Abstract
The molecular rules that govern MHC restriction, and allow T-cells to differentiate between peptides derived from healthy cells and those from diseased cells, remain poorly understood. Here we provide an overview of the structural constraints that enable the T-cell receptor (TCR) to discriminate between self and non-self peptides, and summarize studies that have attempted to correlate the biophysical parameters of TCR/peptide–major histocompatibility complex (pMHC) binding with T-cell activation. We further review how the antigenic origin of peptide epitopes affects TCR binding parameters and the ‘quality’ of a T-cell response. Understanding the principles that govern pMHC recognition by T-cells will unlock pathways to the rational development of immunotherapeutic approaches for the treatment of infectious disease, cancer and autoimmunity.
Keywords: biophysics, crystal structure, peptide-major histocompatibility complex, T-cell activation, T-cell receptor
Introduction
As primary effectors of the adaptive immune response, αβ T-cells act as multi-functional sensors that scan the cellular environment to ensure that foreign invaders and dysregulated homeostatic processes do not compromise host tissues. Direct T-cell-mediated surveillance is critical for the immune integrity of all jawed vertebrates, and is enforced through an elaborate armoury of effector functions that eliminate pathogen-infected and neoplastic cells. In addition, T-cells regulate other cell subsets within both the innate and adaptive compartments of the immune system, enabling immunological memory and physiological homeostasis. Nevertheless, this complex and elegant system is susceptible to malfunction, and aberrant T-cell activity can lead to harmful consequences. For example, T-cell hypersensitivity is the root cause of many autoimmune diseases and allergies, and alloreactive specificities mediate organ rejection and graft-versus-host disease in the transplant setting. Hence, better understanding of the basic mechanisms that underpin T-cell antigen recognition and activation is essential for designing intelligent, next-generation drugs, vaccines and immunotherapies.
At the heart of almost every T-cell response lies the molecular interaction between the clonotypically expressed αβ T-cell receptor (TCR) and cognate peptide–major histocompatibility complex (pMHC) antigen, creating a distinctive interface between the T-cell and the target cell (Fig. 1). In parallel with this MHC component, T-cell antigens are ‘co-received’ by either the CD4 or CD8 T-cell surface glycoproteins. The CD4 and CD8 co-receptors bind to invariant regions of MHC class II and class I, respectively.1,2 These co-receptors can augment antigen recognition through a number of mechanisms, which include: (i) delivery of key signalling molecules to the cytoplasmic side of the TCR/CD3 complex;3,4 (ii) conveyance of the TCR to privileged sites for signal transduction;5,6 and (iii) kinetic enhancement of TCR/pMHC engagement.7,8 Classically, CD8+ T-cells recognize peptides bound to MHC class I (pMHCI) molecules and mediate direct target cell lysis, whereas CD4+ T-cells recognize pMHCII-restricted ligands and play a more complex role in the coordination of adaptive immune responses.
Figure 1.
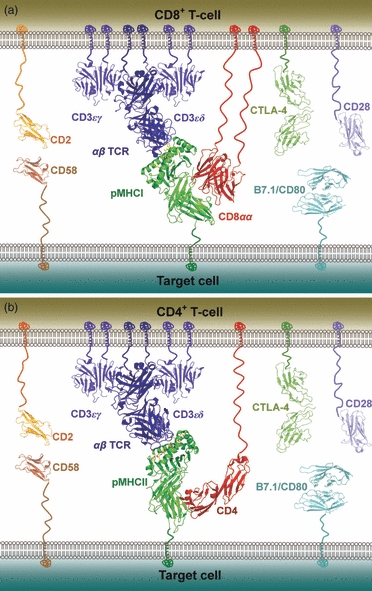
Schematic representation of the molecules involved at the interface between a T-cell and a peptide–MHC (pMHC)-presenting cell during antigen recognition. The co-stimulatory molecule CD28 is shown in mauve and the co-inhibitory molecule CTLA-4 is shown in dark green on the T-cell surface; their cognate ligand, B7.1/CD80, is shown in cyan on the target cell surface. The adhesion molecule CD2 is shown in orange on the T-cell surface and its receptor, CD58, is shown in brown on the target cell surface. The CD3 co-signalling complex is shown in blue. (a) CD8+ T-cell, with the αβ T-cell receptor (αβ TCR) shown in blue, pMHCI shown in green, and the CD8αα co-receptor shown in red. Note that the vast majority of CD8+ T-cells express the CD8αβ heterodimer, but the structure of human CD8αβ is not available for representation. (b) CD4+ T-cell, with the αβ TCR shown in blue, pMHCII shown in green, and the CD4 co-receptor shown in red.
The crystal structures of a number of TCR/pMHC complexes have been solved and are reviewed elsewhere.9,10 Collectively, these structures show that TCRs maintain a relatively conserved mode of binding to cognate pMHC molecules. In this seemingly unwavering docking mode, the TCR adopts a diagonal orientation relative to the MHC peptide-binding groove, with the TCR α-chain contacting the MHCI α2 domain or the MHCII β-chain, and the TCR β-chain contacting the MHCI α1 domain or the MHCII α-chain (Fig. 2). The cellular interface established by this primary binding event is further stabilized by other TCR/pMHC interactions and a number of co-receptor, co-stimulatory and adhesion molecules including CD2, CD58, CTLA-4, CD28 and CD80 (Fig. 1). Hence, a polyvalent link is formed between the T-cell and the target cell that allows a number of different signalling events to trigger a T-cell activation cascade. The affinity range at which TCRs operate is determined during T-cell maturation in the thymus. During thymic maturation, T-cells that bind too strongly or too weakly to pMHC antigen are deleted by negative selection or die by neglect, respectively.11 This affinity editing process results in a preference for TCRs that bind their cognate pMHC ligands relatively weakly (KD = 0·1 to > 500 μm) compared with other immunoglobulin-like proteins such as antibodies (KD = nm to pm).12
Figure 2.
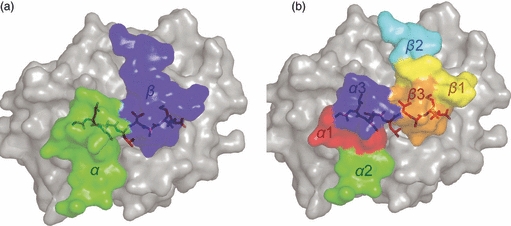
Overview of T-cell receptor (TCR) binding to peptide–MHC (pMHC). The pMHC molecule is shown from above. (a) Surface representation of the footprint of an archetypal αβ TCR (AS01)58, with the α-chain in green and the β-chain in blue. This diagonal orientation, with the TCR α-chain over the MHCI α2 domain or the MHCII β-chain, and the TCR β-chain over the MHCI α1 domain or the MHCII α chain, has been observed in all TCR/pMHC co-complexes. (b) Positions of the CDR loops during antigen recognition. The CDR1 (α chain in red, β chain in yellow) and CDR3 (α chain in blue, β chain in orange) loops are positioned centrally along the axis of the MHC peptide-binding groove, contacting both the peptide and the MHC surface. The CDR2 (α chain in green, β chain in cyan) loops are positioned such that they contact mainly the MHC surface and make limited, or no, contact with the peptide.
