Summary
Viperin is an interferon-inducible protein that inhibits the replication of a variety of viruses by apparently diverse mechanisms. In some circumstances it also plays a role in intracellular signaling pathways. Its expression in mitochondria, revealed by infection with human cytomegalovirus, also affects cellular metabolic pathways. We review here the current status of our understanding of this unusual molecule.
Introduction
The first line of defense against viral infection is the interferon (IFN) response, which triggers the induction of a broad array of antiviral proteins. Although some IFN-inducible proteins, for example protein kinase R (PKR), the GTPase Mx1 (myxovirus resistance 1), ribonuclease L (RNaseL), ISG15 (IFN-stimulated protein of 15 kDa) and IFIT (IFN-induced proteins with tetratricopeptide repeats), have been functionally well characterized as antiviral effectors (Daffis et al., 2010; Pichlmair et al., 2011; Sadler and Williams, 2008), the functions of most IFN-inducible proteins remain unexplored. Viperin (virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible) is an IFN-inducible protein that has recently received increasing attention. It appears to have a number of functions, from being an antiviral protein to modulating signaling events. Here we review these developments and attempt to provide a coherent view of the properties of this unusual protein.
Characterization of viperin
Viperin was identified as the product of an IFN-γ-inducible gene while analyzing the IFN-γ response of human macrophages (Chin and Cresswell, 2001). It is identical to cig5 (cytomegalovirus inducible gene 5) which was isolated by differential display analysis of primary fibroblasts infected with human cytomegalovirus (HCMV) (Zhu et al., 1997). Viperin is highly conserved in evolution. A homologue called BEST5 (bone-expressed sequence tag 5), was described as an IFN-inducible gene expressed during rat osteoblast differentiation, and another called vig-1 (viral hemorrhagic septicemia virus (VHSV)-induced gene 1) was induced in rainbow trout leukocytes infected with VHSV, a fish rhabdovirus. It has since been cloned from species ranging from the mouse to a variety of other fish. A number of experiments involving either over-expression of viperin or its knockdown by short inhibitory RNAs in cell lines have indicated that the protein possesses antiviral activity against a variety of RNA and DNA viruses (see below).
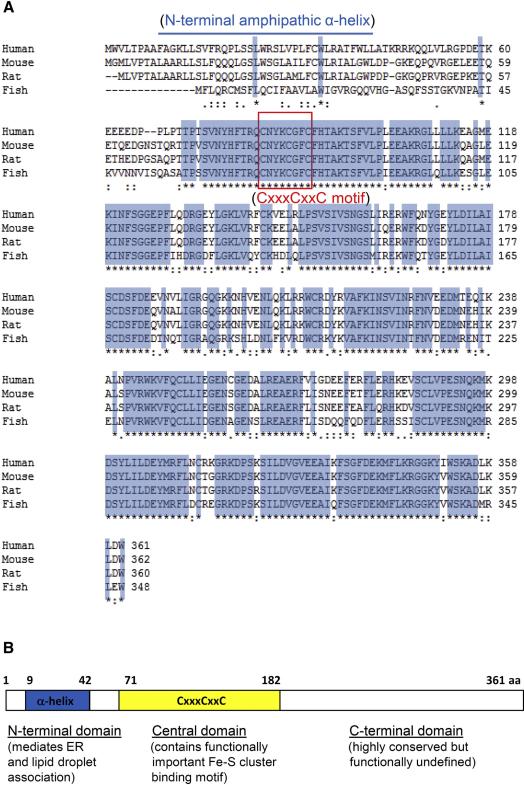
Human viperin is composed of 361 amino acids with a molecular mass of approximately 42 kDa. It is composed of three distinct domains, an N-terminal domain, with length and sequence variability between species, a conserved central domain that contains three cysteine residues organized in a CxxxCxxC motif, and a C-terminal domain that is also highly conserved between species (Figure 1). The N-terminal domain contains an amphipathic α-helix. This sequence may also constitute a leucine zipper, but no zipper-like function, e.g. homodimerization, has been ascribed to it. Amphipathic α-helices are known to bind membranes and induce membrane curvature (McMahon and Gallop, 2005), and the N-terminal α-helix is responsible for viperin association with the cytosolic face of the endoplasmic reticulum (ER) and also with lipid droplets (Hinson and Cresswell, 2009a, b). The interaction of viperin with the former induces structural changes known as crystalloid ER, with closely stacked ER leaflets formed by viperin oligomerization at the cytosolic surface. The amphipathic α-helix is not necessary for viperin