Abstract
The side chain carboxyl groups of acidic proteins found in the extra-cellular matrix (ECM) of mineralized tissues play a key role in promoting or inhibiting the growth of minerals such as hydroxyapatite (HAP), the principal mineral component of bone and teeth. Among the acidic proteins found in the saliva is statherin, a 43-residue tyrosine-rich peptide that is a potent lubricant in the salivary pellicle and an inhibitor of both HAP crystal nucleation and growth. Three acidic amino acids – D1, E4, and E5 – are located in the N-terminal 15 amino acid segment, with a fourth amino acid, E26, located outside the N-terminus. We have utilized 13C{31P} REDOR NMR to analyze the role played by acidic amino acids in the binding mechanism of statherin to the HAP surface by measuring the distance between the δ-carboxyl 13C spins of the three glutamic acid side chains of statherin (residues E4, E5, E26) and 31P spins of the phosphate groups at the HAP surface. 13C{31P} REDOR studies of glutamic-5-13C acid incorporated at positions E4 and E26 indicate a 13C–31P distance of more than 6.5 Å between the side chain carboxyl 13C spin of E4 and the closest 31P in the HAP surface. In contrast, the carboxyl 13C spin at E5 has a much shorter 13C–31P internuclear distance of 4.25±0.09 Å, indicating that the carboxyl group of this side chain interacts directly with the surface. 13C T1ρ and slow-spinning MAS studies indicate that the motions of the side chains of E4 and E5 are more restricted than that of E26. Together, these results provide further insight into the molecular interactions of statherin with HAP surfaces.
Introduction
The remarkable material properties of bone and teeth arise because of the sophisticated crystal engineering capabilities that have evolved in the biological world. Proteins function at the interface between cells and the inorganic phase in these composite hard tissues. The activities of these proteins at the organic-inorganic interface are thus critical not only to the construction of biocomposites with improved mechanical properties but to the maintenance of hard tissue integrity as well. The disruption of these functions has adverse medical and dental ramifications, leading to bone and tooth deterioration, dental calculus formation, atherosclerotic plaque formation, artificial heart valve calcification, kidney and gall stone build-up1–3.
There has been considerable attention focused on the role of acidic proteins in biomineralization processes4–10. It is widely recognized, however, that the molecular details of protein function at the organic-inorganic interface are just beginning to be uncovered. The significance of pushing this frontier will be found in the development of calcification inhibitors and promoters that could impact orthopedics, urology, dentistry, and cardio-vascular fields. A better understanding of how these proteins recognize and assemble in bioactive fashion on inorganic mineral phases could also aid in the development of surface coatings to improve the biocompatibility of implantable biomaterials and tissue engineering scaffolds.
The mineralization of bone occurs by the deposition of hydroxyapatite (HAP, Ca10(PO4)6(OH)2 ) in the extra-cellular matrix. It has long been hypothesized that acidic protein components of the ECM are involved in the initiation and regulation of mineralization. Bone sialoprotein and osteopontin, for example, are extensively phosphorylated and contain regions that are rich in acidic amino acids11,12. Sialoprotein contains two regions consisting of contiguous glutamic acid residues and a RGD cell attachment sequence. The direct involvement of these acidic amino acid regions in HAP nucleation is further supported by studies of the effect of monomeric acidic amino acids and their polymers in HAP nucleation13. Both glutamic acid and aspartic acid markedly inhibit the rate of HAP crystal growth14. Polyglutamic acid nucleates apatites15, and polyaspartate increases the amount of calcium phosphate crystals within assembling collagen fibers16–18.
Acidic ECM proteins play a similarly important role in the regulation of HAP nucleation in the oral cavity. Dentin Matrix Protein 1 is essential in the maturation of odontoblasts and osteoblasts and subsequent dentin mineralization19; amelogenin is crucial to proper formation of enamel20, yet the operations of neither are well understood. Functional studies using NMR measurements carried out on a derivative of amelogenin have shown its structure is largely random coil when adsorbed to hydroxyapatite, and identified 3 residues that are within 7 Å from the surface of HAP21.
Statherin is an acidic human salivary protein of 43 amino acid residues, DpSpSEEKFLRRIGRFGYGYGPYQPVPEQPLYPQPYQPQYQQYTF (pS=phosphoserine), with a molecular weight of 5.38 kDa. It inhibits both the nucleation and growth of HAP. Previous studies of statherin fragments by Nancollas and coworkers22,23 showed that the N-terminal 15 amino acid fragment (SN-15, DpSpSEEKFLRRIGRFG, pS=phosphoserine) is essential for binding to HAP crystal surfaces. Removal of the DpSpSE moiety (SN-11, EKFLRRIGRFG) reduces the HAP binding affinity by a factor of 4.5 compared to SN-15. Mutation of both phosphoserines to simple serine reduces the HAP binding affinity by a factor of almost nine, while mutation to aspartic acid restores the binding affinity to 70% that of SN-15, indicating that the phosphoserine residues and acidic amino acid side chains are important for the binding of statherin to HAP.
Circular dichroism and solution NMR studies indicate that SN-15 is transiently helical in solution24. For this same moiety in HAP-bound statherin, measurement of a number of torsion angles and internuclear distances using solid state NMR indicate that the N-terminus is an α-helix25–26. Solid state NMR constraints and the Rosetta structure prediction algorithm have been used to obtain a model of the structure of HAP-bound statherin27. This model confirms the helicity of the N-terminus and attributes a helical structure to the C-terminus as well. Gray and coworkers28 have developed a novel computational approach for modeling the interactions of statherin with HAP crystal surfaces. Using an all-atom Monte Carlo-minimization search algorithm to optimize rigid body and side chain conformations for binding to the 001 face of the HAP crystal, they predict side chain-surface interactions involving residues pS2, pS3, K6, R9, R10, R13, Y16, Q32, and P33. Solid state NMR36 and calorimetric data29 confirm the existence of some of these computationally-predicted protein-surface interactions including protein surface interactions involving several basic amino acid side chains (e.g. K6, R9, R10, and R13) as well as several non-polar amino acids including L8 and F1430,41.
In this paper, we augment our knowledge of the fundamental nature of protein-surface interactions by using 13C{31P} Rotational Echo DOuble Resonance (REDOR)39 to directly detect distances between the carboxyl 13C spins in glutamic acid side chains in statherin and 31P spins located in the HAP crystal surface. Since dipolar recoupling of 13C and 31P via REDOR is effective over distances as great as 7 Å, the possibility exists that a given side chain carboxyl 13C spin may couple to numerous 31P spins in the HAP surface, which may complicate the analysis. Consistent with previous work involving similar interactions between the ε-15N spin of the side chain of K6 and the HAP surface40, fits between the side chain carboxyl 13C spins in glutamic acid and several 31P spins will be considered. This structural data is further supplemented by variable-temperature measurements of the rotating-frame spin-lattice relaxation time (T1ρ), which are sensitive to the dynamics and solvent mobility near the 13C label.