Genetically conserved interactions between the TCR and the MHC surface
Structurally, the TCR/pMHC interaction is complex because of the large, and conformationally plastic, footprint between the TCR and the pMHC surface. Thus, a number of different models that explain how this interaction leads to T-cell activation have been described (Box 1). For example, Colf et al.13 recently solved the structure of a murine TCR (2C) in complex with both self (Ld-QL9) and non-self (Kb-dEV8) antigens. In this study, it was clear that the 2C TCR could bind to the two different antigens with different conformational modalities. These data demonstrate that a single TCR can drastically modify its binding strategy depending on the antigen encountered. Equally, there are examples of different TCRs that can use distinct docking strategies to bind to the same antigen.14,15 As a result, TCR binding can be flexible, albeit within the constraints of MHC restriction and a relatively fixed diagonal orientation. How TCRs adhere to these ‘rules’ while binding to hyper-variable antigens of both self (MHC) and often non-self (peptide) origins has not been fully resolved. A recent study suggested that germline-encoded TCR/pMHC interaction codons exist, which allow TCRs to use ‘ancient’ MHC anchor points to dock with an invariant modality. In this study, the binding strategies of four murine TCRs, each expressing the same TRB gene (Vβ8.2), were explored in complex with the MHC molecule I-Au.16 In all four examples, a common TCR contact footprint was observed with the MHC surface. The existence of conserved pairwise binding may help explain why the orientation of the TCR during pMHC binding is so well maintained, and how a single TCR can recognize multiple peptides presented by the same MHC molecule. In support of this notion, accumulated structural data have revealed the existence of a number of conserved interactions, termed the ‘restriction triad’, between the TCR and the MHC surface, which are common to the majority of TCR/pMHC structures solved to date.9,17 The discovery of these fixed interactions lends support to the idea that common TCR/pMHC contacts may have an important role in conserving the overall binding conformation of the TCR/pMHC interaction.
Box 1. Models of T-cell activation
| Theory | Definition |
|---|---|
| Kinetic Proofreading45 | The ability of a T-cell to discriminate between agonist and non-specific peptide–MHCs (pMHCs) based on kinetic parameters required for sequential downstream phosphorylation steps |
| Serial Triggering49 | The ability of a small number of pMHCs to achieve a high T-cell receptor (TCR) occupancy via serial engagement of the same TCR |
| Germline-encoded Codon Theory16 | The theory that germline-encoded codons permit specific TCR variable-domain recognition of certain MHC molecules |
| Induced Fit Model94 | A thermodynamic mechanism by which TCRs ‘scan’ the pMHC to ‘search’ for complementarity – this structural reordering may be transmitted along the entire length of the TCR, inducing a conformational change that may trigger TCR signalling |
| Conformational Change Model95,96 | A model to explain TCR activation based on the premise that ligand engagement by the TCR induces a conformational change in the TCR/CD3 complex that permits linkage to T-cell signalling |
| Permissive Geometry Model97 | A refinement of the conformational change model, based on the premise that dimeric/multimeric pMHC binding promotes rotation of the αβ TCR subunits with respect to each other – this rotation induces a scissor-like movement of the CD3 chains, permitting their phosphorylation |
| Aggregation Models96 | Models of T-cell activation based on the premise that pMHC engagement induces aggregation of co-receptor in the vicinity of TCRs (co-receptor heterodimerization) and/or aggregation of TCR complexes (pseudodimer aggregation) |
| Kinetic Segregation Model98 | A model that explains the progression of TCR signalling based on size exclusion of phosphatases from the vicinity of activated TCRs |
MHC restriction guided by the CD4 and CD8 co-receptors
Although genetically conserved interactions between the TCR and MHC surface provide an attractive explanation for MHC restriction, there are examples of TCR/pMHC structures that do not support a genetically fixed recognition signal. For example, in the structure of a TCR bound to MHCI complexed with a 13-mer ‘super-bulged’ peptide, the TCR made extremely few contacts with the MHC surface and recognition was governed almost exclusively through TCR/peptide interactions. In this system, it seems unlikely that fixed TCR/MHC interactions are central to T-cell antigen recognition.17 Furthermore, another recent study challenged the importance of the MHC ‘restriction triad’ by demonstrating that TCR recognition is not drastically altered when these docking points are knocked out by mutagenesis.18 In a groundbreaking report, van Laethem and colleagues provided experimental evidence that T-cells lacking CD4 or CD8 co-receptor expression can recognize antigen in an MHC-independent manner.19 They demonstrated that these co-receptors sequester the tyrosine kinase Lck, which is essential for triggering a T-cell signalling cascade. In this way, only MHC-dependent antigen recognition, in which the co-receptors bind to MHC and release Lck, can result in T-cell maturation. Hence, the CD4 and CD8 molecules only support the development of T-cells bearing TCRs that bind MHC. The offshoot of this focusing event may be the conserved docking mode we presently find in the TCR/pMHC system. Ultimately, however, we have to date only solved an infinitesimally small fraction of the possible TCR/pMHC structures that exist in mice and humans. Hence, it is possible that the next generation of TCR/pMHC structures to be solved will undermine all current models and force a complete re-evaluation of T-cell antigen recognition theory.
