Abstract
The integration of the brain and face and to what extent this relationship constrains or enables evolutionary change in the craniofacial complex is an issue of long-standing interest in vertebrate evolution. To investigate brain-face integration, we studied the covariation between the forebrain and midface at gestational days 10-10.5 in four strains of laboratory mice. We found that phenotypic variation in the forebrain is highly correlated with that of the face during face formation such that variation in the size of the forebrain correlates with the degree of prognathism and orientation of the facial prominences. This suggests strongly that the integration of the brain and face is relevant to the etiology of midfacial malformations such as orofacial clefts. This axis of integration also has important implications for the evolutionary developmental biology of the mammalian craniofacial complex.
Keywords: face development, modularity, integration, mice, geometric morphometrics, computed microtomography, cleft lip
Introduction
Morphological integration, or the tendency for traits to covary, is a central concern in the study of the developmental basis of evolutionary change (Hendrikse et al., 2007; Wagner et al., 2007; Hallgrímsson et al., 2009). Integration results when local developmental influences dominate over global ones in determining the transfer of developmental variance between tissues that give rise to anatomical structures (Mitteroecker and Bookstein, 2008). Direct developmental interactions (e.g., molecular induction, internal subdivision of developmental fields, physical displacements) can impact variation by inducing spatial patterns of relatively high covariance between sets of varying traits. Such sets of traits comprise morphological modules that are more integrated internally than they are with the other parts of the whole (Wagner and Altenberg, 1996). However, covariation between modules also arises through a joint deployment of genetic variation in isolated pathways underlying the growth and differentiation of otherwise separate features that do not interact directly over the course of development (Cheverud, 1996; Klingenberg, 2005). Further, global influences affecting all traits can generate covariance patterns not reflective of regionally dissociated developmental processes (e.g., allometric effects) (Mitteroecker, 2009). It is important to determine the relative roles of the different developmental sources of covariance, as different kinds of covariation-generating mechanisms may have different effects on evolvability (Hallgrímsson et al., 2009).
Morphological integration can occur through two ways. First, via direct developmental interactions between pathways or developing tissues. Second, they can occur through the indirect effects of an extrinsic variable such as the influence of growth hormone on somatic tissues during growth. These two kinds of developmental origins for integration can be distinguished using fluctuating asymmetry (Klingenberg, 2003). As Klingenberg has argued, asymmetry deviations that are correlated among traits reflect the direct transmission of variance between developing components. Unfortunately, asymmetry deviations are normally small and difficult to detect. In mouse embryos, the variation introduced by fixation and handling of the specimens is large relative to these small asymmetry signals (Schmidt et al., 2010). An alternative to the fluctuating asymmetry based method is to focus on variation that arises through known developmental processes (Hallgrimsson et al., 2006; Hallgrimsson et al., 2007b; Hallgrímsson et al., 2009; Klingenberg, 2010). Here, we employ this approach, investigating the integration of the developing brain and face in inbred mouse. The use of inbred mice has the additional advantage of minimizing genetic and environmental variation. While this does not eliminate all the possible parallel sources of variation, it does eliminate parallel effects due to genetic covariance. The covariance that remains in such a sample is due to a combination of direct developmental interactions, the parallel effects of factors acting at regional or systemic levels (e.g. growth hormone) and the effects of whatever environmental variance remains in laboratory conditions.
The mammalian skull figures prominently in evolutionary developmental biology as a study system for testing hypotheses of morphological integration in complex traits (Cheverud, 1982; Marroig and Cheverud, 2001; Hallgrímsson et al., 2009). In the most basic configuration, the mammalian skull is composed of three modules, calvarium, cranial base and face (Cheverud, 1996; Goswami, 2006; Lieberman, 2010). Hypothesized interactions between these three modules have been important in explanations of primate and specifically human craniofacial form. Biegert (1963), for example, proposed the spatial packing hypothesis in which the highly flexed basicranium and foreshortened face of the human skull is a response to enlargement of the brain. This model has been tested in a comparative dataset (Ross and Ravosa, 1993; Ross and Henneberg, 1995; Lieberman et al., 2000) as well as in mouse mutants with altered brain and facial growth (Hallgrimsson and Lieberman, 2008). Enlow (1973) also hypothesized that the neurocranium impacts facial form, noticing that the brain showed “topographic variations which appear to be associated with corresponding characteristic differences in facial structure and topography” (pg. 256). Perhaps because of its importance in human evolution, the effect of brain size and shape on the other modules of the skull in adult humans and non-human primates continues to be an important area of study and debate (Ross and Ravosa, 1993; Lieberman et al., 2000; Hallgrimsson and Lieberman, 2008; Lieberman et al., 2008; Bastir et al., 2010).
Recent work has begun to reveal the complex molecular interactions between the developing brain and face (Hu et al., 2003; Marcucio et al., 2005; Hu and Marcucio, 2009b). The face is formed from four paired prominences (maxillary and mandibular) and a fifth midline prominence (frontonasal, which also gives rise to the lateral and medial nasal prominences). The midface or upper jaw forms from the maxillary and frontonasal prominences (Figure 1). Midfacial outgrowth depends on molecular (Shh) signaling from the forebrain brain which establishes a signaling center in the overlying ectoderm. This center, the Frontonasal Ectodermal Zone (FEZ) maintains outgrowth of the central midface during face formation (Hu et al., 2003; Marcucio et al., 2005; Hu and Marcucio, 2009a). In addition to these molecular interactions, however, there may also be physical or architectural interactions between the brain and the face. The facial prominences grow outwards from the brain which is also growing in size as the facial prominences are integrating to form the face. A narrower or slower growing forebrain, for example, would produce a narrower gap for the facial prominences to cross and would result in them making physical contact at an earlier or smaller stage. Earlier physical contact would then alter the shape of the face that is produced, producing resulting in a more prognathic and narrower face (Boughner et al., 2008b).
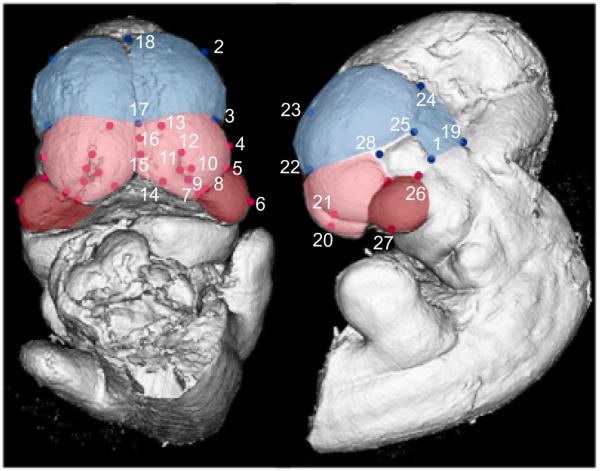
Figure 1.
Surface reconstruction of a micro-CT scan of an E10.5 C57BL6/J with landmarks listed on Tables 4-5. The blue region and landmarks are considered to be part of the forebrain; the red landmarks are part of the face; the pink area is the frontonasal process and the red area is the maxillary process.
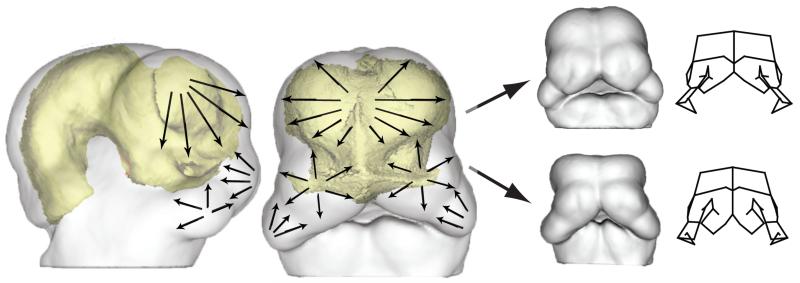
Figure 2 shows a conceptual model for how we hypotheses these physical interactions shape the growing face. In this model, as the brain grows, the facial prominences are pulled apart. The rate of growth within the facial prominences must, therefore, overcome this basic tendency. Rapid growth of the face will produce a prognathic face in which the facial prominences appose earlier. Slower growth will produce a less prognathic face in which the facial prominences appose later, or at extremes, not at all. Secondly, the growth of the facial prominences themselves produces an additional physical interaction. The medial and lateral nasal prominences push outwards as they grow, displacing the maxillary once they are physically apposed. In turn, the growth of the maxillary prominence pushes on the lateral nasal prominence, which contribute to outgrowth or prognathism of the midface.