Materials and Methods
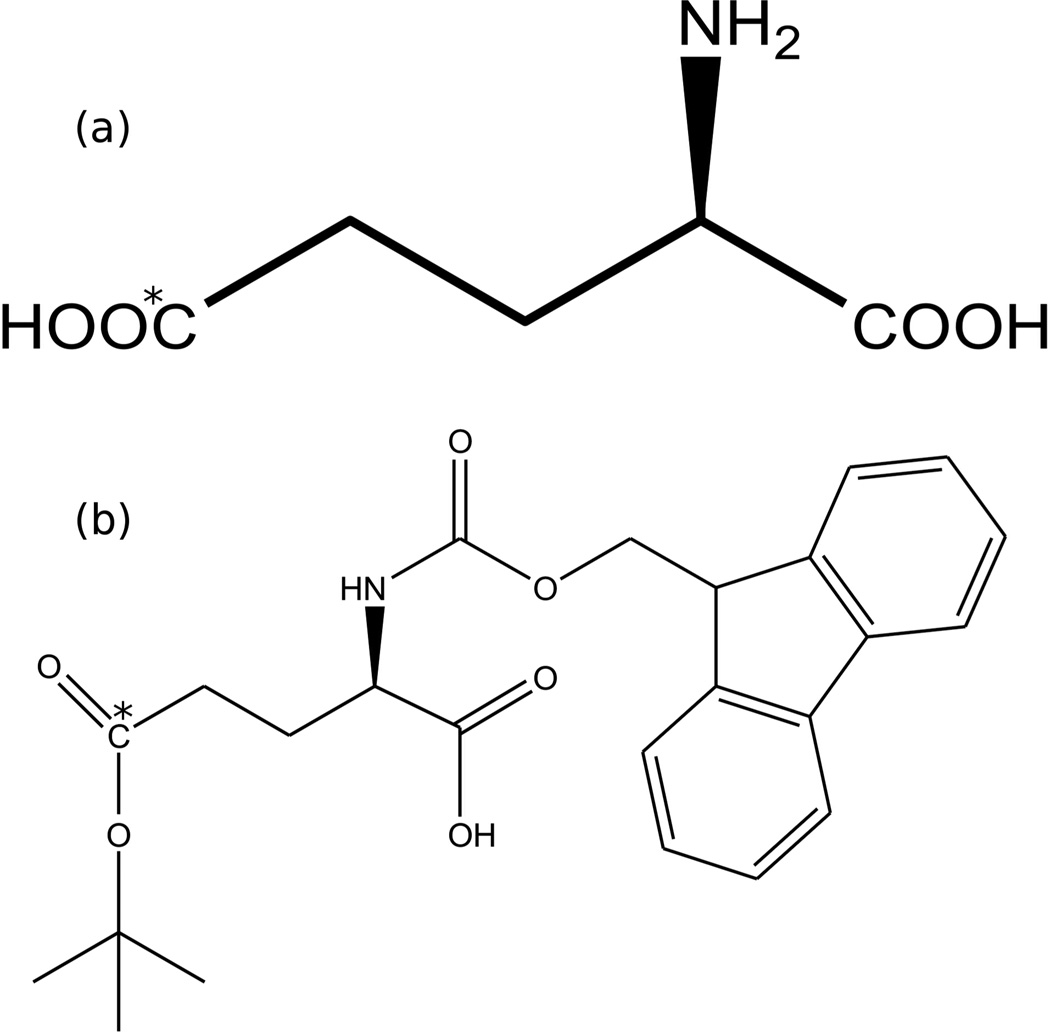
l-Glutamic-5-13C acid, 99% enrichment (Figure 1A) was purchased from Isotec (Sigma-Aldrich). N-Fmoc-l-Glu-5-13C-γ-tert-butyl ester (Figure 1B) was made using a four step protection and de-protection reaction described in the literature31, with variations which resulted in considerably higher yield.
Figure 1.
Structure of (a) glutamic acid, 5-13C, (b) Fmoc-glutamic acid (5-13C) t-butyl ester. 13C labeled site indicated with asterisk.
A: Synthesis of N-Fmoc-l-Glu-5-13C Acid (2)
L-glutamic acid, 99% 5-13C-labeled (0.970 g) was placed in a 100 mL round bottom flask with 15 mL of warm 10% aqueous sodium carbonate (Na2CO3) in which it was dissolved. Fmoc-OSu (9-fluorenyl methyl oxycarbonyl-N-hydroxysuccinimide, 2.70 g) was dissolved in 15 mL of dioxane and added to the amino acid solution. This mixture was stirred for 24 hours at room temperature. The solution was diluted with 100 mL of water and washed thrice with 50 mL of ethyl ether. The aqueous phase was acidified to pH 2 with 2 M HCl. The white solid was then extracted with 100 mL of ethyl acetate. Sodium chloride was added to the aqueous layer and again extracted with 50 mL of ethyl acetate. The combined ethyl acetate extractions were washed with brine, dried over magnesium sulfate, and concentrated. The N-Fmoc-glutamic acid, 5-13C was confirmed with 1H NMR (MeOH-d4) and mass spectrometry. The product mass was 2.42 g (100% yield).
B: Synthesis of N-Fmoc-l-Glu-5-13C γ-Benzyl Ester (3) and γ-Benzyl Ester (3')
N-Fmoc-L-Glu-5-13C (2), (2.42 g, 6.5 mmol) and N,N'-Dicyclohexylcarbodiimide (1.34 g, 6.5 mmol) were dissolved in 5 mL tetrahydrofuran (THF) and mixed for 23 hours. The resulting mixture was filtered and the filtrate was evaporated leaving a crude anhydride. This material was dissolved in 5 mL of THF and the solution was treated with benzyl alcohol (1.40 mL) and potassium carbonate, K2CO3 (1.2 g, 8.96 mmol). After 5 hours, the reaction was filtered and the solid was washed with dichloromethane, and the resultant solution washed with 1N HCl and saturated brine, then dried. The crude residue on evaporation was chromatographed over silica gel (100 g), eluted first with 100% CH2Cl2 and then 3% MeOH / 97% CH2Cl2. The α-benzyl ester (2.0372 g, 75% yield) eluted first: 1H NMR (CDCl3) δ 1.7–2.6 (m, 4H), 4.0–4.6 (m 4H), 5.0–5.2 (s, 2H), 5.6–5.9 (d, 1H), 7.1–7.8 (m, 13H). The γ-benzyl ester (494.5 mg, 18% yield) eluted later, also characterized by 1H NMR (CDCl3).
C: Synthesis of N-Fmoc-l-Glu-5-13C α-Benzyl, γ-tert-Butyl Ester (4)
N-Fmoc-l-Glu-5-13C α-benzyl ester (3), (2.0372 g, 4.4 mmol), concentrated sulfuric acid (0.5 mL), and CH2Cl2 (20 mL) were added to a Parr bottle and cooled to −78°C in a dry ice-acetone bath. To this was added isobutylene (20 mL) condensed at −78°C. The resultant mixture was attached to a Parr apparatus and shaken for 23 hours at 25°C. The reaction was then vented, diluted with methylene chloride and extracted with 5% aqueous sodium bicarbonate (NaHCO3). The organic layer was dried and evaporated. The crude residue was chromatographed over silica gel (200 g) and eluted with 20% ethyl acetate in n-hexane. The product (1.8794 g, 83% yield) was isolated as a colorless solid: 1H NMR (CDCl3) δ1.4 (s, 9H), 1.8–2.4 (m, 4H), 3.9–4.6 (m, 4H), 5.2 (s, 2H), 5.6–5.8 (d, 1H), 7.1–7.8 (m, 13H).
D: Synthesis of N-Fmoc-l-Glu-5-13C γ-tert-Butyl Ester (5)
A hydrogenation reaction using N-Fmoc-l-Glu-5-13C α-benzyl, γ-tert-butyl ester (4), (1.8794 g, 3.6 mmol), 10% Pd/C (200 mg) in methanol (30 mL), and tetrahydrofuran (30 mL) was performed at 50 psi for 5 hours. The reaction mixture was diluted with ethyl acetate and filtered over diatomaceous earth. The residue was washed with additional ethyl acetate, and the combined filtrates were evaporated. The crude product was chromatographed over silica gel (200 g) and eluted with 1:40:59 acetic acid: ethyl acetate: n-hexane, giving the product as a colorless solid (1.3839 g, 89% yield): 1H NMR (CDCl3) δ1.4 (s, 9H), 1.8–2.6 (m, 4H), 4.1–4.7 (m, 4H), 5.6–5.8 (d, 1H), 7.1–7.8 (m, 13H). The overall yield was 56%.