oligomerization and does not induce crystalloid ER when attached to the monomeric fluorescent reporter protein dsRed (Hinson and Cresswell, 2009b). The central domain (residues 71–182 of human viperin) shows significant homology with the MoA/PQQIII motif present in the `Radical SAM' family of enzymes that use S-adenosylmethionine as a cofactor, in which the CxxxCxxC motif is responsible for binding iron-sulfur clusters. Thus, viperin is also known as RSAD2 (radical SAM domain-containing 2). Recently it has been shown that viperin can indeed bind Fe-S clusters (Duschene and Broderick, 2010; Shaveta et al., 2010). Radical SAM enzymes generally use the associated Fe-S cluster to generate a highly oxidizing deoxyadenosyl radical to mediate a variety of reactions. Although no specific enzyme activity has been ascribed to viperin, the Fe-S binding motif is essential for its functional activities in hepatitis C virus (HCV) and human cytomegalovirus (HCMV) infection (Jiang et al., 2008; Seo et al., 2011). The role of the conserved C-terminal domain is unknown but it might be involved in protein-protein interactions and/or substrate recognition required for mediating an enzyme activity. Consistent with this, an aromatic amino acid residue at the C-terminus of viperin is required for its antiviral activity against HCV (Jiang et al., 2008).
Figure 1.
Viperin is evolutionarily highly conserved and contains an N-terminal amphipathic α-helix and binds Fe-S clusters. (A) The sequences for human, mouse, rat and fish viperins are shown and conserved amino acid residues are shaded in blue. The amphipathic helix, shared by the mammalian species, extends from residues 9–42. The CxxxCxxC motif, responsible for coordinating the binding of Fe4-S4 clusters is embedded in a conserved region extending from residues 71 to 182 that identifies viperin as a member of the MoaA/PDQQIII family of radical SAM proteins. (B) Schematic diagram of viperin's known domains. The N-terminal domain is required for its localization to the cytosolic face of the ER and lipid droplets. The central domain which contains an Fe-S cluster binding motif (CxxxCxxC) is essential for its functional activities in hepatitis C virus (HCV) and human cytomegalovirus (HCMV) infection. The C-terminal domain (residues 183–361) is highly conserved but no functions have yet been ascribed to it.
Regulation of viperin expression
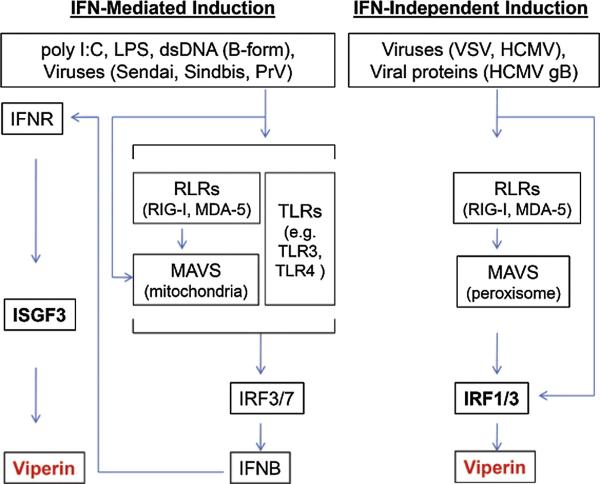
Viperin is induced in a variety of cell types by Type I (α and β), II (γ) and III (λ) IFNs, by double stranded (ds) B-form DNA, the dsRNA analog poly I:C, lipopolysaccharide (LPS) and by infection with a range of viruses. While IFN-γ strongly induces viperin expression in primary macrophages, IFN-α and -β are more effective in inducing its expression in the majority of cell types. Viperin induction by poly I:C, LPS, dsDNA and by many viruses, for example Sendai virus, Sindbis virus and Pseudorabies virus (PrV), is mediated by the classical IFN-stimulated gene (ISG) induction pathways (Bowie and Unterholzner, 2008). IFN-β synthesis is induced by interferon regulatory factors IRF3 and 7 that are activated by pattern recognition receptors (PRRs), such as the Toll-like receptors TLR3 and 4, the cytosolic retinoic acid inducible gene I (RIG-I)-like receptors (RLRs), and cytosolic DNA sensor(s). The secreted IFN-β binds to the type I IFN receptor (IFNR) on the cell surface in autocrine/paracrine manner. This induces the formation of the IFN-stimulated gene factor 3 (ISGF3) complex, which binds to the viperin promoter and induces its expression (Figure 2).