Figure 2.
Conceptual figure of the direct reciprocal, morphological interaction between the forebrain and the face. The arrows depicted on the facial processes indicate that each module grows they could impact each other’s development resulting in pattern of coordinated variation (A). Depending on the covariation structure of the organism, these influences can result in varied phenotypes as depicted in (B).
The model shown in Figure 2, is the basis for our predictions about the physical interactions between the growing brain and face. We predict, first, that the brain and face are tightly integrated during face formation and that variation in the width of the brain is correlated with the shape of the growing face. More specifically, we predict that the size of the forebrain should correspond to variation in the degree of facial prognathism and apposition of the midfacial prominences. We also hypothesize that if the midface and forebrain are integrated via direct developmental interactions during face formation, then the relative levels of shape variance in the forebrain and/or face in each strain should be related to the degree of covariance between modules. This would occur if the covariance between modules reflects the transmission of variance from on to the other. Increasing the variance of one module will produce a corresponding increase in the covariance with another if the interaction between them is direct. Since there is negligible genetic variance in three out of four strains in our study, this result would be unlikely if the covariance among modules is due to the parallel effects of an integrating such as the activity of a gene acting in parallel in both modules.
To test these predictions, we compare the embryonic brains and faces of different mouse strains known to differ both in mean shape and level of variance: A/WySnJ, A/J, C57BL6/J and Crf4 (Young et al., 2007; Boughner et al., 2008a; Parsons et al., 2008). The first three of these mouse lines are highly inbred and so present little to no genetic variance within each line used in our comparisons. Thus, there can be little to no role for genetic covariance between the brain and face in shaping the covariance structure (Klingenberg, 2008). The fourth, Crf4, is a mixed-background line in which there is genetic variance. Thus in addition to direct developmental interactions between craniofacial modules, pleiotropy may also contribute to covariance among modules in this strain. We study the variation that arises in each craniofacial module at an embryonic stage of development, gestational days (GD) 10-10.5, because examining the relationship of variation between multiple modules can help us draw conclusions about the underlying developmental processes (Breuker et al., 2006). We chose this time point, GD 10-10.5, because this is when the paired processes of the presumptive face come together and begin to fuse. We relate our findings to the evolutionary development of the upper jaw and face, and discuss the implications for better understanding the etiology of craniofacial dysmorphologies such as cleft lip in mouse models and humans.
Results
Forebrain and facial shapes differ between mouse strains
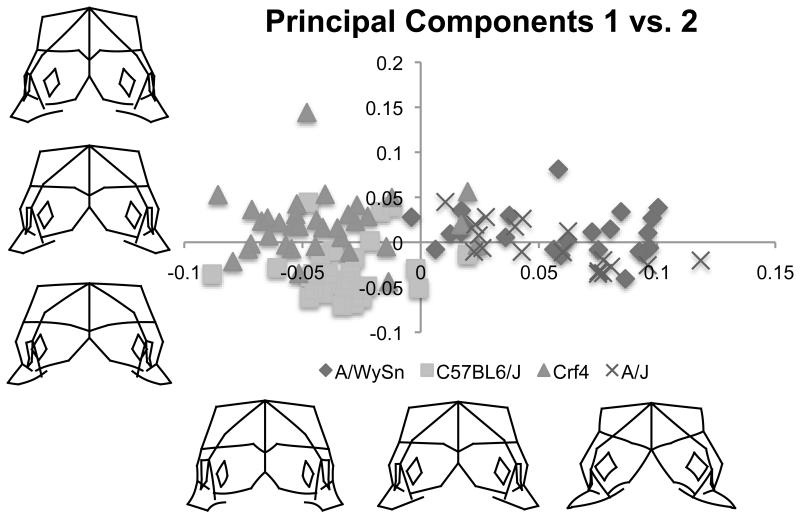
A PCA of combined face and brain landmarks for all four mouse strains revealed that strain distributions separated from each other across PCs 1-3, which together accounted for 46% of the total shape variance. The first PC (29% of the total variance) captured the variance that distinguished the A-strain (A/J and A/WySnJ) mice from the C57BL6/J and Crf4 mice (Figure 3). Compared to both C57BL6/J and Crf4 mice, both A-strains (Figure 3, positive end of PC1) included a wider maxillary process, a less fused medial nasal process, a shorter forebrain in the superior-inferior direction, and reduced maxillary outgrowth. Conversely, relative to the A-strains, C57BL6/J and Crf4 specimen shape described at the negative end of PC1 was an overall taller face and forebrain in the superior-inferior direction, a relatively larger forebrain, greater fusion of the medial nasal processes, and increased maxillary outgrowth. Thus, across PC1, the forebrain and facial shapes of the Crf4 mutant and C57BL6/J wildtype strains pulled apart from those of the A/J and A/WySnJ strains.
Figure 3.
Bi-plot of principal component one versus principal component two and associated shape variation along the x and y axes. Principal component 1 separates the A strains from the other two and seems to represent shapes similar to those seen over the course of facial fusion, even though the shape data has been regressed on tail somite number to eliminate linear shape differences due to developmental stage.
The Crf4 and C57BL6/J strains separated from each other across PC2 (11% of the shape variance). The shapes at the positive end of PC2, where most of the Crf4 specimens sat, included a narrower face with larger nasal processes, medially displaced nasal pits, a deeper face in the anterior-posterior direction, and a taller, yet narrower head in the superior-inferior direction. This shape change corresponded to a relatively enlarged frontonasal mass compared to the forebrain in Crf4 embryos, a finding consistent with Boughner et al. (2008). The negative (C57BL6/J) end of PC2 was characterized by a wider face and forebrain and less merging of the medial nasal processes.
The third PC discriminated between A/WySnJ and A/J strains (not shown). PC3 accounted for 9% of the total shape variance. All of the A/J specimens lay at the positive end of PC3, where head shape was shorter overall in the superior-inferior direction. Here, the forebrain was narrower, the face wider, and the medial nasal and maxillary processes larger. Head shape was also shallower in the anterior-posterior direction but with deeper maxillary processes along the same axis. Head shape at the A/WySnJ or negative end of PC3 was deeper in the anterior-posterior direction, with smaller nasal and maxillary processes and a taller forebrain. Also, the most anterior point of the maxillary process was laterally displaced and the medial nasal processes were further from the midline compared to shapes at the positive (A/J) end of PC3.
The null hypothesis, that the midgestational craniofacial morphologies of all four genotypes are the same was tested using a permuation test. Mahalanobis distances (p-value <0.0001) and Procrustes distances (p-value <0.0001) found that the mean shapes of all four strains were significantly different. The variances of each strain are shown in Table 1.
Table 1.
Shape variances
| C57BL6/J | Crf4 | A/WySnJ | A/J | |
|---|---|---|---|---|
| Variance | 0.00501 | 0.00993 | 0.00959 | 0.00669 |
Forebrain and facial shapes are individually distinct between the strains
For forebrain shape variation alone, the A-strains tended to be pointier and the C57BL6/J and Crf4 mice were wider and longer in the superior-inferior direction. Forebrain shapes exhibited statistically significant differences among strains by permutation for Mahalanobis distance (p-value <0.0001). Procrustes distances were all significantly different by permutation as well: both A/J and A/WySnJ were deemed different from all other strains (including from each other; p-value <0.0001). Crf4 and C57BL6/J were also significantly different, but the p-value was larger (p-value = 0.0022).
The Crf4 strain lies at one end of facial shape variation, with midfaces that were taller in the superior-inferior direction with narrower, nasal pits that are rotated inferiorly and medially, a larger frontonasal mass, more fused medial nasal processes, an anteriorly extended maxillary process, and a deeper face in the anterior-posterior direction. The A-strain faces were less fused, narrower and shorter, with a wider maxillary process. Permutation tests revealed that mean facial shapes were statistically significantly different between all strains for both Mahalanobis distances and Procrustes distances (p-value <0.0001).