Statherin Synthesis
N-Fmoc-l-Glu-5-13C γ-tert-butyl ester was used to synthesize three statherin samples: statherin[4-Glu[5-13C]] (stE4), statherin[5-Glu[5-13C]] (stE5), and statherin[26-Glu[5-13C]] (stE26). Peptide synthesis was conducted on an automated synthesizer (Rainin PS3, Protein Technologies, Tucson, AZ) using standard Fmoc chemistry, with N-methyl-2-pyrrolidinone (NMP) as the solvent, 20% piperidine in NMP as the deprotecting reagent, and 0.4 M N-methylmorpholine in dimethyl formamide as the activating reagent. The solid phase support was Fmoc-Phe-TGA resin from Novabiochem (417 mg, 0.24 mmol/g substitution). Cleavage of the peptide from the solid support was achieved by stirring gently for 2 hours in a covered solution of 95% trifluoroacetic acid, 2.5% triisopropylsilane, and 2.5% water. The protein was precipitated from this solution in a cold (−20°C) solution of tert-butyl methyl ether.
Protein Purification
The protein was purified on a bioCAD SPRINT FPLC using a 10 mm × 100 mm, 7.9 mL, POROS HQ/M perfusion anion-exchange column. The mobile phase consisted of (A) 50 mM Tris-HCl at pH 8.5, (B) 0.5M NaCl and buffer A, (C) acetonitrile, with the latter held at 30% to maintain protein solubility. The crude peptide was dissolved in a mixture of 30% acetonitrile, 70% buffer A solution (10 mg peptide/mL), and 3 mL injection volumes into a 4 mL loop were utilized to load the column. A linear gradient from 0 to 70% B in 14.7 column volumes was used to elute the protein at a flow rate of 3 mL/min. UV absorption at a wavelength of 280 nm was used to detect the desired product, which eluted at 35% buffer B. The protein was then concentrated using a Centriplus 3 kDa MWCO centrifugal filter unit. Following purification, the sample was lyophilized, and then analyzed by mass spectrometry for composition and purity.
Statherin Binding to Hydroxyapatite (HAP, Ca10(H2O)2(PO4)6)
Hydroxyapatite seed crystals were generously provided by W. Shaw and A. Campbell (Battelle Pacific Northwest National Laboratory, Richland, WA). X-ray powder diffraction confirmed the formation of HAP and the absence of any undesired calcium phosphate phases. BET (Quantachrom) measurements showed a specific surface area of 57 m2/g. Crystal morphology, as determined by electron microscopy (SEM and TEM), was as expected for synthetic hydroxyapatite with plate-like morphology terminating predominantly along the [001] face of the crystal.
Pure statherin was dissolved in PBS buffer (100 mM NaCl, 40 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.4) to maintain constant pH conditions at temperatures below the freezing point of water. Approximately 16 mg of protein were adsorbed by mixing 30 mg of the protein with 100 mg of hydroxyapatite in PBS for 4 hours. After centrifugation, the wet pellet containing the complex was washed three times with the phosphate buffer to remove all unbound protein. The mineral-protein complex was then flash-frozen with liquid nitrogen before insertion into a pre-cooled NMR probe to increase cross polarization efficiency while avoiding water crystallization or salt depletion. This approach is consistent with previous studies, which have indicated that the N-terminus of statherin exhibits no substantial changes in the secondary structure upon freezing27. For the remainder of this paper, we will refer to the samples as stE4hap, stE5hap, and stE26hap (after binding of the respective proteins to hydroxyapatite) while the samples of the free protein are referred to as stE4, stE5 and stE26 as defined in the materials and methods section.
NMR Experiments
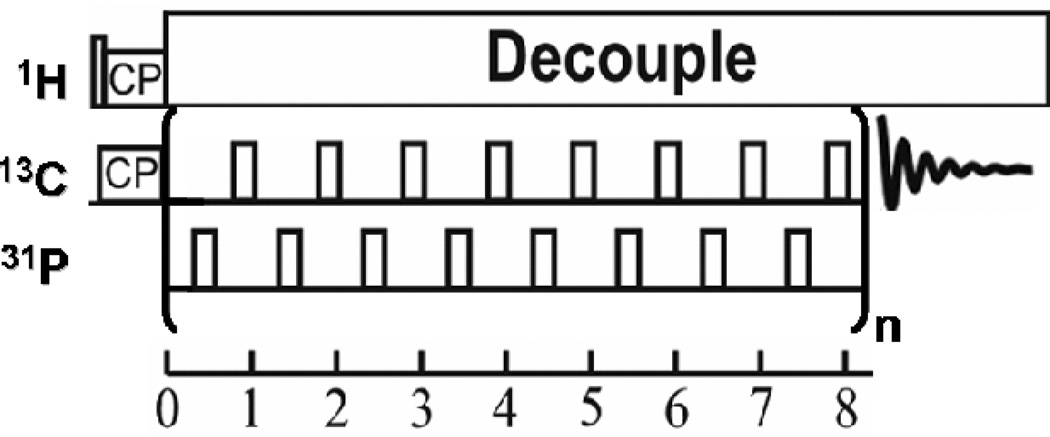
Solid-state NMR experiments were performed on a home-built spectrometer attached to a 11.74 T superconducting magnet and operating at Larmor frequencies of 500.11 MHz for 1H, 202.53 MHz for 31P, and 125.76 MHz for 13C. The 13C NMR signal was enhanced using ramped cross polarization with a 1H–13C contact time of 1 ms. CP/MAS, REDOR, and T1ρ experiments were performed using a 4 mm HXY triple-resonance Varian T3 magic angle spinning probe. Unless otherwise stated, the sample temperature was held at −32° C. An XY8-phase cycling 13C{31P} REDOR sequence with alternating π-pulses in both the 13C-observed and 31P-dephasing channel was utilized (Figure 2). REDOR dephasing times were restricted to 12 ms. Pulse lengths were 3.25 µs for 1H π/2 pulses, 7.0 µs for 13C π pulses, and 8.3 µs for 31P π pulses.
Figure 2.
XY8 phase cycling 13C{31P} REDOR pulse sequence with alternating π pulses.
Simulations and Data Fitting
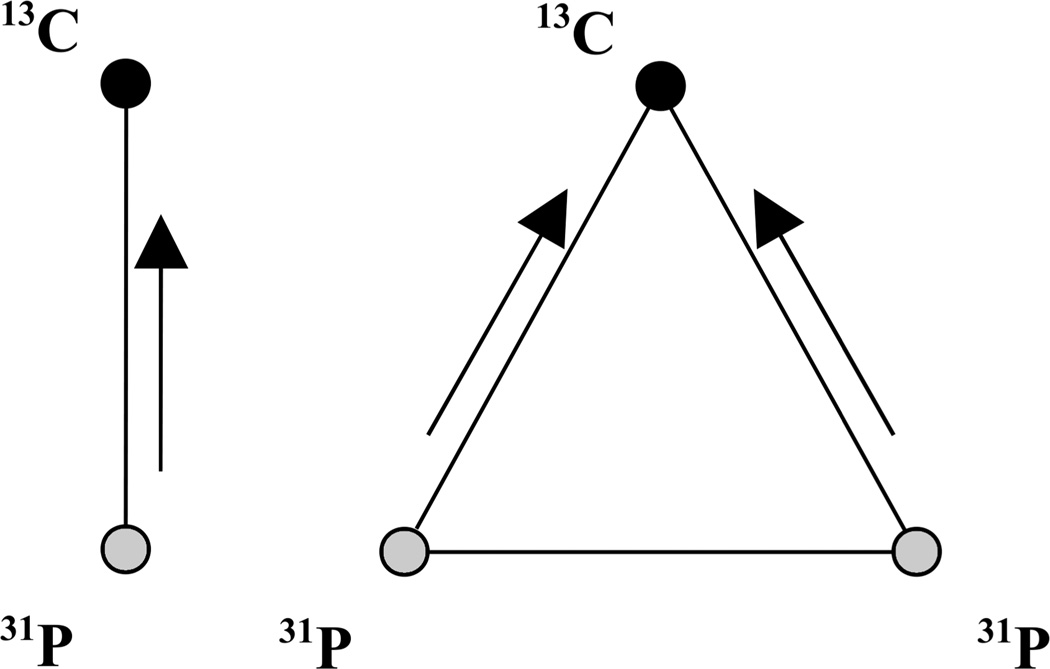
For 13C experiments, the T1ρ value was extracted from a linear fit of the semi-log plot of the signal amplitude as a function of spin-lock time. For the REDOR data, numerical simulations of the experimental data were performed using the NMR simulation software SIMPSON32 employing the direct algorithm without any relaxation. Powder averages were obtained using a minimal set of 232 {α, β} Euler angles specified by the Zaremba-Conroy-Wolfsberg scheme33–34 and 10 γ angles. A chemical shift anisotropy (CSA) of 80 ppm and asymmetry parameter of 0.68 for the side chain carboxylate 13C in glutamic acid, and 11.5 ppm and asymmetry of 0.9 for the phosphate 31P in hydroxyapatite35, were incorporated. Mutual orientations of the phosphorous CSA and dipolar interactions with respect to the carbon CSA tensor were neglected in all simulations. For stE4hap and stE26hap, the REDOR simulations were performed assuming dephasing by a single 31P spin. For the stE5hap sample, where significant dephasing was observed experimentally, additional simulations were performed to account for the multiple phosphorous atoms encountered on the surface of HAP in the form of a pseudo-31P-pair with a larger effective interaction as reported before36. See Figure 3. These spin triad simulations involved a pair of 31P spins with an effective coupling of 600 Hz and one 13C spin coupled to the two 31P spins.