Structural determinants of TCR specificity
The antigen contact zone of the TCR/pMHC interaction occurs between the pMHC surface and six highly flexible TCR hypervariable complementarity determining region (CDR) loops.9 The CDR1α and CDR2α loops are encoded by one of 47 germline-derived TCR-α variable (TRAV) genes, whereas the CDR1β and CDR2β loops are encoded by one of 57 germline-derived TCR-β variable (TRBV) genes. In contrast, the CDR3α and CDR3β loops are somatic and manufactured by variable (V), diversity (D; beta only) and joining (J) gene rearrangement, DNA nuclease activity and random N nucleotide addition at the V(D)J junctions. Generally speaking, the V-gene-encoded CDR2 loops contact mainly the conserved helical region of the MHC surface, the V-gene-encoded CDR1 loops can contact both the MHC and the peptide, and the somatic and hypervariable CDR3 loops contact mainly the antigenic peptide. The TCR recombination machinery can produce 1015–1020 unique αβ receptor structures, which provide a potentially massive range of antigen shape coverage.20
A number of studies have suggested that this binding conformation enables the CDR2 loops to contact the MHC surface in a conserved manner, which is likely to be an important factor that allows TCRs to recognize multiple pMHC ligands.16,21–23 Mechanistically, Wu et al.24, proposed a two-step binding model in which an initial transition state is formed between the CDR2 loops and the MHC surface, enabling the CDR3 loops to scan the antigenic peptide. Conversely, evidence from other studies suggests that the CDR3 loops play the dominant role during MHC binding and that CDR2 loops can contact the antigenic peptide.9,18,25 Indeed, Burrows et al.18 demonstrated that disruption of the conserved interactions between the TCR and MHC surface could result in the formation of compensatory interactions. The classical definition of the roles of the CDR loops during pMHC binding cannot therefore provide a cohesive set of rules that govern TCR specificity.
Structural data for both unbound pMHC molecules and TCR/pMHC complexes have shown that pMHC molecules are relatively rigid, and do not retain a high degree of flexibility when bound to the TCR. However, there are exceptions to this general rule, with reported examples of large conformational changes in either the peptide or the MHC molecule upon TCR docking.26,27 Conversely, paired structures of unbound TCRs and TCR/pMHC complexes have revealed substantial flexibility within the CDR loops of the TCR molecule, with stabilization occurring upon binding to pMHC. Hence, a flexible binding mechanism has been proposed for TCR/pMHC docking.28–30 This so-called TCR ‘plasticity’ is thought to be an important determinant of the ligand degeneracy that has been reported for both MHCI-restricted and MHCII-restricted TCRs.31,32 Thermodynamic experiments have extended our understanding of this phenomenon, showing that TCR/pMHC binding is typically characterized by a negative TΔSo, or a loss of entropy, which suggests that there is a transition from disorder to order during binding.33–36
Although we do not fully understand the molecular rules that govern T-cell specificity, it is clear that the TCR is exquisitely tuned so that it can recognize foreign epitopes with sufficient strength to initiate activation, while remaining largely tolerant and inactive to self tissue. Accumulated biophysical data have complemented structural studies and shed light on the parameters that underpin this fine dichotomy.
TCR binding affinity is governed by antigenic origin
Until the advent of soluble TCR manufacture and experimentation, it was difficult to pair kinetic binding measurements with T-cell activation measurements. This was largely because cellular assays were typically based on competition protocols which, although capable of providing reasonable measures of affinity, were not amenable to kinetic measurements. Sykulev et al.37 previously made use of such competition assays to demonstrate that the functional responses of T-cells expressing the MHCI-restricted 2C TCR correlate well with affinity. Such affinity-based hypotheses (Box 1) were supported by the premise that negative and positive selection in the thymus were also governed by TCR/pMHC affinity.37
Over the last two decades, techniques such as surface plasmon resonance and isothermal titration calorimetry have enabled very accurate and precise analysis of TCR binding affinity, kinetics and thermodynamics. To date, such techniques have been used to study over 30 different TCR interactions with naturally selected antigens (and many more using artificially altered ligands) restricted by both MHCI and MHCII (Table 1). These measurements, which span a variety of systems, have greatly enhanced our understanding of what constitutes a functional TCR/pMHC binding interaction. Notably, using artificially altered ligands, it has been shown that CD8+ T-cell clones can respond to antigen even when the affinity of the TCR/pMHC interaction is extremely low (KD > 500 μm).38,39 The accumulated biophysical data suggest a hierarchy of binding affinities, within which pMHCI-specific TCRs bind with stronger affinities (mean KD = 32 μm) compared with pMHCII-specific TCRs (mean KD = 92 μm). Similarly, self-reactive TCRs bind at the lower end of the affinity range (mean KD = 90 μm) and non-self ligands bind at the higher end of the spectrum (mean KD = 8 μm) (Table 1; Fig. 3).40 These observations are consistent with the demonstration that many tumour-associated antigens recognized by CD8+ T-cells are self-derived and subject to immune tolerance mechanisms designed to prevent autoimmunity.41 Indeed, direct biophysical measurements show that anti-cancer TCRs bind their cognate pMHC ligands with affinities that lie approximately one order of magnitude below the corresponding affinities of TCRs specific for non-self antigens.40 These affinity differences are likely to impair the ability of T-cells to kill tumour cells and potentially explain the disappointing success rates of cancer vaccines. Many studies, including numerous clinical trials, have attempted to remedy this constraint by improving the interaction between anti-cancer TCRs and their cognate pMHCs, either by altering the antigenic peptide or by using gene-transfer to express high-affinity anti-cancer TCRs in T-cells.42–44 This strategy to enhance TCR affinity is supported by a number of recent studies that have shown a correlation between affinity and optimal T-cell function (Table 2).
Table 1.
Affinities and kinetics of human T-cell receptor/peptide–MHC (TCR/pMHC) interactions
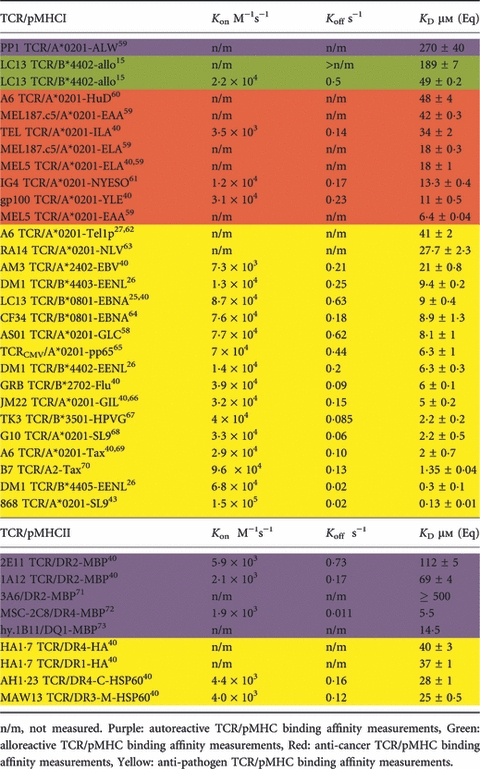 |
Figure 3.