Forebrain and facial shapes covary strongly within each strain
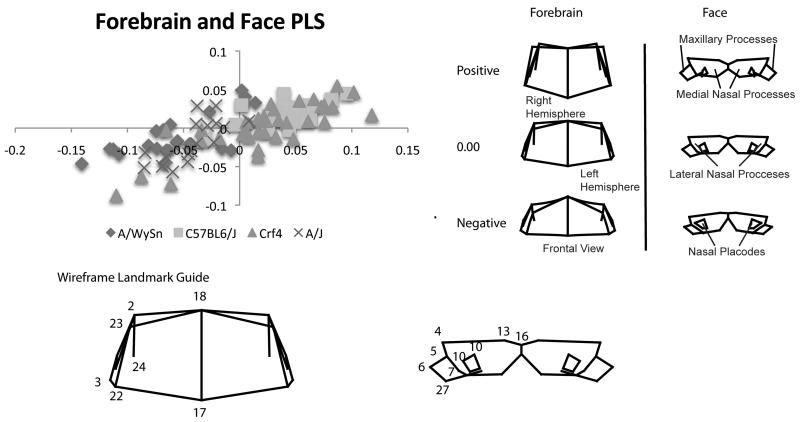
A two-block PLS analysis revealed that forebrain and face shapes of all four strains covaried significantly (RV coefficient = 0.2776; p-value <0.0001). PLS1 represented 37% of the total covariation. As PLS1 describes the greatest amount of covariation of one dimension this is the only one discussed here for all PLS analyses. Subsequent PLS dimensions can be found in Supplementary Material. The Crf4 embryos spread across this entire axis of covariation (Figure 4A). The A-strains congregated at the negative end of PLS1; C57BL6/J individuals mainly clustered at the positive end of PLS1.
Figure 4.
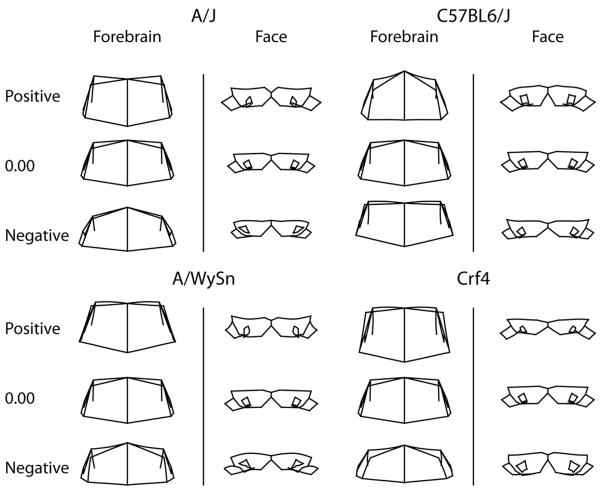
A) Bi-plot of the first singular warps of the partial least squares analysis between the forebrain and face. All strains follow a similar linear trend. B) Shape associations with the first singular warps for forebrain and face. These associations also show an ontogenetic trend from negative to positive, correlating primarily with A-strains C57BL6/J respectively while Crf4, the relatively outbred strain spans the whole range covariation.
Across all strains, certain forebrain and facial shapes covaried with each other (Fig. 4B). At the positive end of PLS1, a longer (anteriorly-posteriorly) and taller (superior-inferior) forebrain was associated with a more angled and thicker frontonasal mass, smaller nasal pits, and a more anteriorly projecting maxillary process. This is similar to the shape variance described by PC1 for C57BL6/J and Crf4 mice. At the negative end of PLS1, forebrain shape was shorter in the superior-inferior direction and slightly wider, with its superior-lateral points noticeably displaced inferiorly. Facial depth was shallow in the anterior-posterior direction, the maxillary process extended more posteriorly and the frontonasal mass was angled inferiorly. This is akin to what PC1 described for A-strain shape variance.
Specific patterns of covariation vary among strains
All strains showed a clear linear trend of covariation between the forebrain and face. The first PLS of only A/J mice accounted for 48% of the total covariation between forebrain and facial shapes (Figure 5). The positive end of PLS1 described a forebrain that, in the midline, was longer, while also taller laterally and posteriorly, narrower laterally, and displaced anteriorly (Figure 5A). The associated facial shape had a larger and anteriorly projecting maxillary process, a longer frontonasal mass, a deeper face in the anterior-posterior direction, and nasal pits that were closer to the midline. The opposite (negative) end of PLS1 described a more domed forebrain that was shorter in the superior-inferior direction and wider. The associated negative facial shape had a thinner maxillary process in the superior-inferior direction, a narrower maxillary process in the anterior-posterior direction, and laterally displaced nasal pits.
Figure 5.
Shape associations of the within-strain PLS analyses. The A/J, A/WySn and Crf4 all show similar patterns of covariation while the C57BL6/J shows the reverse pattern that appears to be less associated with developmental progress than the other three.
The first A/WySnJ PLS accounted for 46% of the total covariation between forebrain and face. A longer and slightly wider forebrain covaried with a narrower maxillary process and a deeper face in the anterior-posterior direction (Figure 5B). At the opposite end, a shorter forebrain was associated with a shorter frontonasal mass, a wider and shorter maxillary process and less fused medial nasal processes.
In the Crf4 PLS analysis PLS1 accounted for 50% of the total covariation between the forebrain and face. A straighter and taller forebrain covaried with a thinner frontonasal mass, and a thinner, shorter and wider maxillary process (Figure 5D). At the opposite end, a shorter forebrain was associated with a thicker frontonasal mass, and a narrower and deeper maxillary process.
For the C57BL6/J analysis, PLS1 accounted for 57% of the total covariation between the forebrain and the face. The positive end of PLS 1 described a forebrain that is domed, taller, narrower, deeper and superiorly larger (Figure 5C). The associated facial shape had a narrower frontonasal mass, medially displaced nasal pits, wider and thicker maxillary processes, and medial nasal processes that were closer to the midline. The opposite (negative) end PLS1 described a flatter forebrain that is both wider and shallower. The associated negative facial shape had laterally displaced nasal pits, a shorter maxillary process and an overall thinner face.
Strength of covariance and correlation among the forebrain and facial processes varies among strains
We conducted three separate PLS analyses: first, comparing the forebrain and medial and lateral process landmark data; second, comparing the forebrain and maxillary process data; and finally, comparing maxillary, medial and lateral nasal process landmarks. The following results are relative to those from our PLS analysis of all forebrain and facial landmark data combined. Because PLS1 explains the most covariation of any one dimension and is most salient to this study, we discuss the results for this axis only.
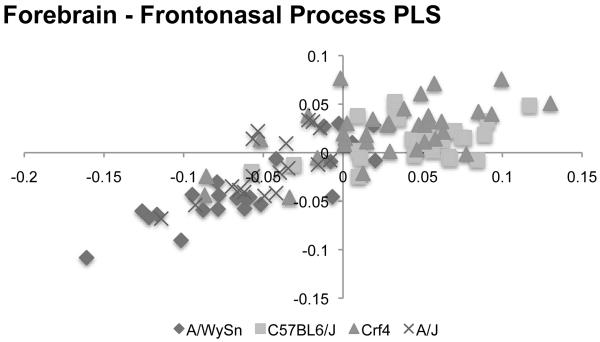
When we excluded all landmarks associated with the maxillary process (Figure 6), the correlation between the forebrain and frontonasal prominence decreased in strength for both the Crf4 and C57BL6/J embryos (Table 2). Conversely, in A/WySnJ mice these two modules correlated more strongly when maxillary process data were omitted. We found no discernable difference in the A/J embryos whether we included the maxillary process data or not. The over-all strength of association between blocks was statistically significant (RV=0.28, p-value<0.0001).
Figure 6.
Bi-plot of the PLS analysis between the forebrain and frontonasal process for all four strains. The A-strains again tend to cluster separately from C57BL6/J while showing the same linear trend.
Table 2.