Figure 3.
31P spin topologies used in simulation of 13C{31P} REDOR data. Arrows in the figure indicate the dipolar couplings that were varied in simulations. Homonuclear coupling of pseudo-31P-pair was fixed at 600 Hz based on previous studies36.
Results
13C CPMAS Spectra of HAP-Bound Statherin
For the side chain carboxyl 13C spins of glutamic acid at positions E4, E5, and E26 in free statherin, the isotropic chemical shift was observed in CPMAS experiments to be 179±1 ppm (data not shown) relative to TMS, which is within experimental error of the value obtained from the same spin species in polyglutamic acid37. Chemical shifts of 13C spins in peptides change upon adsorption to mineral surfaces as a result of conformational changes of the peptide or as a result of protein-surface interactions.
Figures 4a,b show the 13C carboxyl peak of stE4hap and stE5hap. A similar spectrum is shown in Figure 4c for glutamic acid (E). Principal values for the chemical shift anisotropy (CSA) tensors are given in Table 1.
Figure 4.
CPMAS spectra of stE4hap, stE5hap, and polycrystalline glutamic acid (E) acquired at room temperature and a spinning rate of 4 kHz.
Table 1.
Principal values for the 13C CSA tensors of stE4hap, stE5hap, and glutamic acid (E) relative to TMS. The anisotropy and asymmetry calculations assumed the convention observed in Duncan38.
| Sample | δ11 (ppm) |
δ22 (ppm) |
δ33 (ppm) |
δiso (ppm) |
CSA (ppm) |
η |
|---|---|---|---|---|---|---|
| stE4hap | 251 ± 6 | 184 ± 6 | 118 ± 6 | 184 ± 1 | 100 ± 10 | 1.0 ± 0.1 |
| stE5hap | 253 ± 6 | 183 ± 6 | 116 ± 6 | 184 ± 1 | 104 ± 10 | 1.0 ± 0.1 |
| E | 261 ± 3 | 172 ± 3 | 114 ± 3 | 182.0 ± 0.2 | 118 ± 5 | 0.7 ± 0.1 |
While the carboxylate 13C chemical shift in stE26 is invariant upon binding to HAP, remaining at about 179 ppm, the isotropic chemical shift of the carboxylate 13C spins in stE4 and stE5 shift downfield to about 184 ppm upon binding. Assuming that a downfield shift of the carboxylate 13C resonances in stE4hap and stE5hap result from deshielding due to a close approach to a positive charge center in the HAP surface, these chemical shift data indicate that the side chain carboxyl groups of E4 and E5 are closely associated with the HAP surface, while the carboxyl group of E26 is more removed.
13C{31P} REDOR Studies of Interactions between Statherin Glutamic Acid Side Chains and HAP
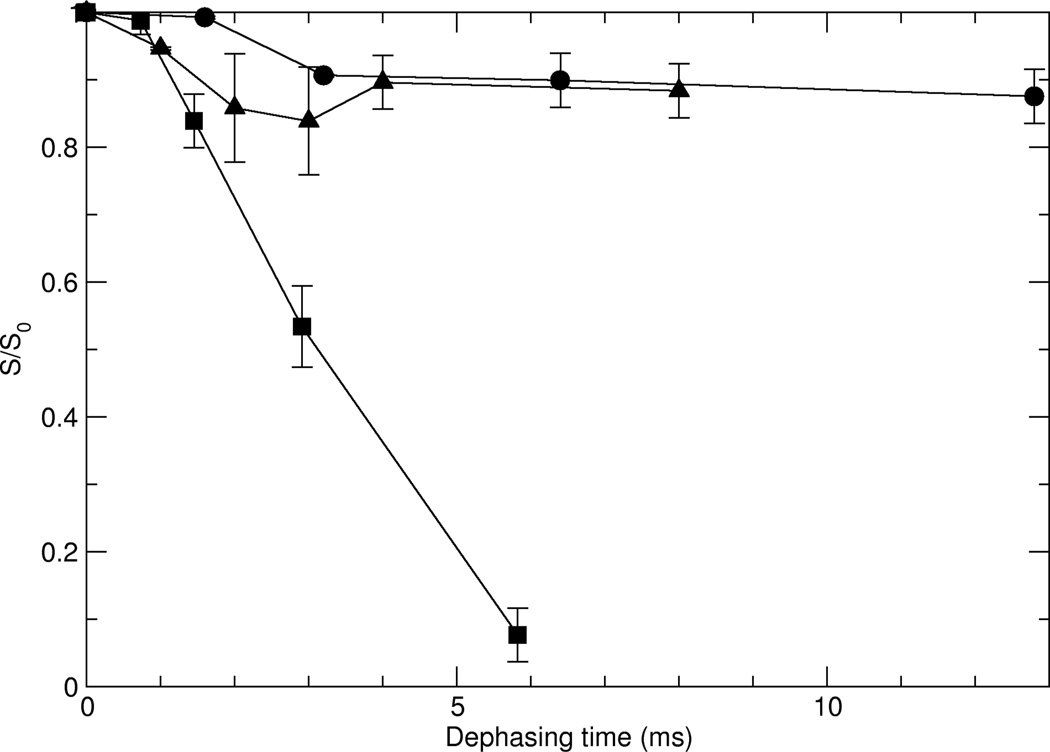
The 13C{31P} REDOR pulse sequence39 shown in Figure 2 was applied to all three HAP-bound samples in order to determine specific distances between individual protein spins and the surface. Results are shown in Figure 4. The REDOR data vary drastically as a function of position of the 5-13C-glutamic acid in the primary sequence of statherin. The behavior observed for stE4hap and stE26hap are essentially identical: for both, the intensity of the side chain carboxylate 13C signal slowly diminishes to about 90% of its S0 intensity as a result of REDOR irradiation for recoupling times in excess of 12 ms. In contrast, for stE5, the intensity of the side chain carboxyl 13C signal rapidly diminishes to about 10% of its S0 intensity after only 5.8 ms. The REDOR data observed in Figure 4 indicate that the side chain carboxyl 13C spins in stE4hap and stE26hap are far more removed from 31P spins than is the case for the corresponding 13C spin in stE5hap.