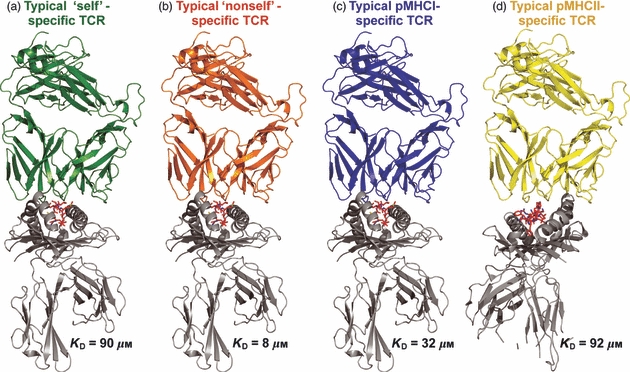
Representation of typical T-cell receptor (TCR) binding affinities. This schematic representation illustrates the mean binding affinities for each type of TCR/peptide–MHC (pMHC) interaction shown. (a) Self-specific TCRs binding to either pMHCI or pMHCII. (b) Non-self-specific TCRs binding to either pMHCI or pMHCII. (c) pMHCI-specific TCRs. (d) pMHCII-specific TCRs. The difference in mean affinity between TCRs specific for self and non-self antigens supports the notion that strongly reactive self-specific T-cells are deleted in the thymus. The weaker mean binding affinity of pMHCII-specific TCRs compared with pMHCI-specific TCRs may reflect the different biological functions of CD4+ and CD8+ T-cells during adaptive immune responses.
Table 2.
Summary of reports that compare T-cell receptor/peptide–MHC complex (TCR/pMHC) biophysical parameters with T-cell activation
| System | TCR/peptide altered | T-cell function and assay time | Biophysical analysis | Binding correlate |
|---|---|---|---|---|
| 2C TCR37 | Peptide | Cytotoxicity assay | Affinity assays | Affinity |
| 2B4 TCR74 | Peptide | 16–24 hr IL-2 | SPR 25°C* | Off-rate |
| 2C TCR75 | Peptide | 2–4 hr cytotoxicity, 24 hr IL-3 | SPR 25°C | Not affinity |
| Model45 | n/a | n/a | n/a | Off-rate |
| KS140 TCR76 | TCR/peptide | 5 hr IFN-γ, < 3 hr TCR down-regulation | n.d. | Off-rate |
| 2B4 TCR77 | Peptide | 24 hr IL-2 | SPR 25°C* | Off-rate |
| Model78 | n/a | n/a | n/a | Off-rate |
| 3.L2 TCR79 | Peptide | 3 min phospho-blot | SPR 25°C | Off-rate |
| PbCS TCR80 | TCR/peptide | 4 hr cytotoxicity, ≤ 15 min Ca2+ flux, 10 min phospho-blot | TCR photo-affinity labelling 26°C | Off-rate |
| 2B4 & 3.L2 TCR81 | TCR/peptide | < 1 hr synapse formation 24 hr proliferation | n.d. | Off-rate |
| OT-1 TCR82 | Peptide | n/a | SPR 25 & 37°C | Off-rate/allosteric effect |
| 5C.C7 TCR83 | TCR | n/a | Tetramer dissociation | Off-rate |
| N30·7 TCR84 | TCR/peptide | 24 hr IL-2 | Tetramer dissociation | Off-rate |
| OT-1 TCR56 | Peptide | ≤ 6 min Ca2+ flux, 30 min phospho-blot, ≤ 24 hr pJun, ≤ 24 hr CD69, 48 hr cytotoxicity | SPR 25 & 37°C, tetramer dissociation | Off-rate/affinity |
| N30·7 TCR85 | TCR | 5 hr TCR down-regulation | n.d. | Off-rate |
| 2B4 TCR86 | Peptide | 24 hr IL-2 | SPR 4-37°C, ITC | Off-rate/allosteric effect |
| OT-1 TCR87 | Peptide | ≤ 3 min ppERK, 3 hr CD69 and IFN-γ | n.d. | Off-rate |
| N30·7 TCR88 | TCR/peptide | 24 hr IL-2 5 hr TCR down-regulation | n.d. | Off-rate |
| P14 TCR89 | Peptide | 4 hr cytotoxicity, 48 hr IFN-γ | SPR 25°C | Affinity |
| P14 TCR47 | Peptide | 4 hr TCR down-regulation | SPR 25°C | Affinity/off-rate |
| 2C TCR90 | TCR/peptide | 30 min pERK, 24 hr IL-2 and IFN-γ | SPR 25°C | Affinity/off-rate |
| B3K506/508 TCR91 | TCR/peptide | 5 hr TNF-α, 16 hr TCR down-regulation, 48 hr proliferation | SPR 25°C | On-rate |
| BV13-clono-1 TCR92 | TCR | ≤ 20 min phospho-blot, 4 hr cytotoxicity ≤ 96 hr proliferation | SPR & cell assays at 25°C | Affinity/off-rate |
| 1G4 TCR93 | Peptide | 4 hr IFN-γ4 hr cytotoxicity | SPR 25°C | Affinity/on-rate/off-rate |
| OT-1 TCR52 | Peptide | 18 hr cytotoxicity | Two-dimensional cell assay 25 & 37°C | Affinity/on-rate/off-rate |
The table details the major studies in which attempts have been made to correlate T-cell function with one or more biophysical parameters. References are arranged chronologically from earliest to latest.
n.d., not determined; *the authors did not carry out measurements for this particular report, but used measurements determined previously.
IL-2, interleukin; IFN-γ, interferon-γ; TNF-α, tumour necrosis factor-α; SPR, surface plasmon resonance; ITC, isothermal titration calorimetry.
TCR/pMHC binding half-life, or off-rate, governs T-cell sensitivity
The spectrum of observed off-rates for different agonist TCR/pMHC interactions lies within a conserved range with a mean of 0·24 s−1 (Table 1). This tight range of off-rate values is consistent with structural data, which show that the number of bonds between different TCR/pMHC complexes is similar.9 The conservation of TCR/pMHC off-rate is also consistent with a kinetic proofreading theory of T-cell activation (Box 1), which states that the TCR must engage pMHC with a sufficient ‘dwell time’ to produce an activation signal, while also disengaging fast enough to allow the cognate pMHC molecule to engage further TCRs in the contact zone.45 This kinetic window of TCR/pMHC binding allows T-cells to be highly sensitive to even very low numbers of pMHC ligands.46 Further support for this notion comes from investigations showing that fast off-rates can lead to T-cell anergy, and that extremely fast off-rates can result in T-cell antagonism (ref. 47 and citations therein). Experimental evidence has also shown that off-rate is important for T-cell sensitivity. Indeed, off-rate has been shown to be the best correlate of T-cell activation in the majority of studies that have measured the biophysical parameters of a TCR/pMHC interaction in conjunction with functional readouts (Table 2).