Figure 2.
Regulation of viperin expression. Viperin induction is mediated by both the classical IFN-stimulated gene induction pathway and the IFN-independent pathway. The IFN-mediated viperin gene expression is regulated by ISGF3 (left panel), while IFN-independent viperin gene expression is regulated by IRF1 and IRF3, which can be activated by viral factors or by the peroxisomal MAVS signaling pathway.
Some viruses, such as HCMV and vesicular stomatitis virus (VSV) directly induce viperin independently of IFN production (Boehme et al., 2004; Boudinot et al., 2000; Chin and Cresswell, 2001; Zhu et al., 1997). In the former case this may be mediated by the HCMV membrane glycoprotein, glycoprotein B (gB) (Boehme et al., 2004). IFN-independent viperin induction is directly regulated by IRF1 or IRF3 (DeFilippis et al., 2006; Stirnweiss et al., 2010). It is known that IRF3 is able to discriminate between different IFN-stimulated responsive element (ISRE)-containing genes that directly respond to virus infection (Grandvaux et al., 2002). Both human and mouse viperin promoters have two ISREs that are directly responsive. A recent study has shown that the IFN-independent viperin induction by VSV appears to use IRFs activated by the mitochondrial anti viral signaling protein (MAVS), in this case localized to peroxisomes rather than mitochondria. Upon viral infection, peroxisomal MAVS induces rapid and transient IFN-independent viperin expression, whereas mitochondrial MAVS activates IFN-mediated viperin expression with delayed kinetics (Dixit et al., 2010). The IFN-mediated viperin gene expression is regulated by ISGF3 and the IFN-independent viperin gene expression is regulated by IRF1 and 3, as described above. The latter also play a direct role in promoting the peroxisomal MAVS signaling pathway.
Antiviral functions of viperin
Viperin has been reported to inhibit a broad spectrum of DNA and RNA viruses, including herpesviruses (HCMV), flaviviruses (HCV, West Nile virus (WNV) and Dengue virus), an alphavirus (Sindbis virus), an orthomyxovirus (Influenza A virus), a paramyxovirus (Sendai virus), a rhabdovirus (VSV) and a retrovirus (HIV-1) (Chin and Cresswell, 2001; Jiang et al., 2008; Jiang et al., 2010; Rivieccio et al., 2006; Stirnweiss et al., 2010; Wang et al., 2007; Zhang et al., 2007). Most experiments involve in vitro infections of cell lines, using stable or transient pre-expression (or over-expression) of viperin to inhibit viral replication, or knockdown of viperin by RNA interference (RNAi) to detect a reversal of viperin-mediated inhibition of viral replication. The sheer range of affected viruses, which use different routes of infection and mechanisms of replication, has made it very difficult to identify a unified mechanism that explains all the data.
Some in vivo data that argue for an antiviral function were provided by studies of Sindbis virus (Zhang et al., 2007). Subcutaneous inoculation of neonatal mice with RNA encoding Sindbis virus that also encoded viperin resulted in a significant attenuation of virulence compared to a similar Sindbis RNA encoding a control protein. Over-expression of viperin in a stable tetracycline-inducible murine fibroblast culture system also resulted in a modest inhibition of Sindbis virus replication. More recently, Szretter et al. have found that mice lacking the viperin gene are significantly more susceptible to WNV infection, and that this correlates with a modest increase in WNV replication in viperin−/− macrophages and dendritic cells in vitro (Szretter et al., 2011). Other investigators showed that knockdown of viperin with RNAi partially reverses poly I:C-induced inhibition of HIV-1 replication in astrocytes (Rivieccio et al., 2006), suggesting that it has a possible role in the innate response against infection in the central nervous system. In all of these cases the underlying molecular mechanism of viperin action remains unknown.