Correlations between the brain and facial processes
| Strain | Forebrain vs. Face |
Forebrain vs. Frontonasal Process |
Forebrain vs. Maxillary Proces |
Frontonasal Process vs. Maxillary Process |
|---|---|---|---|---|
| A/WySnJ | 0.74 | 0.85 | 0.39 | 0.31 |
|
|
||||
| C57BL6/J | 0.59 | 0.41 | 0.59 | 0.38 |
|
|
||||
| Crf4 | 0.81 | 0.6 | 0.79 | 0.61 |
|
|
||||
| A/J | 0.73 | 0.71 | 0.4 | 0.37 |
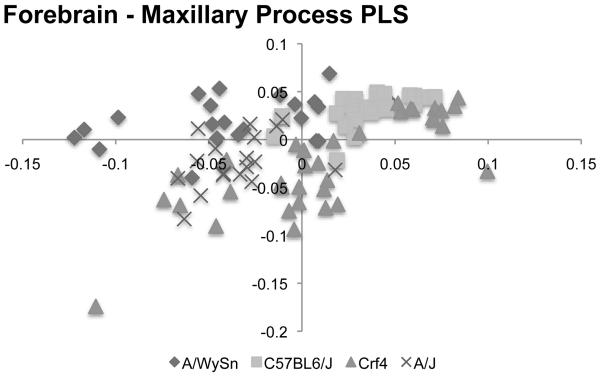
Next, after a PLS analysis of the forebrain and maxillary process data alone (Figure 7), Crf4 embryos showed the strongest correlation between these two modules (Table 2). In C57BL6/J the strength of correlation was about equal to that among the forebrain and entire set of facial landmarks. Correlations between the forebrains and maxillary processes of both A-strains were weak and not significant.
Figure 7.
Bi-plot of the PLS analysis between the forebrain and maxillary process for all four strains. The trend is much different from that seen in the forebrain-frontonasal process analysis.
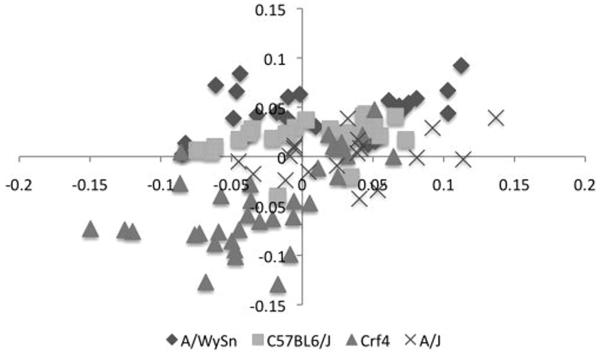
Finally, when we excluded all forebrain data and ran a PLS analysis of maxillary and frontonasal process landmarks (Figure 8), we found that both these two facial modules were weakly correlated in C57BL6/J, A/WySnJ and A/J. The Crf4 strain alone had a stronger correlation between the landmarks on these two structures.
Figure 8.
Bi-plot of the PLS analysis between the frontonasal process and maxillary process for all four strains. This analysis shows the least linear trend and the greatest differences in strain clustering.
Discussion
This study tests the hypothesis that the developing forebrain and face covary signficantly in ways that are consistent with the effects of direct physical interactions between the brain and face during face formation. We used PLS statistical analyses to determine the level of covariation and strain-specific correlations between the forebrain and face. We also used PLS statistical analyses to determine specifically which facial modules are involved. We looked at possible interactions between the forebrain and the frontonasal process and the forebrain and maxillary process. Additionally, we investigated the modular structure of the embryonic midface. We show that the samples with the highest levels of over-all shape variance also exhibit stronger correlations between modules.
We analyzed the craniofacial landmark data sets using a two-block PLS designed to measure morphometric shape covariance between the embryonic forebrain and various parts of the face. We found that all four strains show a significant covariation of the forebrain and face. The correlation between brain shape and face shape along PLS1 is very strong at 0.748. These results indicate that the forebrain and the midface exhibit morphological covariation at the time of embryonic facial formation, mouse E10.5. What these results alone do not tell us is whether this covariation due to direct transmission of variation from one module to another via physical interaction or to covariation due to extrinsic factors.
To generate covariation, two requirements must be met. First, variation must be present, but it arises within traits. Second, some mechanism must create association between traits (e.g. pleiotropy, parallel environmental effects, genetic linkage, direct molecular interaction, partitioning of developmental fields, epigenetic mechanical effects, etc.). Many mechanisms can produce integration and thus lead to covariance in the presence of variation. One of these possibilities is direct developmental interaction among modules. To determine if this is the case here, we needed to tease direct developmental interaction out from at least three other candidate causes (Klingenberg, 2008; Mitteroecker, 2009). The first of these is environmental variation, which was minimized, if not eliminated, because all embryos developed within mothers housed under the same conditions. The second possible cause that we eliminated in three out of the four strains is genetic variation. A/J, A/WySnJ and C57BL6/J are inbred lines while there is segregating genetic variation in the Crf4 line. The third cause may be allometry, or the correlated shape effects of size. This has been removed from the dataset via regression of shape variables against tail somite number. With these three candidate causes removed, remaining explanations for the covariation of the forebrain and face are parallel variation due to developmental factors acting either systemically (e.g. Insulin-like Growth Factor 1 [IGF1] (Spaventi et al., 1990)).or at a more local anatomical level via direct developmental interaction (Klingenberg, 2008). The direct interaction could take the form of either molecular or physical interaction between the growing tissues of the forebrain and face.
An explanation for the covariation of the forebrain and face that cannot be fully eliminated by our study design is the parallel effects of systemic or local factors. Variation in IGF level, for example, would produce correlated responses in various growing tissues and structures. That particular factor is unlikely to be the cause of the pattern of covariation that we see because systemic variation in size tends to produce allometric variation which has been removed in our analysis through regression on tail somite number. Local factors, however, such as variation in the expression of a particular gene in both the brain and the face could account for the covariation that we see. However, given that there is no genetic variation in three out of the four samples, this could only occur via a factor that is secreted within the head and is acting directly on both the brain and the face. While no such factor is known, this possibility cannot be excluded in our analysis. However a more likely explanation is that the integration is due to direct physical interactions between growing tissues.
Additional evidence for this direct interaction model comes from examining the patterns of variance and covariance among the modules and strains. If the covariation in the forebrain and midface is due to direct integration of the modules (that is, if the forebrain and midface are reciprocally influencing each other’s phenotypic variation) then as variance of the forebrain and facial shapes increase then so should the strength of their covariation. This would not necessarily occur if the covariance was due simply to the parallel variation in some locally acting factor. A relationship between within module variance and among-module covariance is exactly what we see in our data. Of the inbred strains, the strain with the greatest amount of shape variance is A/WySn, followed by A/J and C57BL6/J has the least. On PLS 1, which captures the greatest single amount of covariation between the brain and face, A/WySn has the highest correlation, followed by A/J and then C57BL6/J. This result also points towards a direct mode of integration for all three strains. If the forebrain and face did not directly interact, then it would be highly unusual to see this kind of coordinated variation.
The Crf4 case, however, is different. This strain has higher genetic and phenotypic variances than the other three. Unlike the other strains, it is of mixed background, and thus, has segregating genetic variation derived from its founder strains. The Crf4 strain also shows the highest correlation between the shapes of the forebrain and face. Some of the covariation seen in Crf4 is also likely due to the direct interactions between the brain and the face, but other factors such as genetic covariance may contribute as well. The fact that that pattern of covariation in this strain differs somewhat from the others may reflect this heterogeneity. On the other hand, the higher correlations between brain and face shape in this strain suggest that the genetic variation is acting through some of the same covariance generating processes as in the inbred strains. For example, a higher variance in brain size, due in part to increased genetic variance, may produce correlated effects in face shape through a combination of direct and indirect effects. Although the patterns of integration are different, all four strains show the basic pattern that would be expected from physical interaction between the brain and the face. This pattern is that a larger brain is associated with more divergent and less apposed prominences while a smaller brain is associated with more convergent and more advanced appearing faces. This is strong circumstantial evidence for direct physical interactions as the primary cause of the pattern of covariation between the brain and face in all four strains.