TCR/pMHC interactions with extremely fast or extremely slow off-rates can be agonistic
The idea that off-rate controls T-cell sensitivity implies that a TCR/pMHC dwell time threshold exists for a T-cell activation signal to be initiated. However, Boulter et al.47 have shown that a murine TCR, P14, binds with an extremely fast off-rate. Furthermore, the sensitivity of T-cells expressing this TCR was directly correlated with TCR affinity in this study, with weakly binding ligands acting as weak agonists, or antagonists. This observation, as well as the discovery that fewer than 10 pMHCs are sufficient to activate T-cells46,48, suggests that multiple binding and serial triggering events may contribute to the process of T-cell activation.49 Other recent investigations have also demonstrated that TCR binding affinity can be the best correlate of T-cell sensitivity, with others favouring on-rate only (Table 2). Studies using a number of enhanced affinity TCRs, some with off-rates in the range of many hours, have been used to show that substantially extended off-rates can still result in T-cell activation.43,50,51 Although such modified TCRs are non-natural, these findings may require a modification of existing serial triggering and kinetic proofreading theories. Interestingly, Huang et al.52 recently developed a novel two-dimensional cell surface binding assay to re-investigate the binding affinity and kinetics of the OT-1 TCR to a range of agonist and antagonist ligands. Using this approach, they found that binding affinity, on-rate and off-rate could all correlate well with T-cell sensitivity. Hence, although the accumulated data suggest that TCR/pMHC off-rate is the most consistent correlate of T-cell sensitivity, on-rate and affinity can also play significant roles depending on the T-cell system under investigation, the binding assay implemented and, perhaps most importantly, the T-cell assay used (Table 2). These differences may also reflect the dynamic events that occur at the T-cell surface during antigen engagement, differences in ligand density, and the local synapse environment. It is, therefore, important to recognize that the described correlation between TCR/pMHC off-rate and T-cell sensitivity is not a universal phenomenon and that further experimental evidence is required for a full understanding of T-cell activation.
Fast TCR on-rates are related to fast TCR off-rates
Compared with the theoretical on-rate, or association, of TCR/pMHC binding (105–109 M−1s−1), which takes into account random collisions and electrostatic interactions, TCRs bind to their cognate pMHC molecules remarkably slowly (103–105 M−1s−1). This observation suggests that, for a TCR to bind to pMHC, it must undergo a structural shuffling, or re-ordering, to achieve the desired conformation. As mentioned above, this feature of TCR binding has been revealed experimentally by comparing the structures of complexed and uncomplexed TCRs, and by using thermodynamic experiments. The latter show that TCR/pMHC binding is normally characterized by a negative TΔSo, or a loss of entropy, suggesting a binding-induced transition from disorder to order.35,53 Interestingly, the range of TCR on-rates (1·9 × 103 to 1·5 × 105 M−1s−1) is similar to the range of TCR off-rates (0·01–0·73 s−1). Furthermore, TCR/pMHC interactions that have on-rates at the faster end of the spectrum also tend to have faster off-rates (Table 1). Hence, the range of binding affinities for individual TCR/pMHC complexes (KD = 0·13 to 500 μm) is governed by differences in the corresponding ratios between on-rate and off-rate, with neither parameter playing a leading role.
Two-dimensional measurements of TCR/pMHC binding affinity and on-rate exceed the corresponding three-dimensional values
Although TCR off-rate is currently the most favoured correlate of T-cell activation, some recent investigations favour TCR on-rate, or a combination of on-rate, off-rate, affinity and heat capacity (ΔCpo) (Table 2).35 How can we reconcile these disparate findings? First, as mentioned above, events at the T-cell surface are more complex than a simple 1:1 interaction between a TCR and a pMHC molecule. For example, TCRs migrate into supramolecular activation clusters, forming part of the immune synapse at the cell surface during T-cell activation.54 Second, a number of different T-cell systems and functional outputs have been used to generate the current body of data. Differences in the experimental set-up, the duration of the T-cell activation assay, and how the data were interpreted have probably generated substantial differences between studies. Third, a limitation of attempts to correlate biophysical parameters with T-cell function could be associated with the intrinsic nature of the three-dimensional kinetic measurements obtained. Hence, the question arises as to whether measurements made in three dimensions are pertinent to the two-dimensional nature of TCR/pMHC interactions in the membrane-constrained state on the surface of opposing cells? Recent attempts to address this issue have thrown light on what constitutes optimal binding of TCR to pMHC. Using a micropipette and biomembrane-force probe, Huang et al.52 were able to measure the displacement of an MHC-functionalized bead/red blood cell when contacted with a T-cell bearing OT-1 TCRs. Measurements of affinity in two dimensions were found to be higher than the corresponding three-dimensional measurements because of a faster on-rate in the two-dimensional setting. Furthermore, two-dimensional off-rates were quicker compared with three-dimensional measurements. In support of these observations, Huppa et al.55 recently used a novel fluorescence resonance energy transfer-based cell surface assay to investigate TCR/pMHC binding in situ with similar conclusions.
Conclusions
The structural and biophysical characteristics of the TCR/pMHC interaction are central to the process of T-cell activation. Overall, off-rate is the most favoured correlate of T-cell sensitivity. However, there is considerable disagreement surrounding this notion and many studies, including measurements of TCR/pMHC binding affinity and kinetics in situ, have generated different conclusions. Cross-comparison of datasets between studies is challenging because of methodological and systematic differences. As an example of how such factors may impinge on the results obtained, Rosette et al.56 and Thomas et al.57 have shown that different ovalbumin and Tax specific TCRs, respectively, can generate different activation kinetics, with the former study reporting observable differences only in short-term analyses. Thus, assay time can substantially impact activation readouts and subsequent data interpretation.
By cataloguing and comparing data in this review, we observe a clear relationship between binding affinity and antigenic origin (Table 1). Similarly, off-rate and on-rate values show some consistencies with respect to the source of cognate antigen. Interestingly, TCRs that bind to potentially dangerous non-self epitopes tend to have higher affinities, slower off-rates and faster on-rates. On the other hand, self-specific TCRs tend to bind with weaker affinities, faster off-rates and slower on-rates. This distinction could explain, at least in part, why most anti-cancer T-cell responses are sub-optimal. Hence, engineering anti-cancer TCR/pMHC binding parameters towards those of TCRs that bind to foreign antigens, either by mutating the TCR or the antigenic peptide, may be a useful therapeutic strategy for future drug and vaccine design.
Disclosures
The authors declare no conflict of financial interests.
