectable, and the Gata4 G9 enhancer was active, in the allantois (Fig 2D–F, Fig 4B–D), which contains FLK1+ vasculogenic tissue (Downs et al., 1998), suggesting the endocardium and allantois may share common regulatory pathways.
Using embryonic stem cells bearing an Nfatc1-lacZ BAC transgene to track endocardial cell production, Misfeldt et al., demonstrated that culturing ES cells with Wnt3a gave rise to endothelial cells, while culturing ES cells with Noggin and Dickkopf-1 gave rise to endocardial and myocardial cells (Misfeldt et al., 2009), suggesting developing endocardium responds to cardiogenic signals. Several studies have demonstrated the close association between endocardial and myocardial progenitors (Kattman et al., 2006; Masino et al., 2004) and the data presented here, that the Gata4 G9 enhancer is briefly active in both myocardial and endocardial cell types prior to restricting to the endocardium and endocardial cushions, also supports the close relationship between endocardial and myocardial cells.
Transcriptional pathways in congenital heart disease
Analyses of the specific mutations in human GATA4 and mechanistic studies in model systems suggest that disruption of GATA4 transactivation function or interaction with other transcription factors may be the underlying reason for the defects associated with GATA4 mutations (Garg et al., 2003; Moskowitz et al., 2011; Schluterman et al., 2007). Together, these studies suggest that a greater understanding of the transcription factor interactions and pathways involving GATA4 may lead to a clearer understanding of the mechanisms underlying endocardially-derived heart defects. Consistent with this idea, ETS1 mutations have also recently been associated with endocardium-derived defects in humans (Ye et al., 2010).
An ultimate goal of defining transcriptional pathways of cardiac development is to clarify the mechanisms underlying cardiac defects. GATA4 mutations in humans are strongly associated with valve and septation defects (Butler et al., 2010; Garg et al., 2003; Hirayama-Yamada et al., 2005; Zhang et al., 2008). The valve leaflets and the membranous portions of the septa are largely derived from endothelial cells of endocardial origin (Harris and Black, 2010; Person et al., 2005), suggesting that GATA4 function in endocardial derivatives is essential for proper valve and septal development in humans. Inactivation of Gata4 in endothelial cells, including the endocardium, results in embryonic lethality and profound endocardial cushion and valve leaflet defects that mimic the heritable human diseases observed in families with GATA4 mutations (Rivera-Feliciano et al., 2006). Our work implicates ETS1 and ERG as direct regulators of Gata4 via the Gata4 G9 enhancer, and as such, suggests disturbances in ETS1 or ERG function in the endocardium could underlie cardiac diseases. This notion is supported by the recent report that deletion of ETS1 was observed in patients with Jacobsen syndrome, and targeted disruption of Ets1 in mice led to malformations in cardiac structures derived from endocardial cells (Ye et al., 2010). It would interesting to determine whether mutations in other ETS factors that are expressed in the heart, including ERG, or in ETS-dependent noncoding regulatory regions, such as the Gata4 G9 enhancer described in these studies, are also associated with endocardial malformations in humans.
Highlights.
-
>
A distal enhancer of Gata4 is active in cardiac lineages in transgenic mouse embryos.
-
>
Activity of the Gata4 enhancer restricts to the endocardium by E9.5 in the mouse.
-
>
The Gata4 cardiac enhancer is dependent on 4 ETS transcription factor binding sites.
-
>
The Gata4 cardiac enhancer is bound by Ets1 and Erg.
-
>
We examine the expression of all ETS family members in the mouse embryonic heart at E8.5.