Because the facial processes are developmentally heterogeneous (Hu et al., 2003; Creuzet et al., 2005; Szabo-Rogers et al., 2010), they might be expected to exhibit different relationships with the brain. To explore this, we divided the facial landmarks into a maxillary process module and a frontonasal process module and performed separate PLS analyses for these regions. Both analyses resulted in over-all significant covariation values (forebrain-frontonasal process RV=0.24, p-value <0.0001; forbrain-maxillary process RV=0.21, p-value<0.0001) however the PLS1 plots for each analysis look quite distinct. This leads us to suspect that while some aspect of the forebrain covaries with the frontonasal process, some other, possibly independent, aspect is covarying with the maxillary process. The way in which the forebrain and the facial modules covary for each specific strain provides us with some insight into what these different aspects might be.
In a plot of PLS1of forebrain vs. frontonasal process (Figure 7), the Crf4 embryos sit at the one end of the trend line whereas the A-strain mice sit at the opposite end. This sample distribution is meaningful since, compared to C57BL6/J, Crf4 neonates and adults have shorter faces and more globular skulls (Boughner and Hallgrimsson, 2008) while A/WySnJ adults have longer skulls and faces (Parsons et al., 2008). Thus, the pattern of forebrain-face covariation in the embryos correlates well with postnatal skull and face lengths, based on 3D morphometric data. This suggests that PLS1 is describing embryonic shape variance linked to adult phenotype, specifically length. In past studies we have shown that strain-specific variation is detectable in embryos at least as early as GD10 (Boughner et al., 2008a; Parsons et al., 2008). Thus the template for adult face and skull length may be set by this stage of embryonic development, and integration via the direct developmental interaction of forebrain and face modules may contribute to it.
In the case of the forebrain-maxillary process correlation, both AWS and A/J had lower values (0.39 and 0.4 respectively) than the other two strains. These two longer-faced strains thus show less covariation between the forebrain and maxillary process as the face is forming. In contrast, the embryonic Crf4 forebrain shape correlates more strongly with the shape of the maxillary process shape than with that of the frontonasal mass. Strength of correlation between forebrain and face modules in C57BL6/J embryos was in between the Crf4 values on one hand and the A/WySnJ and A/J values on the other. The smaller forebrain of the Crf4 mouse places the maxillary process into physical contact with the frontonasal mass at an earlier stage because the distance between them is smaller. We speculate that this earlier physical contact produces an earlier and tighter relationship between brain size and maxillary prominence morphology. As the brain varies in size relative to the face, the maxillary prominences are pulled away from or pushed towards the frontonasal mass, deforming their shape and producing covariation between the shape of the maxillary prominence and the brain.
An alternative explanation for the covariance of the brain and maxillary prominences is direct developmental interaction via molecular signaling (Marcucio et al., 2005). This could occur in addition to physical interaction. A third non-exclusive possibility is that signals are transmitted to the maxillary process from the eye (Marcucio et al., 2005; Hu and Marcucio, 2009a), which abuts the facial process, instruct the facial process in some way(s). The eye cup has long been thought to affect skull development, even prior to chondrification (De Beer, 1937). The molecular data to test this last possibility are being developed further. If this hypothesis of separate causes of craniofacial covariation is true then it is provocative because it would mean that while brain and face modules covary, the various parts may do so for different reasons. It is interesting that different parts of the face may be integrated with the forebrain in different ways. This possibility deserves further investigation since it may get at the route processes that structure craniofacial integration.
Since the facial prominences of the face show different patterns of covariation with the forebrain, we tested for covariation among the prominences themselves via a PLS analysis. This revealed the presence of interesting strain-specific patterns. The Crf4 strain again shows a different pattern compared to the inbred strains as it is the only strain that shows a strong correlation between frontonasal and maxillary shape variations. Again, this could reflect the smaller brain placing the two prominences into physical contact at an earlier stage.
The A/WySnJ strain stands out the most in this analysis. In the other PLS analyses that involved the forebrain, this plot in Figure 8, however, shows the A strains and the A/WySnJ in particular standing out from the other strains. A- strains exhibit a dramatically elevated frequency of clefting of the primary palate compared to C57BL6/J and Crf4. Several investigators have hypothesized that the brain might be implicated in etiology of cleft lip (Diewert et al., 1993; Weinberg et al., 2009). Our results suggest that a large brain relative to the facial prominences will create a larger gap for the prominences to close as they grow together. Within the context of an expanding head, this would reduce the time window of opportunity for strain followed the same linear trend as the other strains. The maxillary vs. frontonasal process PLS fusion of the primary palate. Perturbing the integration of face and brain development may thus shift a developmental system in the direction of a threshold for cleft-lip formation. It is important to note, however, that the formation of the seam itself does not appear to be delayed in A strain mice, although the formation of the mesenchymal bridge is (Wang et al., 1995). The etiology of facial clefting is complex and multifactorial: brain development could be one cause, either alone or in concert with other factors, but further work is necessary to understand its relative causal role both more generally and in A strain mice.
This study also has important implications for understanding the modular organization of variation in the head. How variation is structured in complex phenotypes has important implications for evolvability and for evolutionary explanation (Cheverud, 1996; Hendrikse et al., 2007; Wagner et al., 2007). Our results show how variation in the face and developing brain are integrated in the absence and presence of genetic variation. Selection on the growth of the brain is likely to produce a correlated response in the morphology of the developing face which may translate to morphological variation at later developing stages. Several evolutionary questions emerge from this analysis. These include questions such as to what extent is the flattened face of human embryos related to brain growth or to what extent does that translate to morphology at later stages? A third related question is to what extent is the high prevalence of cleft in humans compared to other mammals, such as mice, due to the relationship between brain and face growth during face formation? It is possible that one of the many evolutionary costs of enlargement of the human brain is increased susceptibility to clefting.
A limitation of this study is that it does not conclusively reveal the direction in which variation is transferred. Does the face deform as the result of variation in brain growth or are there interactions that go the other way? We think that the former possibility is more likely for several reasons. For one, the brain is larger than the face during the developmental period investigated here and the prominences grow forward from the brain with the floor of the forebrain forming a platform for face development. Second, known molecular signals such as SHH emanate from signaling centers in the brain and influence the face (Marcucio et al., 2005; Hu and Marcucio, 2009a). Abnormal brain growth is also known to have dysmorphic effects on the developing face (Vermeij-Keers et al., 1983; Diewert and Lozanoff, 1993). Admittedly, the direction of integration may change with the developmental time-point and it is not impossible that variance is transferred in both directions.
Conclusion
We have shown that morphological variation in the brain has a direct effect of the variation on face (and/or vice versa) during the development and fusion of the facial processes. Phenotypic variation in the embryonic face may be a factor in the etiology of certain malformations like cleft lip (Parsons et al., 2008). It is also possible that structural variation in the brain may be implicated in the cause specific facial shapes linked with congenital conditions (Bergsma and Brown, 1975; Sulik and Johnston, 1982; Weinberg et al., 2009). Our work shows that, first, variation in the size of the brain is correlated with variation in the shape of the face. Second, that this covariation is closely related to the morphological transformation that accompanies fusion of the facial prominences and formation of the mid-face. These results both suggest strongly that the integration of the brain and face is relevant to the etiology of cleft lip and palate, which is a failure of fusion of the primary palate.
This study has uncovered an important part of the epigenotype of the mammalian craniofacial complex. The epigenotype is composed of all the interactions between different structures throughout ontogeny (Waddington, 1942). The forebrain and face are each the products of complex developmental processes. We show here that each contributes to the epigenetic context of the other. We suggest strongly that brain and face development is characterized by a physical interaction that influences their morphogenesis. This direct integration between the forebrain and face as illustrated in this study is a classical epigenetic interaction sensu Waddington (Jamniczky et al., 2010). This integration is both a source of variation and a constraint on the variation that is possible within the head. Transmission of variation from one module to another influences has obvious implications for evolvability (Wagner and Altenberg, 1996; Hansen and Houle, 2008). The existence of such interactions, however, also implies that not all combinations of face and brain shapes are developmentally possible. The integration of the brain and face morphology is thus a key feature of the organization of the craniofacial complex that is important for understanding both the generation of normal and dysmorphic variation within species and the evolution of the diversity of craniofacial form.