References
- 1.Gao GF, Rao Z, Bell JI. Molecular coordination of αβ T-cell receptors and coreceptors CD8 and CD4 in their recognition of peptide-MHC ligands. Trends Immunol. 2002;23:408–13. doi: 10.1016/s1471-4906(02)02282-2. [DOI] [PubMed] [Google Scholar]
- 2.Janeway CA., Jr The T cell receptor as a multicomponent signalling machine: CD4/CD8 coreceptors and CD45 in T cell activation. Annu Rev Immunol. 1992;10:645–74. doi: 10.1146/annurev.iy.10.040192.003241. [DOI] [PubMed] [Google Scholar]
- 3.Barber EK, Dasgupta JD, Schlossman SF, Trevillyan JM, Rudd CE. The CD4 and CD8 antigens are coupled to a protein-tyrosine kinase (p56lck) that phosphorylates the CD3 complex. Proc Natl Acad Sci U S A. 1989;86:3277–81. doi: 10.1073/pnas.86.9.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veillette A, Bookman MA, Horak EM, Bolen JB. The CD4 and CD8 T cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell. 1988;55:301–8. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- 5.Arcaro A, Gregoire C, Bakker TR, et al. CD8β endows CD8 with efficient coreceptor function by coupling T cell receptor/CD3 to raft-associated CD8/p56(lck) complexes. J Exp Med. 2001;194:1485–95. doi: 10.1084/jem.194.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arcaro A, Gregoire C, Boucheron N, Stotz S, Palmer E, Malissen B, Luescher IF. Essential role of CD8 palmitoylation in CD8 coreceptor function. J Immunol. 2000;165:2068–76. doi: 10.4049/jimmunol.165.4.2068. [DOI] [PubMed] [Google Scholar]
- 7.Gakamsky DM, Luescher IF, Pramanik A, Kopito RB, Lemonnier F, Vogel H, Rigler R, Pecht I. CD8 kinetically promotes ligand binding to the T-cell antigen receptor. Biophys J. 2005;89:2121–33. doi: 10.1529/biophysj.105.061671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wooldridge L, Van den Berg HA, Glick M, et al. Interaction between the CD8 coreceptor and major histocompatibility complex class I stabilizes T cell receptor-antigen complexes at the cell surface. J Biol Chem. 2005;280:27491–501. doi: 10.1074/jbc.M500555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–66. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 10.Rudolph MG, Wilson IA. The specificity of TCR/pMHC interaction. Curr Opin Immunol. 2002;14:52–65. doi: 10.1016/s0952-7915(01)00298-9. [DOI] [PubMed] [Google Scholar]
- 11.Jameson SC, Hogquist KA, Bevan MJ. Positive selection of thymocytes. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- 12.Van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–84. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- 13.Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–46. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 14.Ding YH, Smith KJ, Garboczi DN, Utz U, Biddison WE, Wiley DC. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity. 1998;8:403–11. doi: 10.1016/s1074-7613(00)80546-4. [DOI] [PubMed] [Google Scholar]
- 15.Macdonald WA, Chen Z, Gras S, et al. T cell allorecognition via molecular mimicry. Immunity. 2009;31:897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 16.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–83. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 17.Tynan FE, Burrows SR, Buckle AM, et al. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat Immunol. 2005;6:1114–22. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 18.Burrows SR, Chen Z, Archbold JK, et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc Natl Acad Sci U S A. 2010;107:10608–13. doi: 10.1073/pnas.1004926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Laethem F, Sarafova SD, Park JH, et al. Deletion of CD4 and CD8 coreceptors permits generation of αβ T cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–50. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Miles JJ, Douek DC, Price DA. Bias in the αβ T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol. 2011;89:375–87. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- 21.Dai S, Huseby ES, Rubtsova K, Scott-Browne J, Crawford F, Macdonald WA, Marrack P, Kappler JW. Crossreactive T cells spotlight the germline rules for αβ T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–34. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–7. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the αβ T-cell receptor control thymic selection. Nature. 2009;458:1043–6. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418:552–6. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- 25.Borg NA, Ely LK, Beddoe T, et al. The CDR3 regions of an immunodominant T cell receptor dictate the ‘energetic landscape’ of peptide-MHC recognition. Nat Immunol. 2005;6:171–80. doi: 10.1038/ni1155. [DOI] [PubMed] [Google Scholar]
- 26.Archbold JK, Macdonald WA, Gras S, et al. Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition. J Exp Med. 2009;206:209–19. doi: 10.1084/jem.20082136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borbulevych OY, Piepenbrink KH, Gloor BE, Scott DR, Sommese RF, Cole DK, Sewell AK, Baker BM. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity. 2009;31:885–96. doi: 10.1016/j.immuni.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia KC, Degano M, Pease LR, Huang M, Peterson PA, Teyton L, Wilson IA. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 1998;279:1166–72. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 29.Garcia KC, Teyton L, Wilson IA. Structural basis of T cell recognition. Annu Rev Immunol. 1999;17:369–97. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 30.Reiser JB, Darnault C, Gregoire C, et al. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat Immunol. 2003;4:241–7. doi: 10.1038/ni891. [DOI] [PubMed] [Google Scholar]
- 31.Ishizuka J, Stewart-Jones GB, Van der Merwe A, Bell JI, McMichael AJ, Jones EY. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its Vβ domain. Immunity. 2008;28:171–82. doi: 10.1016/j.immuni.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 32.Wilson DB, Wilson DH, Schroder K, Pinilla C, Blondelle S, Houghten RA, Garcia KC. Specificity and degeneracy of T cells. Mol Immunol. 2004;40:1047–55. doi: 10.1016/j.molimm.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong KM, Baker BM. A comprehensive calorimetric investigation of an entropically driven T cell receptor-peptide/major histocompatibility complex interaction. Biophys J. 2007;93:597–609. doi: 10.1529/biophysj.107.104570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cole DK, Yuan F, Rizkallah PJ, et al. Germline-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. J Biol Chem. 2009;284:27281–9. doi: 10.1074/jbc.M109.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armstrong KM, Insaidoo FK, Baker BM. Thermodynamics of T-cell receptor-peptide/MHC interactions: progress and opportunities. J Mol Recognit. 2008;21:275–87. doi: 10.1002/jmr.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ely LK, Beddoe T, Clements CS, Matthews JM, Purcell AW, Kjer-Nielsen L, McCluskey J, Rossjohn J. Disparate thermodynamics governing T cell receptor-MHC-I interactions implicate extrinsic factors in guiding MHC restriction. Proc Natl Acad Sci U S A. 2006;103:6641–6. doi: 10.1073/pnas.0600743103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sykulev Y, Brunmark A, Jackson M, Cohen RJ, Peterson PA, Eisen HN. Kinetics and affinity of reactions between an antigen-specific T cell receptor and peptide-MHC complexes. Immunity. 1994;1:15–22. doi: 10.1016/1074-7613(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 38.Huseby ES, Crawford F, White J, Marrack P, Kappler JW. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat Immunol. 2006;7:1191–9. doi: 10.1038/ni1401. [DOI] [PubMed] [Google Scholar]