ACKNOWLEDGEMENTS
The authors thank Deepak Srivastava and Benoit Bruneau for helpful comments. This work was supported by grants R01 HL64658 and P01 HL89707 from the NHLBI to BLB. WS was supported by a Predoctoral Fellowship from the American Heart Association, Western States Affiliate.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson JP, Dodou E, Heidt AB, De Val SJ, Jaehnig EJ, Greene SB, Olson EN, Black BL. HRC is a direct transcriptional target of MEF2 during cardiac, skeletal, and arterial smooth muscle development in vivo. Mol Cell Biol. 2004;24:3757–3768. doi: 10.1128/MCB.24.9.3757-3768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries LJ, Brutsaert DL. Endocardial endothelium in the rat: cell shape and organization of the cytoskeleton. Cell Tissue Res. 1993;273:107–117. doi: 10.1007/BF00304617. [DOI] [PubMed] [Google Scholar]
- Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–498. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- Arceci RJ, King AA, Simon MC, Orkin SH, Wilson DB. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- Brutsaert DL, Andries LJ. The endocardial endothelium. Am J Physiol. 1992;263:H985–H1002. doi: 10.1152/ajpheart.1992.263.4.H985. [DOI] [PubMed] [Google Scholar]
- Butler TL, Esposito G, Blue GM, Cole AD, Costa MW, Waddell LB, Walizada G, Sholler GF, Kirk EP, Feneley M, Harvey RP, Winlaw DS. GATA4 mutations in 357 unrelated patients with congenital heart malformation. Genet Test Mol Biomarkers. 2010;14:797–802. doi: 10.1089/gtmb.2010.0028. [DOI] [PubMed] [Google Scholar]
- Charron F, Nemer M. GATA transcription factors and cardiac development. Semin Cell Dev Biol. 1999;10:85–91. doi: 10.1006/scdb.1998.0281. [DOI] [PubMed] [Google Scholar]
- Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circ Res. 2009;105:408–421. doi: 10.1161/CIRCRESAHA.109.201566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, Anderson JP, Heidt AB, Khiem D, Xu SM, Black BL. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev Biol. 2004;275:424–434. doi: 10.1016/j.ydbio.2004.08.016. [DOI] [PubMed] [Google Scholar]
- De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, Pennacchio LA, Dubchak I, Krieg PA, Stainier DY, Black BL. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Dodou E, Xu SM, Black BL. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech Dev. 2003;120:1021–1032. doi: 10.1016/s0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- Downs KM, Gifford S, Blahnik M, Gardner RL. Vascularization in the murine allantois occurs by vasculogenesis without accompanying erythropoiesis. Development. 1998;125:4507–4520. doi: 10.1242/dev.125.22.4507. [DOI] [PubMed] [Google Scholar]
- Drake CJ, Fleming PA. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- Fehling HJ, Lacaud G, Kubo A, Kennedy M, Robertson S, Keller G, Kouskoff V. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- Flamme I, Frolich T, Risau W. Molecular mechanisms of vasculogenesis and embryonic angiogenesis. J Cell Physiol. 1997;173:206–210. doi: 10.1002/(SICI)1097-4652(199711)173:2<206::AID-JCP22>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Galang CK, Muller WJ, Foos G, Oshima RG, Hauser CA. Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem. 2004;279:11281–11292. doi: 10.1074/jbc.M311887200. [DOI] [PubMed] [Google Scholar]
- Gao Z, Kim GH, Mackinnon AC, Flagg AE, Bassett B, Earley JU, Svensson EC. Ets1 is required for proper migration and differentiation of the cardiac neural crest. Development. 2010;137:1543–1551. doi: 10.1242/dev.047696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Garrett-Sinha LA, Dahl R, Rao S, Barton KP, Simon MC. PU.1 exhibits partial functional redundancy with Spi-B, but not with Ets-1 or Elf-1. Blood. 2001;97:2908–2912. doi: 10.1182/blood.v97.9.2908. [DOI] [PubMed] [Google Scholar]
- Gory S, Dalmon J, Prandini MH, Kortulewski T, de Launoit Y, Huber P. Requirement of a GT box (Sp1 site) and two Ets binding sites for vascular endothelial cadherin gene transcription. J Biol Chem. 1998;273:6750–6755. doi: 10.1074/jbc.273.12.6750. [DOI] [PubMed] [Google Scholar]
- Graves BJ, Petersen JM. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- Harris IS, Black BL. Development of the endocardium. Pediatr Cardiol. 2010;31:391–399. doi: 10.1007/s00246-010-9642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiya N, Jo N, Aoki M, Matsumoto K, Nakamura T, Sato Y, Ogata N, Ogihara T, Kaneda Y, Morishita R. In vivo evidence of angiogenesis induced by transcription factor Ets-1: Ets-1 is located upstream of angiogenesis cascade. Circulation. 2004;109:3035–3041. doi: 10.1161/01.CIR.0000130643.41587.DB. [DOI] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Huber RM, Ladle DR, Murphy K, Arber S. ETS transcription factor Erm controls subsynaptic gene expression in skeletal muscles. Neuron. 2007;55:726–740. doi: 10.1016/j.neuron.2007.07.028. [DOI] [PubMed] [Google Scholar]