Experimental Procedures
Sample Composition
We used four distinct laboratory mouse strains in this study, including Crf4, C57BL6/J, A/WySnJ, and A/J. Sample sizes are listed in Table 3. The Crf4 mice are homozygous Crf4 (crf4/crf4) male and female mice came from the Baylor Mouse Mutagenesis Resource. Crf4 was derived from outcrosses between 129S6, C57BL6/J and C3H. The mutation is unmapped but is known to sit outside chromosome 11 (Monica Justice and Kathyrn Hentges, personal communication), the specific region of interest for the mutagenic screen from which the line was derived (Kile et al, 2003). The Crf4 line was backcrossed to 129S6 an unknown number of times before we acquired it. Our colony was been maintained through brother-sister matings without continued backcrossing to 129S for the generation of the sample used in this study but appears never to have been fully inbred for the short-faced trait. The short-faced Crf4 phenotype, which is clearly evident to be present in the sample used here and in previous studies (Hallgrimsson et al., 2007a; Boughner et al., 2008b) was lost in this line more recently due to technician error in our animal care facility. C57BL6/J is a common inbred laboratory strain.
Table 3.
Sample sizes
| Strain | C57BL6 /J |
Crf4 | AWySn /J |
A/J |
|---|---|---|---|---|
|
Embryos
10-15 TS |
27 | 35 | 23 | 18 |
The A/J and A/WySnJ are sister strains displaying different degrees of cleft lip penetrance. Both lines share two components and differ at a third component of the genetic liability for cleft lip. An epistatic interaction between the alleles of Wnt9b and a unknown gene mapped to chromosome 13 (clf2) is further modified by a third unmapped locus that differs between A/J and A/WySnJ (Juriloff et al., 2006). Variation at this latter locus acts through a maternal effect that depresses cleft lip penetrance in A/J relative to A/WySnJ.
Data Collection
We collected mouse embryos at GD 10-10.5 based on initial detection of a vaginal plug. We assumed that a litter was aged GD 0.5 the morning of plug detection. The embryos of the A/WySn/J line were fixed in utero with Bouin’s solution, and later dissected from the uterus and rinsed and stored 70% ethanol (Ciriani and Diewert, 1986). The remaining lines were dissected from the uterus in ice-cold phosphate buffered saline. A/J embryos were fixed overnight in 4% paraformaldehyde and secondarily fixed with Bouin’s solution. C57BL6/J and Crf4 embryos were fixed immediately into and stored in Bouin’s solution. Embryos were staged by tail somite (TS) number, which afforded an accurate and fine-scaled indication of developmental stage (Young et al., 2007; Parsons et al., 2008).
3D Computed Microtomography and Landmarking
To collect 3D landmark data and facilitate three-dimensional (3D) morphometric analysis of our embryos, we micro-CT (μ-CT) scanned each embryo head using a Skyscan 1072 X-Ray Micro-Tomograph as described elsewhere (Boughner et al., 2008a; Parsons et al., 2008). The reconstructed image stacks were then imported into ANALYZE 5.0 software (Mayo Foundation for Medical Education and Research, Rochester, MN, USA) for landmarking.
Figure 1 and Tables 4-5 show 28 3D landmarks that we used to define the forebrains and faces of our four groups of embryos at GD10-10.5. We chose Type 1 and Type 2 landmarks (Bookstein, 1991) according to the guidelines presented by (Zelditch, 2004). Type 1 landmarks (points at discrete juxtapositions of tissues; limited on three axes) were more challenging to locate on the unossified embryonic heads. Thus, we used a greater proportion of Type 2 landmarks (local minima and maxima of curvatures of local structures), the use of which has been validated (Valeri et al., 1998) for sampling and analyzing anatomical size and shape in 3D. To increase landmarking precision, we took points on a curve in a single consistent view. To standardize the x-axis, we positioned the superior border of the frontonasal mass perpendicular to the plane of view. We standardized the y-axis by centering the midline of the head in the frontal view. We exactly superimposed the medial nasal processes in the lateral views.
Table 4.
Brain landmarks.
| Number | Landmark Description |
|---|---|
| 1(R/L) | Most posterior point of the eye cup (lateral view |
| 2 (R/L) | Maximum of curvature of cerebral hemispheres (frontal view) |
| 17 (M) | Midline point marking the eventual place of separation between medial nasal processes and the cerebral hemisphere |
| 18 (M) | Junction of the two cerebral hemispheres and the midbrain |
| 19 (M) | Maximum of curvature of the hindbrain extending above the cerebral hemispheres (lateral view) |
| 22 (R/L) | Junction between medial and lateral nasal processes and the cerebral hemispheres (lateral view) |
| 23 (R/L) | Maximum of curvature of the cerebral hemispheres (lateral view) |
| 24 (R/L) | Maximum of curvature and most posterior point of the cerebral hemispheres (or the most superior-anterior point of the midbrain flexure |
| 25 (R/L) | Most posterior-superior point at the junction of the eye cup and cerebral hemisphere (lateral view) |
| 28 (R/L) | Junction between anterior-superior point of primordial eye and the cerebral hemisphere and lateral nasal process |
Table 5.
Face landmarks.
| Number | Landmark Description |
|---|---|
| 1 (R/L) | Future site of primary choanae |
| 3 (R/L) | Maximum of superior-lateral curvature of lateral nasal process (frontal view) |
| 4 (R/L) | Most lateral point (maximum of curvature) on lateral nasal process (frontal view) |
| 5 (R/L) | Maximum of inferior-lateral curvature of lateral nasal process (frontal view) |
| 6 (R/L) | Most lateral point (maximum of curvature) on maxillary process (frontal view) |
| 7 (R/L) | Most inferior-medial point of lateral nasal process |
| 8 (R/L) | Most inferior-lateral point of medial nasal process |
| 9 (R/L) | Most inferior point on medial nasal process |
| 10 (R/L) | Maximum of curvature of lateral nasal process along lateral edge of nasal pit |
| 11 (R/L) | Minimum of curvature of medial nasal process along medial edge of nasal pit |
| 12 (R/L) | Most superior point of nasal placode/pit (junction of medial and lateral nasal processes) |
| 13 (R/L) | Maximum of superior-medial curvature of medial nasal process (frontal view) |
| 14 (R/L) | Maximum of inferior-medial curvature of medial nasal process (frontal view) |
| 15 (M) | Most inferior point of junction of the medial nasal processes |
| 16 (M) | Most superior point of junction of the medial nasal processes |
| 20 (R/L) | Maximum of curvature of medial nasal process (lateral view) |
| 21 (R/L) | Maximum of curvature of lateral nasal process (lateral view) |
| 26 (R/L) | Most posterior point of the junction between the maxillary process and eye cup |
| 27 (R/L) | Maximum of the anterior-inferior curvature of the maxillary process |
| 29(R/L) | Junction of the maxillary process, lateral nasal process and eye cup |
Internal features other than spaces are not discernible in our scans using the fixation protocol that has been validated for morphometric analysis (Schmidt et al., 2010). This means that internal features of the brain cannot be landmarked and that brain shape is represented by landmarks placed on the external surface of the embryo around the telencephalon (Figure 1). A resulting limitation is that the ventral surface of the forebrain, including the region that is immediately deep to the face, is not included in the analysis. This limitation will make it more difficult to pick up covariation resulting from direct physical interactions between the developing forebrain and face.
Statistical Shape Analyses
Our statistical procedures tested the null hypothesis that the developing brain and face do not covary during face formation. We used geometric morphometric methods to quantify and compare size and shape from landmark-based data. Procrustes coordinates were calculated using Generalized Procrustes Analysis (GPA) (Goodall, 1991) and the software packages MorphoJ (Klingenberg, 2011).