- 39.Laugel B, Van den Berg HA, Gostick E, et al. Different T cell receptor affinity thresholds and CD8 coreceptor dependence govern cytotoxic T lymphocyte activation and tetramer binding properties. J Biol Chem. 2007;282:23799–810. doi: 10.1074/jbc.M700976200. [DOI] [PubMed] [Google Scholar]
- 40.Cole DK, Pumphrey NJ, Boulter JM, et al. Human TCR-binding affinity is governed by MHC class restriction. J Immunol. 2007;178:5727–34. doi: 10.4049/jimmunol.178.9.5727. [DOI] [PubMed] [Google Scholar]
- 41.Boon T, Coulie PG, Van den Eynde BJ, Van der Bruggen P. Human T cell responses against melanoma. Annu Rev Immunol. 2006;24:175–208. doi: 10.1146/annurev.immunol.24.021605.090733. [DOI] [PubMed] [Google Scholar]
- 42.Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10:909–15. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varela-Rohena A, Molloy PE, Dunn SM, et al. Control of HIV-1 immune escape by CD8 T cells expressing enhanced T-cell receptor. Nat Med. 2008;14:1390–5. doi: 10.1038/nm.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, Bennett AD, Zheng Z, et al. High-affinity TCRs generated by phage display provide CD4+ T cells with the ability to recognize and kill tumor cell lines. J Immunol. 2007;179:5845–54. doi: 10.4049/jimmunol.179.9.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci U S A. 1995;92:5042–6. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manz BN, Jackson BL, Petit RS, Dustin ML, Groves J. T-cell triggering thresholds are modulated by the number of antigens within individual T-cell receptor clusters. Proc Natl Acad Sci U S A. 2011;108:9089–94. doi: 10.1073/pnas.1018771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boulter JM, Schmitz N, Sewell AK, Godkin AJ, Bachmann MF, Gallimore AM. Potent T cell agonism mediated by a very rapid TCR/pMHC interaction. Eur J Immunol. 2007;37:798–806. doi: 10.1002/eji.200636743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. T cell killing does not require the formation of a stable mature immunological synapse. Nat Immunol. 2004;5:524–30. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- 49.Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature. 1995;375:148–51. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Moysey R, Molloy PE, et al. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat Biotechnol. 2005;23:349–54. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- 51.Stone JD, Chervin AS, Kranz DM. T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity. Immunology. 2009;126:165–76. doi: 10.1111/j.1365-2567.2008.03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, Zhu C. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature. 2010;464:932–6. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Armstrong KM, Piepenbrink KH, Baker BM. Conformational changes and flexibility in T-cell receptor recognition of peptide-MHC complexes. Biochem J. 2008;415:183–96. doi: 10.1042/BJ20080850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–6. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 55.Huppa JB, Axmann M, Mortelmaier MA, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–7. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosette C, Werlen G, Daniels MA, et al. The impact of duration versus extent of TCR occupancy on T cell activation: a revision of the kinetic proofreading model. Immunity. 2001;15:59–70. doi: 10.1016/s1074-7613(01)00173-x. [DOI] [PubMed] [Google Scholar]
- 57.Thomas S, Xue SA, Bangham C, Jakobsen BK, Morris E, Stauss HJ. Human T cells expressing affinity matured TCR display accelerated responses but fail to recognise low density of MHC/peptide antigen. Blood. 2011;118:319–29. doi: 10.1182/blood-2010-12-326736. [DOI] [PubMed] [Google Scholar]
- 58.Miles JJ, Bulek AM, Cole DK, et al. Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein–Barr virus infection. PLoS Pathog. 2010;6:e1001198. doi: 10.1371/journal.ppat.1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cole DK, Edwards ES, Wynn KK, et al. Modification of MHC anchor residues generates heteroclitic peptides that alter TCR binding and T cell recognition. J Immunol. 2010;185:2600–10. doi: 10.4049/jimmunol.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borbulevych OY, Piepenbrink KH, Baker BM. Conformational melding permits a conserved binding geometry in TCR recognition of foreign and self molecular mimics. J Immunol. 2011;186:2950–8. doi: 10.4049/jimmunol.1003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen JL, Stewart-Jones G, Bossi G, et al. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J Exp Med. 2005;201:1243–55. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laugel B, Boulter JM, Lissin N, et al. Design of soluble recombinant T cell receptors for antigen targeting and T cell inhibition. J Biol Chem. 2005;280:1882–92. doi: 10.1074/jbc.M409427200. [DOI] [PubMed] [Google Scholar]
- 63.Gras S, Saulquin X, Reiser JB, et al. Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope. J Immunol. 2009;183:430–7. doi: 10.4049/jimmunol.0900556. [DOI] [PubMed] [Google Scholar]
- 64.Gras S, Burrows SR, Kjer-Nielsen L, et al. The shaping of T cell receptor recognition by self-tolerance. Immunity. 2009;30:193–203. doi: 10.1016/j.immuni.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 65.Gakamsky DM, Lewitzki E, Grell E, Saulquin X, Malissen B, Montero-Julian F, Bonneville M, Pecht I. Kinetic evidence for a ligand-binding-induced conformational transition in the T cell receptor. Proc Natl Acad Sci U S A. 2007;104:16639–44. doi: 10.1073/pnas.0707061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Willcox BE, Gao GF, Wyer JR, Ladbury JE, Bell JI, Jakobsen BK, Van der Merwe PA. TCR binding to peptide-MHC stabilizes a flexible recognition interface. Immunity. 1999;10:357–65. doi: 10.1016/s1074-7613(00)80035-7. [DOI] [PubMed] [Google Scholar]