The data in Figure 5 have at least three explanations. We can conclude from it that in contrast to stE4hap and stE26hap, the side chain carboxyl 13C spin in stE5hap is: 1) dephased by one or both of the 31P spins located in the phosphoserine side chains; 2) dephased by at least one 31P spin located in the HAP surface; 3) dephased by a combination of phosphoserine and HAP 31P spins.
Figure 5.
REDOR decay plots for the three statherin samples bound to HAP. Circles indicate stE4hap, squares indicate stE5hap, and triangles indicate stE26hap.
Neither stE4 nor stE5 dephased under 13C{31P} REDOR irradiation, indicating that when not adsorbed onto HAP, the distances between the 31P spins in the side chains of pS2 and pS3 are too great (>>7 Å) to result in observable dephasing. These observations, however, do not indicate with certainty that the phosphoserine side chains do not contribute to the dephasing of the carboxylate 13C in E5, as the phosphoserine and glutamate side chains could change conformation upon adsorption to HAP, a possibility that invalidates the REDOR study of stE5 as a control.
The possible involvement of 31P spins from phosphoserine residues of statherin was also encountered in the analysis of 15N{31P} REDOR data acquired from the ε-15N in the K6 side chain of statherin adsorbed onto HAP crystals40. In that case, the likely proximity of a pS side chain to the K6 side chain was obtained from a statistical analysis of 31 low energy structures predicted using de novo energy minimization techniques from the Rosetta program41–43 The possibility of a 15N{31P} REDOR effect between the ε-15N of K6 and the 31P in the side chains of either pS2 or pS3 was excluded because only 1 of 31 predicted structures show a distance even as close as 4.5 Å between the K6 15N and the pS3 oxygen in an unphosphorylated SN-15 sample. All the other structures showed distances greater than 5 Å. Insofar as none of the computationally predicted distances between the ε-15N and phosphoserine 31P spins resulted in a dipolar coupling large enough to simulate the 15N{31P} REDOR data, it was concluded that HAP 31P spins were responsible for most of the dephasing observed in the 15N{31P} REDOR experiment.
A similar but more rigorous analysis was performed to determine the possibility of a 13C{31P} REDOR effect between the 13C carboxylate spin of E5 and the side chains of pS2 and/or pS3. The minimum energy structure obtained for statherin on 001 HAP reported recently by Gray and coworkers using the RosettaSurface program27 yields predicted distances between glutamic acid carboxylates and phosphoserine phosphate groups, which along with their predicted dipolar coupling constants are summarized in Table 2.
Table 2.
Simulated distances between glutamic acid carboxylate sites and phosphoserine phosphate sites in statherin based on lowest-energy Rosetta structure, with dipolar coupling for those distances.
| Residue Pair | Distance | Coupling |
|---|---|---|
| E4–pS2 | 12.0 Å | 7.1 Hz |
| E4–pS3 | 8.5 Å | 20 Hz |
| E5–pS2 | 12.0 Å | 7.1 Hz |
| E5–pS3 | 6.5 Å | 45 Hz |
A 6.5 Å 13C–31P internuclear distance results in a 45 Hz dipolar coupling constant. This indicates that the 31P spins in phosphoserines 2 and 3 cannot account for the amount of 13C{31P} REDOR dephasing observed for the carboxylate 13C spin in stE5hap.
Assuming 31P spins from HAP dominate the 13C{31P} REDOR observed for the stE5hap sample, the size of the 31P dephasing system must be determined in order to simulate a REDOR curve. In 15N{31P} REDOR studies of diammonium hydrogen phosphate (DHP) and of HAP-bound SN-1536 with the ε amino group in the side chain of K6 enriched with 15N, it was found that accurate nearest neighbor 15N–31P heteronuclear couplings could be determined even when the 31P dephasing system was truncated to two 31P spins. Simulations of known distributions of 15N and 31P spins in DHP, for example, show that accurate heteronuclear dipolar couplings to the two nearest neighbor 31P spins can be extracted from a truncated 31P dephasing system assuming a 31P–31P dipolar coupling of 600 Hz.
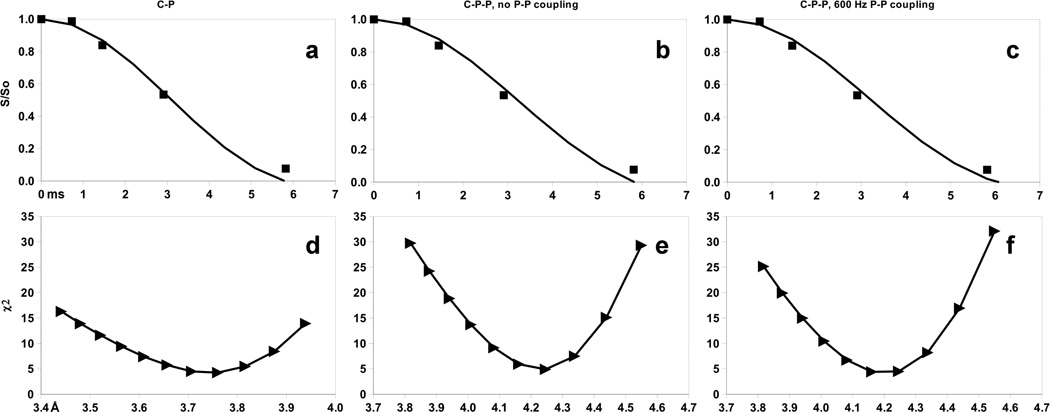
Following this strategy, simulations of the REDOR data using both an isolated 13C–31P spin pair model, and a 13C–31P–31P triad (see Figure 3), are shown in Figure 6a–c. χ2 plots of the family of fits for the three models shown are shown below as Figure 6d–f. Figure 6a shows results for the isolated 13C–31P pair model for 13C–31P dipolar coupling values ranging from 220 to 275 Hz. Close agreement with the data is obtained for a 13C–31P dipolar coupling of 245 Hz, corresponding to a 13C–31P internuclear distance of about 3.7 Å. The χ2 plot in Figure 6d shows a minimum at about 3.75 ± 0.10 Å for the isolated 13C–31P spin pair model. Figures 6b and 6c show simulation scenarios for the 3 spin CPP clusters assuming 31P–31P dipolar couplings of 0 Hz and 600 Hz. For a 31P–31P dipolar coupling of 0 Hz, the best fit to the REDOR data is achieved assuming two equivalent 13C–31P dipolar couplings of 170 Hz, corresponding to a 13C–31P distance of about 4.2 ± 0.1 Å each, as the χ2 plot in Figure 6e shows. A more realistic model using a truncated 31P system, represented as two 31P spins with a 600 Hz 31P–31P coupling, also achieves a best fit for two equivalent 13C–31P couplings, but the χ2 plot in Figure 6f shows the best fit 13C–31P distance for the 600 Hz 31P–31P dipolar coupling case is shifted to a slightly longer distance of 4.25 ± 0.09 Å.
Figure 6.
Simulated 13C{31P} REDOR curves for three possible spin systems: (a) 2-spin 13C–31P coupling, (b) 3-spin coupling of one 13C to two 31P nuclei without any 31P–31P coupling, (c) 3-spin 13C–31P–31P coupling with fixed 600 Hz 31P–31P coupling. Experimental stE5hap data (square symbols) are depicted in (a) – (c) as well. (d) through (f) are the corresponding χ2 values calculated and plotted based on simulations carried at each value of the C–P internuclear distance (Å) shown by the triangle symbols.