Embryo shape varies significantly with age or stage. We quantified and removed this variation in order to conduct stage-standardized comparisons across strains (Boughner et al., 2008b; Parsons et al., 2008). To do this we regressed Procrustes coordinates against TS number using the pooled within-sample multivariate regression of TS on shape in MorphoJ. It is the residuals of this regression that we use here in our subsequent morphometric analyses. This procedure removes any ontogenetic variation that changes linearly with stage but some nonlinear ontogenetic variation may remain in the dataset after this procedure.
To visualize the variation in the entire sample, we used Principal Components Analysis (PCA). To visualize variation among strains, we used Canonical Variates Analysis (CVA). PCA finds the set of orthogonal axes of covariation that explain total variation in the sample and orders these from largest to smallest. CVA scales the variation among groups to the pooled within-group covariance matrix and thus reveals the axes of covariation that best separate groups in the sample. For statistical comparisons of groups, we use the shape permutation test in MorphoJ for both the Mahalanobis and Procrustes distances.
To quantify and test for significant covariation among regions, we used Partial Least Squares (PLS) Analyses. PLS analysis predicts linear combinations of multivariate relationships between two blocks of shape variables (Rohlf and Corti, 2000). The resulting singular warps represent axes of covariation between two blocks (Bookstein and F., 2003) and a correlation coefficient, RV, can be used as a statistic of covariation (Klingenberg, 2009). For our PLS analyses of forebrain, maxillary and frontonasal prominence modules we regressed PLS scores for both blocks and evaluated strengths of correlation “r” for each strain. Finally, to quantify shape variance within each strain, we used the variance of Procrustes Distances around the mean and compared those using the permutation test in IMP Simple3D (Sheets, 2004).
Supplementary Material
Acknowledgements
We thank Wei Liu for assistance with micro-CT scanning. We thank the Alberta Children’s Hospital Foundation and the University of Calgary Faculty of Medicine, NSERC grant #238992-06, NIH grants 1R01DE01963 and 1U01DE020054 to BH and NIH grant 1R01DE01963 to RSM.
Grant information: grant #238992-06 to BH and National Institutes of Health (USA) 1R01DE01963 to RSM and BH.
Literature Cited
- Bastir M, Rosas A, Stringer C, Cuetara JM, Kruszynski R, Weber GW, Ross CF, Ravosa MJ. Effects of brain and facial size on basicranial form in human and primate evolution. J Hum Evol. 2010;58:424–431. doi: 10.1016/j.jhevol.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Bergsma DR, Brown KS. Assessment of ophthalmologic, endocrinologic and genetic findings in the Bardet-Biedl syndrome. Birth Defects Orig Artic Ser. 1975;11:132–136. [PubMed] [Google Scholar]
- Biegert J. The evaluation of characteristics of the skull, hands and feet for primate taxonomy. In: Washburn SL, editor. Classification and human evolution. Aldine; Chicago: 1963. pp. 116–145. [Google Scholar]
- Bookstein F. Cranial integration in Homo: singular warps analysis of the midsagittal plane in ontogeny and evolution. Journal of Human Evolution. 2003;44:167–187. doi: 10.1016/s0047-2484(02)00201-4. [DOI] [PubMed] [Google Scholar]
- Bookstein FL. Morphometric tools for landmark data. Cambridge University Press; Cambridge: 1991. [Google Scholar]
- Boughner JC, Hallgrimsson B. Biological spacetime and the temporal integration of functional modules: a case study of dento-gnathic developmental timing. Dev Dyn. 2008;237:1–17. doi: 10.1002/dvdy.21383. [DOI] [PubMed] [Google Scholar]
- Boughner JC, Wat S, Diewert VM, Young NM, Browder LW, HallgrÃ-msson B. Short-faced mice and developmental interactions between the brain and the face. Journal of Anatomy. 2008a;213:646–662. doi: 10.1111/j.1469-7580.2008.00999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughner JC, Wat S, Diewert VM, Young NM, Browder LW, Hallgrimsson B. Short-faced mice and developmental interactions between the brain and the face. J Anat. 2008b;213:646–662. doi: 10.1111/j.1469-7580.2008.00999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuker CJ, Patterson JS, Klingenberg CP. A single basis for developmental buffering of Drosophila wing shape. PLoS ONE. 2006;1:e7. doi: 10.1371/journal.pone.0000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM. Phenotypic, genetic, and environmental integration in the cranium. Evolution. 1982;36:499–516. doi: 10.1111/j.1558-5646.1982.tb05070.x. [DOI] [PubMed] [Google Scholar]
- Cheverud JM. Developmental integration and the evolution of pleiotropy. Am Zool. 1996;36:44–50. [Google Scholar]
- Creuzet S, Couly G, Le Douarin NM. Patterning the neural crest derivatives during development of the vertebrate head: insights from avian studies. J Anat. 2005;207:447–459. doi: 10.1111/j.1469-7580.2005.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer G. The development of the vertebrate skull. xxiii. Clarendon Press; Oxford: 1937. p. 552. [Google Scholar]
- Diewert VM, Lozanoff S. Growth and morphogenesis of the human embryonic midface during primary palate formation analyzed in frontal sections. J Craniofac Genet Dev Biol. 1993;13:162–183. [PubMed] [Google Scholar]
- Diewert VM, Wang K-Y, Tait B. A new threshold model for cleft lip in mice. Annuals of the New York Academy of Sciences. 1993;678:341–343. doi: 10.1111/j.1749-6632.1993.tb26139.x. [DOI] [PubMed] [Google Scholar]
- Enlow DH, McNamara JA. The neurocranial basis for facial form and pattern. The Angle Orthodontist. 1973;43:256–270. doi: 10.1043/0003-3219(1973)043<0256:TNBFFF>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Goodall C. Procrustes Methods in the Statistical Analysis of Shape. Journal of the Royal Statistical Society Series B. 1991;53:285–339. [Google Scholar]
- Goswami A. Morphological integration in the carnivoran skull. Evolution Int J Org Evolution. 2006;60:169–183. [PubMed] [Google Scholar]
- Hallgrimsson B, Brown JJ, Ford-Hutchinson AF, Sheets HD, Zelditch ML, Jirik FR. The brachymorph mouse and the developmental-genetic basis for canalization and morphological integration. Evol Dev. 2006;8:61–73. doi: 10.1111/j.1525-142X.2006.05075.x. [DOI] [PubMed] [Google Scholar]
- Hallgrímsson B, Jamniczky H, Young N, Rolian C, Parsons T, Boughner J, Marcucio R. Deciphering the Palimpsest: Studying the Relationship Between Morphological Integration and Phenotypic Covariation. Evolutionary Biology. 2009;36:355–376. doi: 10.1007/s11692-009-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrimsson B, Lieberman DE. Mouse models and the evolutionary developmental biology of the skull. Integrative and Comparative Biology. 2008;48:373–384. doi: 10.1093/icb/icn076. [DOI] [PubMed] [Google Scholar]
- Hallgrimsson B, Lieberman DE, Liu W, Ford-Hutchinson AF, Jirik FR. Epigenetic interactions and the structure of phenotypic variation in the cranium. Evol Dev. 2007a;9:76–91. doi: 10.1111/j.1525-142X.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- Hallgrimsson B, Lieberman DE, Young NM, Parsons T, Wat S. Evolution of covariance in the mammalian skull. Novartis Found Symp. 2007b;284:164–185. doi: 10.1002/9780470319390.ch12. discussion 185-190. [DOI] [PubMed] [Google Scholar]
- Hansen TF, Houle D. Measuring and comparing evolvability and constraint in multivariate characters. Journal of Evolutionary Biology. 2008;21:1201–1219. doi: 10.1111/j.1420-9101.2008.01573.x. [DOI] [PubMed] [Google Scholar]
- Hendrikse JL, Parsons TE, Hallgrimsson B. Evolvability as the proper focus of evolutionary developmental biology. Evol Dev. 2007;9:393–401. doi: 10.1111/j.1525-142X.2007.00176.x. [DOI] [PubMed] [Google Scholar]