- Hirayama-Yamada K, Kamisago M, Akimoto K, Aotsuka H, Nakamura Y, Tomita H, Furutani M, Imamura S, Takao A, Nakazawa M, Matsuoka R. Phenotypes with GATA4 or NKX2.5 mutations in familial atrial septal defect. Am J Med Genet A. 2005;135:47–52. doi: 10.1002/ajmg.a.30684. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and Biochemical Insights into the Specificity of ETS Transcription Factors. Annu Rev Biochem Annu Rev Biochem. 2011;80:437–71. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin Cell Dev Biol. 2007;18:101–110. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Kouskoff V, Lacaud G, Schwantz S, Fehling HJ, Keller G. Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. Proc Natl Acad Sci U S A. 2005;102:13170–13175. doi: 10.1073/pnas.0501672102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, Janknecht R, Lim DS, Choi K. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore JJ, Mericko PA, Cheng L, Lu MM, Morrisey EE, Parmacek MS. GATA-6 regulates semaphorin 3C and is required in cardiac neural crest for cardiovascular morphogenesis. J Clin Invest. 2006;116:929–939. doi: 10.1172/JCI27363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, Henley KJ, Ellis S, Voss AK, Metcalf D, Hilton DJ, Alexander WS, Kile BT. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol. 2008;9:810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- Lugus JJ, Chung YS, Mills JC, Kim SI, Grass J, Kyba M, Doherty JM, Bresnick EH, Choi K. GATA2 functions at multiple steps in hemangioblast development and differentiation. Development. 2007;134:393–405. doi: 10.1242/dev.02731. [DOI] [PubMed] [Google Scholar]
- Markwald RR, Norris RA, Moreno-Rodriguez R, Levine RA. Developmental basis of adult cardiovascular diseases: valvular heart diseases. Ann N Y Acad Sci. 2010;1188:177–183. doi: 10.1111/j.1749-6632.2009.05098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masino AM, Gallardo TD, Wilcox CA, Olson EN, Williams RS, Garry DJ. Transcriptional regulation of cardiac progenitor cell populations. Circ Res. 2004;95:389–397. doi: 10.1161/01.RES.0000138302.02691.be. [DOI] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA : visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- Melax H, Leeson TS. Fine structure of the endocardium in adult rats. Cardiovasc Res. 1967;1:349–355. doi: 10.1093/cvr/1.4.349. [DOI] [PubMed] [Google Scholar]
- Misfeldt AM, Boyle SC, Tompkins KL, Bautch VL, Labosky PA, Baldwin HS. Endocardial cells are a distinct endothelial lineage derived from Flk1+ multipotent cardiovascular progenitors. Dev Biol. 2009;333:78–89. doi: 10.1016/j.ydbio.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Molkentin JD. The zinc finger-containing transcription factors GATA-4, −5, and −6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Wang J, Peterson MA, Pu WT, Mackinnon AC, Oxburgh L, Chu GC, Sarkar M, Berul C, Smoot L, Robertson EJ, Schwartz R, Seidman JG, Seidman CE. Cardiac-specific transcription factor genes Smad4 and Gata4 cooperatively regulate cardiac valve development. Proc Natl Acad Sci U S A. 2011;108:4006–4011. doi: 10.1073/pnas.1019025108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova-Krstevski V, Yuan L, Le Bras A, Vijayaraj P, Kondo M, Gebauer I, Bhasin M, Carman CV, Oettgen P. ERG is required for the differentiation of embryonic stem cells along the endothelial lineage. BMC Dev Biol. 2009;9:72. doi: 10.1186/1471-213X-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Maillet M, Watt AJ, Schwartz RJ, Aronow BJ, Duncan SA, Molkentin JD. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98:837–845. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates) Curr Opin Genet Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Person AD, Klewer SE, Runyan RB. Cell biology of cardiac cushion development. Int Rev Cytol. 2005;243:287–335. doi: 10.1016/S0074-7696(05)43005-3. [DOI] [PubMed] [Google Scholar]
- Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, Weinstein BM. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimanda JE, Chan WY, Wilson NK, Smith AM, Kinston S, Knezevic K, Janes ME, Landry JR, Kolb-Kokocinski A, Frampton J, Tannahill D, Ottersbach K, Follows GA, Lacaud G, Kouskoff V, Gottgens B. Endoglin expression in blood and endothelium is differentially regulated by modular assembly of the Ets/Gata hemangioblast code. Blood. 2008;112:4512–4522. doi: 10.1182/blood-2008-05-157560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimanda JE, Ottersbach K, Knezevic K, Kinston S, Chan WY, Wilson NK, Landry JR, Wood AD, Kolb-Kokocinski A, Green AR, Tannahill D, Lacaud G, Kouskoff V, Gottgens B. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci U S A. 2007;104:17692–17697. doi: 10.1073/pnas.0707045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristevski S, O’Leary DA, Thornell AP, Owen MJ, Kola I, Hertzog PJ. The ETS transcription factor GABPalpha is essential for early embryogenesis. Mol Cell Biol. 2004;24:5844–5849. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Feliciano J, Lee KH, Kong SW, Rajagopal S, Ma Q, Springer Z, Izumo S, Tabin CJ, Pu WT. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133:3607–3618. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, Black BL. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development. 2005;132:3405–3417. doi: 10.1242/dev.01913. [DOI] [PubMed] [Google Scholar]
- Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, Schachterle W, Xu SM, Black BL. An endoderm-specific transcriptional enhancer from the mouse Gata4 gene requires GATA and homeodomain protein-binding sites for function in vivo. Dev Dyn. 2009;238:2588–2598. doi: 10.1002/dvdy.22091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, Schachterle W, Xu SM, Martin F, Black BL. Direct transcriptional regulation of Gata4 during early endoderm specification is controlled by FoxA2 binding to an intronic enhancer. Dev Biol. 2010;346:346–355. doi: 10.1016/j.ydbio.2010.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronicke V, Risau W, Breier G. Characterization of the endothelium-specific murine vascular endothelial growth factor receptor-2 (Flk-1) promoter. Circ Res. 1996;79:277–285. doi: 10.1161/01.res.79.2.277. [DOI] [PubMed] [Google Scholar]
- Sato Y. Role of ETS family transcription factors in vascular development and angiogenesis. Cell Struct Funct. 2001;26:19–24. doi: 10.1247/csf.26.19. [DOI] [PubMed] [Google Scholar]
- Schlaeger TM, Bartunkova S, Lawitts JA, Teichmann G, Risau W, Deutsch U, Sato TN. Uniform vascular-endothelial-cell-specific gene expression in both embryonic and adult transgenic mice. Proc Natl Acad Sci U S A. 1997;94:3058–3063. doi: 10.1073/pnas.94.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluterman MK, Krysiak AE, Kathiriya IS, Abate N, Chandalia M, Srivastava D, Garg V. Screening and biochemical analysis of GATA4 sequence variations identified in patients with congenital heart disease. Am J Med Genet A. 2007;143A:817–823. doi: 10.1002/ajmg.a.31652. [DOI] [PubMed] [Google Scholar]
- Snarr BS, Kern CB, Wessels A. Origin and fate of cardiac mesenchyme. Dev Dyn. 2008;237:2804–2819. doi: 10.1002/dvdy.21725. [DOI] [PubMed] [Google Scholar]
- Song H, Suehiro J, Kanki Y, Kawai Y, Inoue K, Daida H, Yano K, Ohhashi T, Oettgen P, Aird WC, Kodama T, Minami T. Critical role for GATA3 in mediating Tie2 expression and function in large vessel endothelial cells. J Biol Chem. 2009;284:29109–29124. doi: 10.1074/jbc.M109.041145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, Watson DK. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol. 2000;20:5643–5652. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, Chang CP. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoudi S, Bee T, Hilton A, Knezevic K, Scott J, Willson TA, Collin C, Thomas T, Voss AK, Kile BT, Alexander WS, Pimanda JE, Hilton DJ. ERG dependence distinguishes developmental control of hematopoietic stem cell maintenance from hematopoietic specification. Genes Dev. 2011;25:251–262. doi: 10.1101/gad.2009211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Siddiqui MA. Signal transduction in early heart development (II): ventricular chamber specification, trabeculation, and heart valve formation. Exp Biol Med (Maywood) 2007;232:866–880. [PubMed] [Google Scholar]
- Wang LC, Kuo F, Fujiwara Y, Gilliland DG, Golub TR, Orkin SH. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. Embo J. 1997;16:4374–4383. doi: 10.1093/emboj/16.14.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Srinivasan R, Cantemir-Stone CZ, Sharma SM, Santhanam R, Weinstein M, Muthusamy N, Man AK, Oshima RG, Leone G, Ostrowski MC. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009;114:1123–1130. doi: 10.1182/blood-2009-03-211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Davis CA, Molkentin JD, Lien CL, Duncan SA, Richardson JA, Olson EN. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc Natl Acad Sci U S A. 2006;103:11189–11194. doi: 10.1073/pnas.0604604103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Coldren C, Liang X, Mattina T, Goldmuntz E, Benson DW, Ivy D, Perryman MB, Garrett-Sinha LA, Grossfeld P. Deletion of ETS-1, a gene in the Jacobsen syndrome critical region, causes ventricular septal defects and abnormal ventricular morphology in mice. Hum Mol Genet. 2010;19:648–656. doi: 10.1093/hmg/ddp532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Sacharidou A, Stratman AN, Le Bras A, Zwiers PJ, Spokes K, Bhasin M, Shih SC, Nagy JA, Molema G, Aird WC, Davis GE, Oettgen P. RhoJ is an endothelial cell-restricted Rho GTPase that mediates vascular morphogenesis and is regulated by the transcription factor ERG. Blood. 2011 doi: 10.1182/blood-2010-10-315275. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg EM, Ma Q, Juraszek AL, Moses K, Schwartz RJ, Izumo S, Pu WT. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J Clin Invest. 2005;115:1522–1531. doi: 10.1172/JCI23769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li X, Shen A, Jiao W, Guan X, Li Z. GATA4 mutations in 486 Chinese patients with congenital heart disease. Eur J Med Genet. 2008;51:527–535. doi: 10.1016/j.ejmg.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, Duncan SA. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol. 2008;317:614–619. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]