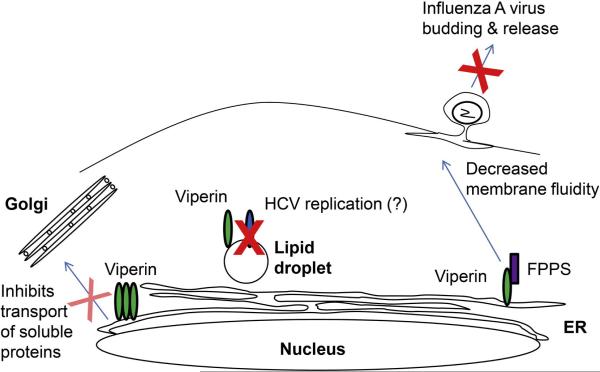
Pre-expression of viperin in human fibroblasts significantly inhibits the replication of HCMV, reducing expression of several HCMV structural proteins, such as gB, pp65 and pp28, that are known to be indispensable for HCMV maturation and/or assembly (Chin and Cresswell, 2001). This suggests that viperin may exert its antiviral effect by inhibiting the synthesis or function of virally-encoded components critical for productive infection. Pre-expressed viperin co-localized with gB in the ER during the attenuated viral infection, while viperin induced by HCMV infection moved together with gB from the ER, first to the Golgi and eventually to the virus assembly compartment (Chin and Cresswell, 2001). It was suggested that this reflected a viral evasion mechanism, i.e. its antiviral effects required viperin to be associated with the ER. Viperin expression has been shown to slow the rate of transport of soluble proteins from the ER (Hinson and Cresswell, 2009b), suggesting that it might interfere with transport of critical viral components. However, the transport rate of membrane-associated glycoproteins was not affected by viperin, making unlikely the otherwise attractive explanation that reduced transport of viral membrane proteins, including gB, to the assembly compartment affects HCMV assembly.
Viperin expression in HeLa cells using a tetracycline-inducible system was found to inhibit budding and release of influenza A virus by disrupting lipid raft microdomains on the plasma membrane (Figure 3). Viperin was found to bind and inhibit farnesyl diphosphate synthase (FPPS), an enzyme involved in the synthesis of multiple isoprenoid-derived lipids, including cholesterol. Over-expression of FPPS reversed the inhibition of virus production and restored normal membrane fluidity (Wang et al., 2007). The data suggest that viperin may inhibit viruses that use lipid rafts for their entry or for budding and release. However, the most appealing idea, that the inhibition of FPPS activity reduced cholesterol synthesis to affect lipid raft formation, could not be substantiated, and the precise mechanism by which viperin affects lipid raft formation and membrane fluidity remains unresolved.
Figure 3.
The effects of viperin expression on cellular processes. Viperin association with the cytosolic face of the ER reduces the secretion rate of soluble molecules, a function ascribed to the N-terminal amphipathic α-helix. The α–helix also can target viperin to lipid droplets, where an activity dependent on Fe-S cluster binding may affect the replication of viruses such as HCV. Viperin expression also affects lipid raft formation, reducing membrane fluidity and inhibiting the budding of influenza A virus. This effect depends on viperin interaction with the enzyme FPPS.
Expression of full-length viperin, but not a mutant form lacking the N-terminal amphipathic α-helix, was found to decrease HCV replication (Jiang et al., 2008). The N-terminal amphipathic α-helix of viperin is essential for its capacity to localize to lipid droplets, a site of HCV replication. It is therefore tempting to suggest that for HCV the antiviral mechanism requires lipid droplet association. The HCV nonstructural 5A protein (NS5A) has a similar N-terminal amphipathic α-helix responsible for its lipid droplet association. Dengue virus also replicates on lipid droplets, suggesting that this may be a characteristic site for flavivirus replication, and the effects of viperin on HCV, Dengue virus and WNV may reflect this common link. This remains speculation until the precise mechanism of viperin action is clarified, although the fact that the Fe-S cluster motif is also required for the inhibitory effect on all three flaviviruses suggests that a radical SAM activity focused on a lipid droplet might be involved (Jiang et al., 2008; Jiang et al., 2010).