13C Side Chain Carboxyl Spin Dynamics
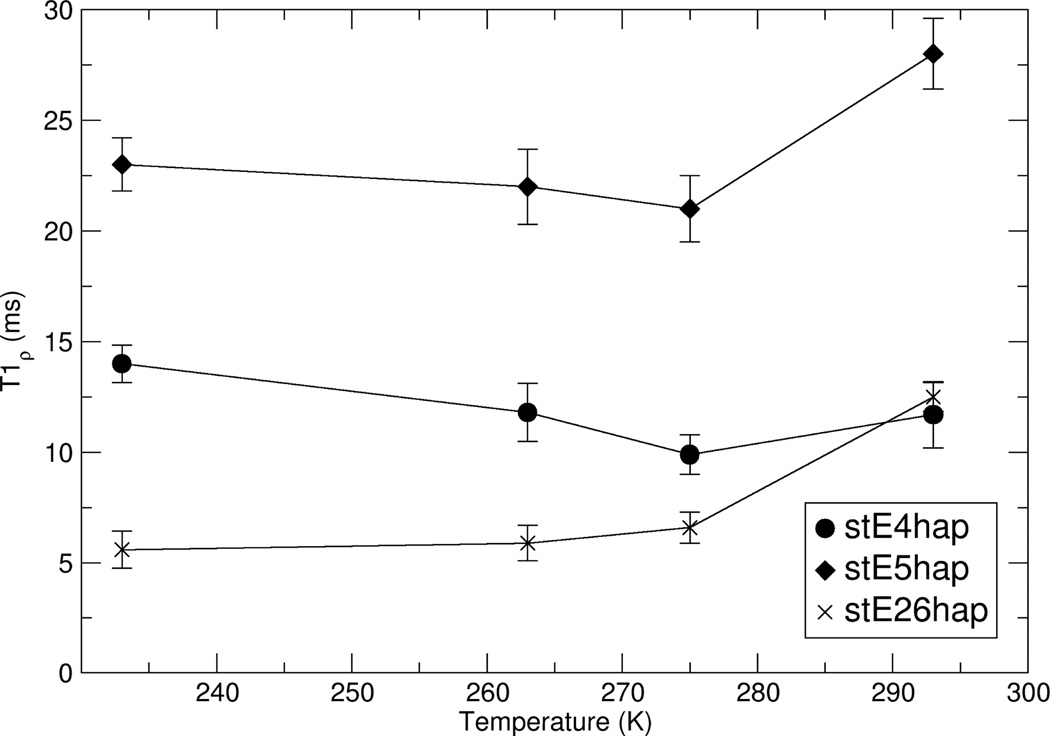
Although the large extent and indeterminate nature of the 31P dephasing network does not allow unique determination of the number and magnitude of 13C–31P dipolar couplings between the side chain carboxyl 13C spins in statherin and the 31P spins of the HAP surface, comparison of the 13C{31P} REDOR data for stE4hap, stE5hap, and stE26hap show that only the side chain of E5 likely lies within 4.0–4.5 Å of the HAP surface. It is therefore interesting to directly compare the trend in 13C{31P} REDOR data with the trend in glutamic acid side chain dynamics, as probed by 13C rotating frame (T1ρ) relaxation measurements. T1ρ measurements, when applied during MAS, can site-specifically elucidate polymer motions on the time scale of the r.f. field amplitude applied to the carboxyl 13C spins. In the present study, the amplitude is 71 kHz.
Figure 7 shows the dependence of side chain carboxyl 13C T1ρ on temperature and position of the labeled glutamic acid side chain in the primary sequence of statherin. For temperatures between 220K and 280K, T1ρ varies with primary sequence position as:
stE26hap < stE4hap < stE5hap.
Figure 7.
HAP-bound statherin T1ρ values for E4, E5, and E26 as functions of sample temperature.
At ambient temperature, the T1ρs of stE4hap and stE26hap are both about 12 ms, while T1ρ≈28 ms for stE5hap. For temperatures above 273K, T1ρ for all three sites increases as the temperature increases, placing all three 13C spins in a dynamic regime where ω1τ < 1, where ω1 is the amplitude of the r.f. field and τ is a generalized correlation time of the 13C–1H vector motion. At temperatures lower than 273K, T1ρ increases as the temperature decreases, indicating that ω1 τ > 1.
Because at ambient temperature all three glutamic acid sites are in the fast motion regime, i.e. 1/τ>>ω1, the rotating frame relaxation rate will depend most strongly on dynamic amplitudes as opposed to the rates of motion. Therefore, in the fast regime, we expect differences in rotating frame relaxation rates to be reflected by differences in the principal values of magnetic interaction tensors that have been pre-averaged by fast side chain motions. Inspection of the CSA data for the side chain carboxyl groups of stE4hap and stE5hap versus polycrystalline glutamic acid elucidate the origins of the T1ρ trend for these amino acid side chains. First, the principal values of both stE4hap and stE5hap are identical to within experimental error. The chemical shift tensors of carboxylate 13C spins are some of the best studied and most thoroughly reported in the literature38. For example, 13C NMR studies of single crystals of oxalic acid dehydrate (HOX) and diammonium oxalate monohydrate (DAOX)44 report not only the principal values of the CSA tensors of those compounds, but also the orientation of the principal axis systems relative to molecule-fixed frames. In DAOX, δ11 is within 3 degrees of the C–C bond axis and nearly bisects the O–C–O angle. Although the carboxylate CSA tensor orientation can vary from compound to compound as a result of local structural variations, the general trend in many carboxylate compounds is that δ22 is oriented close to the O–C–O bond, while δ33 is to varying degrees oriented most closely to the normal vector to the O–C–O plane.
If the observed T1ρ difference between stE4hap and stE5hap were attributable to differential side chain motion occurring at rates fast enough to effect rotating frame relaxation, the rate of such motions would exceed the CSAs for both sites, and we would expect to observe predictable differences in the motionally pre-averaged CSA principal values for stE4hap and stE5hap. For example, one of the best studied instances of large amplitude side chain motions in solid proteins is the dynamics of the isopropyl groups in the side chains of leucine amino acids in collagen fibers45. The exchange of the leucine side chain isopropyl groups between two rotational bond conformers produces a change in the orientation of the Cγ–Cδ1 and Cγ–Cδ2 bond axes by about 109°. If the side chains of stE4hap and stE5hap displayed differential mobility, we would expect profound differences in the pre-averaged principal values. However, their CSA tensors are virtually identical. This argues that the motions of these side chains are similar and thus the modulations of dipolar couplings between the carboxylate 13C spins and nearest neighbor protons on the Cγ carbons are similar in the side chains of stE4hap and stE5hap and are unlikely to account for the large T1ρ difference between the stE4hap and stE5hap sites.
Although the data in Table 1 indicate that the motions of the glutamic acid side chains of statherin do not differ significantly, there is every reason to believe that the proximity of water to these side chains is quite different. Using an isothermal titration calorimetry study, Goobes et al.46, found that the adsorption of the statherin N-terminal fragment SN-15 is nearly thermal-neutral and that the binding of SN-15 to the HAP surface is entropically driven. A small enthalpy of adsorption and a large positive entropy of adsorption occur when protein side chains expel water and ions from the HAP surface. In the present case this means that while stE4hap and stE26hap, both of which are oriented away from the HAP surface and into the solvent, are surrounded by water; but stE5HAP, which is oriented close to the HAP surface, is very likely more remote from water. We conclude that at least for stE4hap and stE5hap, for which we have accurate CSA values and thus some quantitative probe of local side chain motional differences, it is unlikely that the observed T1ρ difference arises from differential side chain mobility alone if at all.