- Hu D, Marcucio RS. A SHH-responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development. 2009a;136:107–116. doi: 10.1242/dev.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS. Unique organization of the frontonasal ectodermal zone in birds and mammals. Dev Biol. 2009b;325:200–210. doi: 10.1016/j.ydbio.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS, Helms JA. A zone of frontonasal ectoderm regulates patterning and growth in the face. Development. 2003;130:1749–1758. doi: 10.1242/dev.00397. [DOI] [PubMed] [Google Scholar]
- Jamniczky HA, Boughner JC, Rolian C, Gonzalez PN, Powell CD, Schmidt EJ, Parsons TE, Bookstein FL, Hallgrimsson B. Rediscovering Waddington in the post-genomic age: operationalising Waddington’s epigenetics reveals new ways to investigate the generation and modulation of phenotypic variation. Bioessays. 2010;32:553–558. doi: 10.1002/bies.200900189. [DOI] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, McMahon AP, Carroll TJ, Lidral AC. Wnt9b is the mutated gene involved in multifactorial nonsyndromic cleft lip with or without cleft palate in A/WySn mice, as confirmed by a genetic complementation test. Birth Defects Res A Clin Mol Teratol. 2006;76:574–579. doi: 10.1002/bdra.20302. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. Developmental instability as a research tool: Using patterns of fluctuating asymmetry to infer the developmental origins of morphological integration. In: Polak M, editor. Developmental Instability: Causes and Consequences. Oxford University Press; Oxford: 2003. pp. 427–442. [Google Scholar]
- Klingenberg CP. Developmental constraints, modules and evolvability. In: Hallgrimsson B, Hall B, editors. Variation. Elsevier Academic Press; Amsterdam: 2005. pp. 219–248. [Google Scholar]
- Klingenberg CP. Morphological Integration and Developmental Modularity. Annu Rev Ecol Syst. 2008;39:115–132. [Google Scholar]
- Klingenberg CP. Morphometric integration and modularity in configurations of landmarks: tools for evaluating a priori hypotheses. Evol Dev. 2009;11:405–421. doi: 10.1111/j.1525-142X.2009.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP. Evolution and development of shape: integrating quantitative approaches. Nat Rev Genet. 2010;11:623–635. doi: 10.1038/nrg2829. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. MORPHOJ: an integrated software package for geometric morphometrics. Molecular Ecology Resources. 2011 doi: 10.1111/j.1755-0998.2010.02924.x. [DOI] [PubMed] [Google Scholar]
- Lieberman D. The evolution of the human head. Belknap Press of Harvard University Press; Cambridge: 2010. [Google Scholar]
- Lieberman DE, Hallgrimsson B, Liu W, Parsons TE, Jamniczky HA. Spatial packing, cranial base angulation, and craniofacial shape variation in the mammalian skull: testing a new model using mice. J Anat. 2008;212:720–735. doi: 10.1111/j.1469-7580.2008.00900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman DE, Ross CF, Ravosa MJ. The primate cranial base: ontogeny, function, and integration. Am J Phys Anthropol. 2000;(Suppl):117–169. doi: 10.1002/1096-8644(2000)43:31+<117::aid-ajpa5>3.3.co;2-9. [DOI] [PubMed] [Google Scholar]
- Marcucio RS, Cordero DR, Hu D, Helms JA. Molecular interactions coordinating the development of the forebrain and face. Dev Biol. 2005;284:48–61. doi: 10.1016/j.ydbio.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Marroig G, Cheverud JM. A comparison of phenotypic variation and covariation patterns and the role of phylogeny, ecology, and ontogeny during cranial evolution of new world monkeys. Evolution. 2001;55:2576–2600. doi: 10.1111/j.0014-3820.2001.tb00770.x. [DOI] [PubMed] [Google Scholar]
- Mitteroecker P. The developmental basis of variational modularity: Insights from quantitative genetics, morphometrics, and developmental biology. Evolutionary Biology. 2009 [Google Scholar]
- Mitteroecker P, Bookstein F. The Ontogenetic Trajectory of the Phenotypic Covariance Matrix, with Examples from Craniofacial Shape in Rats and Humans. Evolution. 2008 doi: 10.1111/j.1558-5646.2008.00587.x. [DOI] [PubMed] [Google Scholar]
- Parsons TE, Kristensen E, Hornung L, Diewert VM, Boyd SK, German RZ, Hallgrimsson B. Phenotypic variability and craniofacial dysmorphology: increased shape variance in a mouse model for cleft lip. J Anat. 2008;212:135–143. doi: 10.1111/j.1469-7580.2007.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf FJ, Corti M. Use of two-block partial least-squares to study covariation in shape. Systematic Biology. 2000;49:740–753. doi: 10.1080/106351500750049806. [DOI] [PubMed] [Google Scholar]
- Ross C, Henneberg M. Basicranial flexion, relative brain size, and facial kyphosis in Homo sapiens and some fossil hominids. Am J Phys Anthropol. 1995;98:575–593. doi: 10.1002/ajpa.1330980413. [DOI] [PubMed] [Google Scholar]
- Ross CF, Ravosa MJ. Basicranial flexion, relative brain size, and facial kyphosis in nonhuman primates. Am J Phys Anthropol. 1993;91:305–324. doi: 10.1002/ajpa.1330910306. [DOI] [PubMed] [Google Scholar]
- Schmidt EJ, Parsons TE, Jamniczky HA, Gitelman J, Trpkov C, Boughner JC, Logan CC, Sensen CW, Hallgrimsson B. Micro-computed tomography-based phenotypic approaches in embryology: procedural artifacts in assessments of embryonic craniofacial growth and development. BMC Dev Biol. 2010;10:18. doi: 10.1186/1471-213X-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets HD. IMP ThreeDStand6. 2004. http://www3.canisius.edu/~sheets/moremorph.html. [Google Scholar]
- Spaventi R, Antica M, Pavelic K. Insulin and insulin-like growth factor I (IGF I) in early mouse embryogenesis. Development. 1990;108:491–495. doi: 10.1242/dev.108.3.491. [DOI] [PubMed] [Google Scholar]
- Sulik KK, Johnston MC. Embryonic origin of holoprosencephaly: interrelationship of the developing brain and face. Scan Electron Microsc. 1982:309–322. [PubMed] [Google Scholar]
- Szabo-Rogers HL, Smithers LE, Yakob W, Liu KJ. New Directions in craniofacial morphogenesis. Developmental Biology. 2010;341:84–94. doi: 10.1016/j.ydbio.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Valeri CJ, Cole TM, III, Lele S, Richtsmeier JT. Capturing data from three-dimensional surfaces using fuzzy landmarks. American Journal of Physical Anthropology. 1998;107:113–124. doi: 10.1002/(SICI)1096-8644(199809)107:1<113::AID-AJPA9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Vermeij-Keers C, Mazzola RF, Van der Meulen JC, Strickler M. Cerebro-craniofacial and craniofacial malformations: an embryological analysis. Cleft Palate J. 1983;20:128–145. [PubMed] [Google Scholar]
- Waddington CH. The canalisation of development and the inheritance of acquired characters. Nature. 1942;150:563. [Google Scholar]
- Wagner GP, Altenberg L. Complex adaptations and the evolution of evolvability. Evolution. 1996;50:967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Pavlicev M, Cheverud J. The Road to Modularity. Nat Genet. 2007;8:921–931. doi: 10.1038/nrg2267. [DOI] [PubMed] [Google Scholar]
- Wang KY, Juriloff DM, Diewert VM. Deficient and delayed primary palatal fusion and mesenchymal bridge formation in cleft lip-liable strains of mice. J Craniofac Genet Dev Biol. 1995;15:99–116. [PubMed] [Google Scholar]
- Weinberg SM, Andreasen NC, Nopoulos P. Three-dimensional morphometric analysis of brain shape in nonsyndromic orofacial clefting. J Anat. 2009;214:926–936. doi: 10.1111/j.1469-7580.2009.01084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, Wat S, Diewert VM, Browder LW, Hallgrimsson B. Comparative morphometrics of embryonic facial morphogenesis: implications for cleft-lip etiology. Anat Rec (Hoboken) 2007;290:123–139. doi: 10.1002/ar.20415. [DOI] [PubMed] [Google Scholar]
- Zelditch ML. Geometric morphometrics for biologists : a primer. Elsevier Academic Press; Boston, MA: 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.