- 67.Gras S, Chen Z, Miles JJ, et al. Allelic polymorphism in the T cell receptor and its impact on immune responses. J Exp Med. 2010;207:1555–67. doi: 10.1084/jem.20100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee JK, Stewart-Jones G, Dong T, et al. T cell cross-reactivity and conformational changes during TCR engagement. J Exp Med. 2004;200:1455–66. doi: 10.1084/jem.20041251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding YH, Baker BM, Garboczi DN, Biddison WE, Wiley DC. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 1999;11:45–56. doi: 10.1016/s1074-7613(00)80080-1. [DOI] [PubMed] [Google Scholar]
- 70.Davis-Harrison RL, Armstrong KM, Baker BM. Two different T cell receptors use different thermodynamic strategies to recognize the same peptide/MHC ligand. J Mol Biol. 2005;346:533–50. doi: 10.1016/j.jmb.2004.11.063. [DOI] [PubMed] [Google Scholar]
- 71.Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24:2968–79. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yin Y, Li Y, Kerzic MC, Martin R, Mariuzza RA. Structure of a TCR with high affinity for self-antigen reveals basis for escape from negative selection. EMBO J. 2011;30:1137–48. doi: 10.1038/emboj.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sethi DK, Schubert DA, Anders AK, et al. A highly tilted binding mode by a self-reactive T cell receptor results in altered engagement of peptide and MHC. J Exp Med. 2011;208:91–102. doi: 10.1084/jem.20100725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsui K, Boniface JJ, Steffner P, Reay PA, Davis MM. Kinetics of T-cell receptor binding to peptide/I-Ek complexes: correlation of the dissociation rate with T-cell responsiveness. Proc Natl Acad Sci U S A. 1994;91:12862–6. doi: 10.1073/pnas.91.26.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.al-Ramadi BK, Jelonek MT, Boyd LF, Margulies DH, Bothwell AL. Lack of strict correlation of functional sensitization with the apparent affinity of MHC/peptide complexes for the TCR. J Immunol. 1995;155:662–73. [PubMed] [Google Scholar]
- 76.Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature. 1995;375:148–51. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 77.Lyons DS, Lieberman SA, Hampl J, Boniface JJ, Chien Y, Berg LJ, Davis MM. A TCR binds to antagonist ligands with lower affinities and faster dissociation rates than to agonists. Immunity. 1996;5:53–61. doi: 10.1016/s1074-7613(00)80309-x. [DOI] [PubMed] [Google Scholar]
- 78.Rabinowitz JD, Beeson C, Lyons DS, Davis MM, McConnell HM. Kinetic discrimination in T-cell activation. Proc Natl Acad Sci USA. 1996;93:1401–5. doi: 10.1073/pnas.93.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kersh GJ, Kersh EN, Fremont DH, Allen PM. High- and low-potency ligands with similar affinities for the TCR: the importance of kinetics in TCR signaling. Immunity. 1998;9:817–26. doi: 10.1016/s1074-7613(00)80647-0. [DOI] [PubMed] [Google Scholar]
- 80.Hudrisier D, Kessler B, Valitutti S, Horvath C, Cerottini JC, Luescher IF. The efficiency of antigen recognition by CD8+ CTL clones is determined by the frequency of serial TCR engagement. J Immunol. 1998;161:553–62. [PubMed] [Google Scholar]
- 81.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–7. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 82.Alam SM, Davies GM, Lin CM, et al. Qualitative and quantitative differences in T cell receptor binding of agonist and antagonist ligands. Immunity. 1999;10:227–37. doi: 10.1016/s1074-7613(00)80023-0. [DOI] [PubMed] [Google Scholar]
- 83.Savage PA, Davis MM. A kinetic window constricts the T cell receptor repertoire in the thymus. Immunity. 2001;14:243–52. doi: 10.1016/s1074-7613(01)00106-6. [DOI] [PubMed] [Google Scholar]
- 84.Kalergis AM, Boucheron N, Doucey MA, Palmieri E, Goyarts EC, Vegh Z, Luescher IF, Nathenson SG. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat Immunol. 2001;2:229–34. doi: 10.1038/85286. [DOI] [PubMed] [Google Scholar]
- 85.Coombs D, Kalergis AM, Nathenson SG, Wofsy C, Goldstein B. Activated TCRs remain marked for internalization after dissociation from pMHC. Nat Immunol. 2002;3:926–31. doi: 10.1038/ni838. [DOI] [PubMed] [Google Scholar]
- 86.Krogsgaard M, Prado N, Adams EJ, He XL, Chow DC, Wilson DB, Garcia KC, Davis MM. Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation. Mol Cell. 2003;12:1367–78. doi: 10.1016/s1097-2765(03)00474-x. [DOI] [PubMed] [Google Scholar]
- 87.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gonzalez PA, Carreno LJ, Coombs D, Mora JE, Palmieri E, Goldstein B, Nathenson SG, Kalergis AM. T cell receptor binding kinetics required for T cell activation depend on the density of cognate ligand on the antigen-presenting cell. Proc Natl Acad Sci U S A. 2005;102:4824–9. doi: 10.1073/pnas.0500922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tian S, Maile R, Collins EJ, Frelinger JA. CD8+ T cell activation is governed by TCR-peptide/MHC affinity, not dissociation rate. J Immunol. 2007;179:2952–60. doi: 10.4049/jimmunol.179.5.2952. [DOI] [PubMed] [Google Scholar]
- 90.Chervin AS, Stone JD, Holler PD, Bai A, Chen J, Eisen HN, Kranz DM. The impact of TCR-binding properties and antigen presentation format on T cell responsiveness. J Immunol. 2009;183:1166–78. doi: 10.4049/jimmunol.0900054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Govern CC, Paczosa MK, Chakraborty AK, Huseby ES. Fast on-rates allow short dwell time ligands to activate T cells. Proc Natl Acad Sci U S A. 2010;107:8724–9. doi: 10.1073/pnas.1000966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schmid DA, Irving MB, Posevitz V, et al. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. J Immunol. 2010;184:4936–46. doi: 10.4049/jimmunol.1000173. [DOI] [PubMed] [Google Scholar]
- 93.Aleksic M, Dushek O, Zhang H, Shenderov E, Chen JL, Cerundolo V, Coombs D, Van der Merwe PA. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity. 2010;32:163–74. doi: 10.1016/j.immuni.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boniface JJ, Reich Z, Lyons DS, Davis MM. Thermodynamics of T cell receptor binding to peptide-MHC: evidence for a general mechanism of molecular scanning. Proc Natl Acad Sci U S A. 1999;96:11446–51. doi: 10.1073/pnas.96.20.11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gil D, Schamel WW, Montoya M, Sanchez-Madrid F, Alarcon B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell. 2002;109:901–12. doi: 10.1016/s0092-8674(02)00799-7. [DOI] [PubMed] [Google Scholar]
- 96.Van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2011;11:47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- 97.Minguet S, Schamel WW. Permissive geometry model. Adv Exp Med Biol. 2008;640:113–20. doi: 10.1007/978-0-387-09789-3_11. [DOI] [PubMed] [Google Scholar]
- 98.Davis SJ, Van der Merwe PA. The structure and ligand interactions of CD2: implications for T-cell function. Immunol Today. 1996;17:177–87. doi: 10.1016/0167-5699(96)80617-7. [DOI] [PubMed] [Google Scholar]