Discussion
Solid state NMR has been used in several prior cases to study the spin dynamics of glutamic acid-rich proteins adsorbed onto HAP crystals. For example, Fernandez and coworkers37 used both CP/MAS and 13C T1ρ measurements to study the adsorption of polyglutamic acid onto HAP and silica particles. In a series of papers by Duer and coworkers, 13C{31P} REDOR was used to elucidate the role of glutamic acid side chains in the binding of proteins to HAP particles47 as well as the interactions between the protein and mineral components of bone35. While these prior studies succeeded in identifying and characterizing properties unique to glutamic acid residues in proximity to a HAP surfaces, these studies also generally applied solid state NMR techniques to 13C spins at natural abundance and therefore were not site selective. This present study differs from prior work on acidic proteins adsorbed onto HAP in that here the glutamic acid sites are individually interrogated using 13C{31P} REDOR to detect specific surface contacts, and 13C T1ρ relaxation measurements are used to comparatively study the dynamics of individual glutamic acid side chains. How do the results of the present study differ from these other studies and to what degree does it extend our knowledge of how proteins interact and recognize biomineral surfaces in general and HAP in particular?
There are two scenarios that would cause variation of T1ρ between different carboxyl side chains in statherin. Both scenarios originate with the degree of proximity of the given carboxylate side chain to the HAP surface. First, differences in local motions of glutamic acid side chains would result in differential modulation of dipolar couplings between the carboxylate 13C spins and protons located on the glutamic acid side chain. Therefore, T1ρ differences between carboxylate 13C spins of E4, E5, and E26 may reflect differences in local motions of the glutamic acid side chains. Second, differences in the structure and modulation of the proton bath near to each side chain carboxyl 13C spin may also modulate T1ρ relaxation. This situation would arise if the distribution and or dynamics of water molecules near to the unprotonated carboxylate 13C spins varied between E5 and the other glutamic acid side chains. Prior studies of the dynamics and hydration of statherin on HAP surfaces indicate that both scenarios are plausible and likely occur with the glutamic acid side chains.
Prior solid state REDOR and T1ρ studies of protonated 13C spins in the phenyl side chains of F7 and F14 in SN-15 adsorbed onto HAP48 indicated differences in the rate of side chain dynamics between a phenyl side chain adjacent to the HAP surface versus a phenyl side chain oriented away from the surface In that study, 13C{31P} REDOR was also used to determine proximity to the HAP surface. The REDOR study showed that the F14 phenyl ring is about 4 Å from the nearest 31P spin in the HAP surface, while the F7 phenyl showed no measureable dephasing during 13C{31P} REDOR irradiation and thus was determined to be greater than 6.5–7.0 Å from the HAP surface. Because the 13C spins in the side chains of F7 and F14 that were observed under cross polarization conditions are covalently protonated, the interpretation of T1ρ data is less ambiguous than in the case of the carboxyl 13C spins of this study, because modulation of the dipolar coupling between the phenyl ring 13C spins and their directly bonded protons would dominate T1ρ relaxation. A study of T1ρ measured as a function of temperature showed that the F7 13C spins lie on the fast side of the T1ρ minimum (ω1τ < 1) while F14 was on the slow side (ω1τ < 1), indicating that proximity to the HAP surface perturbs the rates of side chain dynamics in F14 versus F7.
In the case of E4 and E5, which are both situated on a segment of statherin that is largely immobilized on HAP, the CSAs are identical to within experimental error and unlike F7 and F14, the motions of both side chains are in the fast limit. Both observations indicate that the local motions of E4 and E5 are similar in amplitude amplitude and rate. The possible role of solvent motions in T1ρ relaxation of the glutamic acid side chains is elucidated by a isothermal titration calorimetric study46 by Goobes et al. This ITC study showed that while HAP adsorption of statherin is accompanied by a very small enthalpy of adsorption at low protein coverages and is virtually thermal-neutral at higher coverages, there also occurs a large positive entropy of adsorption. The positive entropy change does not reflect changes in the structural state of statherin upon adsorption because the protein is unstructured in solution and only folds into a permanent structure upon adsorption onto HAP. Rather, the positive entropy of adsorption reflects the expulsion of water and ions from the hydrophilic surface upon adsorption of protein. This means that E5, which is oriented close to the surface, is likely more remote from water than are E4 and E26, which are oriented away from the surface and thus are solvent-exposed. Regardless of which mechanism is responsible for the difference between the T1ρ relaxation of E5 versus E4/E26, the origin undoubtedly lies with the difference between the environment of the protein side chain near to the HAP surface versus the environment remote from the surface.
It is also interesting to compare our results to those of Duer and coworkers who also have performed 13C{31P}REDOR on proteins adsorbed onto HAP particles47 and on bone35. One of the model peptides studied by Duer was poly-aspartic acid (PD) adsorbed onto HAP particles. The strong dephasing of the carbonyl and side chain carboxyl 13C signals of PD during 13C{31P} REDOR irradiation indicated that PD displaces enough water from the HAP surface to enable it to bind closely itself to the surface. In another 13C{31P} REDOR study of bone, they observed strong dephasing of the carboxyl 13C signal arising from the protein component of bone during 13C{31P} REDOR irradiation. From these REDOR data the average distance between the carboxyl 13C spins of the protein component and the 31P spins of the mineral component was estimated to be 4.5–5.0 Å.
Our 13C{31P} REDOR data for stE5hap bear that out to the extent that we also detect a carboxyl 13C–31P distance of about 4.2 Å, which is close to that found in bone. However, the aggregate results obtained from DRAWS studies of SN-15 and 15N{31P} and 13C{31P} of interactions between basic and nonpolar side chains with HAP, in addition to calorimetric studies of the effect of Arg to Ala mutations on the binding of SN-15 to HAP, indicate an intricate mode of interaction between statherin and HAP. This may involve some nonpolar amino acid side chains as well as the phosphoserines, the basic side chains, and some acidic side chains in the N-terminus of statherin. When these ssNMR and calorimetric data are combined with the results of computational studies of statherin on HAP27, a model emerges, shown in Figure 8, which explains how the secondary structure of the N-terminus enables exposure of selected side chains in the N-terminus to the HAP surface. According to the distorted α–helical secondary structure that we have proposed, the side chains of E5, K6, and F14 are within 4.0–4.2 Å of the HAP surface, while the side chains of E4 and F7 are more than 6–7 Å from the surface. In addition, isothermal titration calorimetric studies indicate a reduced binding affinity between statherin mutants in which R9, R10, and R13 are replaced with alanines. Such mutants do not show marked changes in secondary structure, so the implication is that these arginine side chains are also involved in interactions with the HAP surface.
Figure 8.
Model of N-terminal region of statherin on HAP surface, combining new and previously established distance constraints.
Conclusion
We have used REDOR NMR to show that the side chain of E5 are in close contact with the HAP surface and have estimated this distance to be 4.25 Å. In contrast, the side chains of E4 and E26 show weak REDOR dephasing, indicating that they are further removed from the surface. We have also used 13C T1ρ measurements to demonstrate a slower rotating-frame relaxation rate for E5 compared to E4 and E26. This slower relaxation is attributed to restriction in side chain dynamics and/or displacement of water near the HAP surface. Based on this and prior data, we have proposed a model of the binding of statherin to HAP in which the N terminus of statherin adopts a distorted α-helical structure. Further studies involving other side chains in statherin and the role of water at the protein-HAP interface are underway.
Acknowledgments
We thank Dr. Allison Campbell and Dr. Wendy Shaw for the hydroxyapatite sample. This research was supported by National Institutes of Health grants RO1 DE12554-06 and RO1-GM074511-03.
References
- 1.Schoen FJ, Levy RJ. Ann. Thorac. Surg. 2005;79:1072–1080. doi: 10.1016/j.athoracsur.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 2.Coe FL, Parks JH, Asplin JR. N. Engl. J. Med. 1992;327:1141–1152. doi: 10.1056/NEJM199210153271607. [DOI] [PubMed] [Google Scholar]
- 3.McCarty DJ. DM-Dis. Mon. 1994;40:259–299. doi: 10.1016/0011-5029(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 4.Giachelli CM. J. Am. Soc. Nephrol. 2004;15:2959–2564. doi: 10.1097/01.ASN.0000145894.57533.C4. [DOI] [PubMed] [Google Scholar]
- 5.Margolis HC, Beniash E, Fowler CE. J. Dent. Res. 2006;85:775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- 6.He G, Gajjeraman S, Schultz D, Cookson D, Qin CL, Butler WT, Hao JJ, George A. Biochemistry. 2005;44:16140–16148. doi: 10.1021/bi051045l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto T, Okazaki M, Inoue M, Sasaki J, Hamada Y, Takahashi J. Dent. Mater. J. 2006;25:360–364. doi: 10.4012/dmj.25.360. [DOI] [PubMed] [Google Scholar]
- 8.Addadi L, Joester D, Nudelman F, Weiner S. Chem.-Eur. J. 2006;12:981–987. doi: 10.1002/chem.200500980. [DOI] [PubMed] [Google Scholar]
- 9.Cha JN, Shimizu K, Zhou Y, Christiansen SC, Chmelka BF, Stucky GD, Morse DE. Proc. Natl. Acad. Sci. U. S. A. 1999;96:361–365. doi: 10.1073/pnas.96.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beniash E, Simmer JP, Margolis HC. J. Struct. Biol. 2005;149:182–190. doi: 10.1016/j.jsb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Alford AI, Hankenson KD. Bone. 2006;38:749–757. doi: 10.1016/j.bone.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 12.Giachelli CM, Steitz S. Matrix Biol. 2000;19:615–622. doi: 10.1016/s0945-053x(00)00108-6. [DOI] [PubMed] [Google Scholar]
- 13.Palmer LC, Newcomb CJ, Kaltz SR, Spoerke ED, Stupp SI. Chem. Rev. 2008;108:4754–4783. doi: 10.1021/cr8004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koutsopoulos S, Dalas E. J. Crystal Growth. 2000;217:410–415. [Google Scholar]
- 15.Sugino A, Miyazaki T, Ohtsuki C. J. Mater. Sci: Mater. Med. 2008;19:2269–2274. doi: 10.1007/s10856-007-3327-8. [DOI] [PubMed] [Google Scholar]
- 16.Olszta MJ, Douglas EP, Gower LB. Calcif. Tissue Int. 2003;72:583–591. doi: 10.1007/s00223-002-1032-7. [DOI] [PubMed] [Google Scholar]
- 17.Olszta MJ, Odom DJ, Douglas EP, Gower LB. Connect. Tissue Res. 2003;44:326–334. [PubMed] [Google Scholar]
- 18.Bradt JH, Mertig M, Teresiak A, Pompe W. Chem. Mater. 1999;10:2694–2701. [Google Scholar]
- 19.Quin C, D’Souzar R, Feng JQ. J. Dent. Res. 2007;86:1134–1141. doi: 10.1177/154405910708601202. [DOI] [PubMed] [Google Scholar]
- 20.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. J. Biol. Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 21.Shaw WJ, Ferris K, Tarasevich B, Larson JL. Biophys. J. 94;8:3247–3257. doi: 10.1529/biophysj.107.119636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wikiel K, Burke EM, Perich JW, Reynolds EC, Nancollas GH. Arch. Oral Biol. 1994;39:715–721. doi: 10.1016/0003-9969(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 23.Raj PA, Johnsson M, Levine MJ, Nancollas GH. J. Biol. Chem. 1992;267:5968–5976. [PubMed] [Google Scholar]
- 24.Douglas WH, Reeh ES, Ramasubbu N, Raj PA, Bhandary KK, Levine MJ. Biochem. Biophys. Res. Commun. 1991;180:91–97. doi: 10.1016/s0006-291x(05)81259-8. [DOI] [PubMed] [Google Scholar]
- 25.Shaw WJ, Long JR, Dindot JL, Campbell AA, Stayton PS, Drobny GP. J. Amer. Chem. Soc. 2000;122:1709–1716. [Google Scholar]
- 26.Long JR, Shaw WJ, Stayton PS, Drobny GP. Biochemistry. 2001;40:15451–15455. doi: 10.1021/bi010864c. [DOI] [PubMed] [Google Scholar]
- 27.Goobes G, Goobes R, Schueler-Furman O, Baker D, Stayton PS, Drobny GP. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16083–16088. doi: 10.1073/pnas.0607193103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makrodimitris K, Masica DL, Kim ET, Gray JJ. J. Am. Chem. Soc. 2007;129:13713–13722. doi: 10.1021/ja074602v. [DOI] [PubMed] [Google Scholar]
- 29.Goobes R, Goobes G, Shaw WJ, Drobny GP, Campbell CT, Stayton PS. Biochemistry. 2007;46:4725–4733. doi: 10.1021/bi602345a. [DOI] [PubMed] [Google Scholar]
- 30.Goobes G, Goobes R, Gibson JM, Long JR, Paranji R, Popham JM, Raghunathan V, Shaw WJ, Campbell CT, Stayton PS, Drobny GP. Mag. Res. Chem. 2007;45:S32–S47. doi: 10.1002/mrc.2123. [DOI] [PubMed] [Google Scholar]
- 31.Kelly RC, Gebhard I, Wicnienski N. J. Org. Chem. 1986;51:4590–4594. [Google Scholar]
- 32.Bak M, Rasmussen JT, Nielsen NC. J. Magn. Reson. 2000;147:296–330. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- 33.Conroy H. J. Chem. Phys. 1967;47:5307. [Google Scholar]
- 34.Cheng VB, Suzukawa HH, Wolfsberg M. J. Chem. Phys. 1973;59:3992–3999. [Google Scholar]
- 35.Best SM, Duer MJ, Reid DG, Wise ER, Zou S. Magn. Reson. Chem. 2008;46:323–329. doi: 10.1002/mrc.2168. [DOI] [PubMed] [Google Scholar]
- 36.Raghunathan V, Gibson JM, Goobes G, Popham JM, Louie EA, Stayton PS, Drobny GP. J. Phys. Chem. B. 2006;110:9324–9332. doi: 10.1021/jp056644g. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez VL, Reimer JA, Denn MM. J. Am. Chem. Soc. 1992;114:9634–9642. [Google Scholar]
- 38.Duncan TM. Compilation of Chemical Shift Anisotropies. Chicago: The Farragut Press; 1990. p. 3. [Google Scholar]
- 39.Gullion T, Schaefer J. J. Magn. Reson. 1989;81:196–200. [Google Scholar]
- 40.Gibson JM, Raghunathan V, Popham JM, Stayton PS, Drobny GP. J. Amer. Chem. Soc. 2005;127:9350–9351. doi: 10.1021/ja050910m. [DOI] [PubMed] [Google Scholar]
- 41.Rohl CA, Strauss CEM, Misura KMS, Baker D. Methods Enzym. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 42.Schueler-Furman O, Wang C, Bradley P, Misura K, Baker D. Science. 2005;310:638–642. doi: 10.1126/science.1112160. [DOI] [PubMed] [Google Scholar]
- 43.Bradley P, Misura KMS, Baker D. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 44.Griffin RG, Pines A, Pausak S, Waugh JS. J. Chem. Phys. 1975;63:1267–1271. [Google Scholar]
- 45.Batchelder LS, Sullivan CE, Jelinski LW, Torchia DA. Proc. Natl. Acad. Sci. USA. 1982;79:386–389. doi: 10.1073/pnas.79.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goobes R, Goobes G, Campbell CT, Stayton PS. Biochemistry. 2006;45:5576–5586. doi: 10.1021/bi052321z. [DOI] [PubMed] [Google Scholar]
- 47.Jaeger C, Groom NS, Bowe EA, Horner A, Davies ME, Murray RC, Duer MJ. Chem. Mater. 2005;17:3059–3061. [Google Scholar]
- 48.Gibson JM, Popham JM, Raghunathan V, Stayton PS, Drobny GP. J. Am. Chem. Soc. 2006;128:5364–5370. doi: 10.1021/ja056731m. [DOI] [PubMed] [Google Scholar]