Roles for viperin in signaling and the immune response
Recently, it has been suggested that viperin serves as a mediator in signaling pathways, and again lipid droplet association appears to play a key role. A comparison of viperin negative and positive plasmacytoid dendritic cells (pDCs) suggested that viperin mediates TLR7- and TLR9-induced production of Type I IFN by this cell type, which has a critically important role as the major producer of Type I IFNs in viral infections (Saitoh et al., 2011). Viperin was found to recruit the signaling mediators IRAK1 and TRAF6 to lipid droplets in pDCs, apparently by direct interaction. This facilitates ubiquitination of IRAK1, which in turn induces the nuclear translocation of IRF7 and stimulates type I IFN expression. While a direct connection between the two scenarios is not obvious, a signaling function of viperin may underlie the impaired Th2 cell development observed in viperin knockout mice (Qiu et al., 2009). In vitro experiments indicated that viperin facilitates T cell receptor-mediated GATA-3 activation and Th2 cell development by modulating NF-κB and AP-1 activities.
A virus that eliminates viperin and one that co-opts it
Like the adaptive immune system, IFN-inducible proteins commonly force the evolution of viral strategies to counteract their effects and viperin is no exception. Japanese Encephalitis Virus (JEV) infection induces viperin expression but simultaneously causes its proteasome-dependent degradation. Over-expression of viperin or knockdown of viperin expression by RNAi does not substantially influence JEV replication in vitro, but the addition of the proteasome inhibitor MG132 to JEV-infected cells sustains high levels of viperin expression and reveals an antiviral effect (Chan et al., 2008). The mechanism by which viperin degradation is induced is currently unknown.
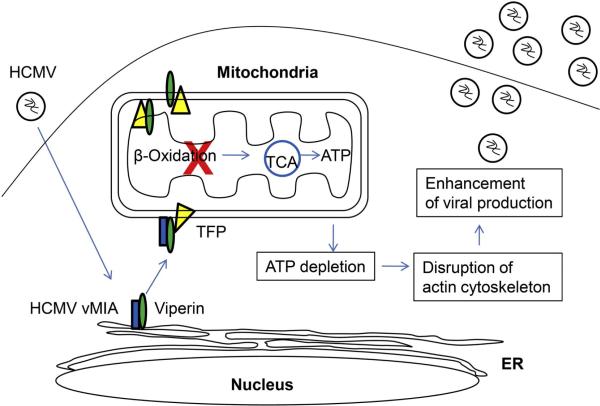
As described earlier, viperin that is induced by HCMV infection moves from its normal location at the cytosolic face of the ER to the Golgi and then the viral assembly compartment (Chin and Cresswell, 2001). This was also interpreted as an evasion strategy, with the assumption that the antiviral effect requires viperin to reside at the ER. Although we have no reason to suppose that lipid droplets are important in the HCMV infection process, the currently unknown mechanism used by the virus to divert viperin would presumably also remove it from lipid droplets, which are ER-derived organelles. However, the story is more complex than it initially appeared. We recently found that, before viperin is transferred to the Golgi and the HCMV assembly compartment, it is transported to mitochondria where it has a profound effect on cellular metabolism (Figure 4) (Seo et al., 2011). The redistribution of viperin to mitochondria is mediated by its interaction with an HCMV-encoded protein, the viral mitochondrial inhibitor of apoptosis (vMIA), which is targeted to mitochondria by an N-terminal mitochondrial localization signal. Viperin then interacts with the β-subunit (HADHB) of the mitochondrial trifunctional protein (TFP) that catalyzes the final steps of fatty acid β-oxidation to generate adenosine triphosphate (ATP). This interaction inhibits TFP activity and therefore reduces cellular ATP generation. A major consequence is that the actin cytoskeleton is disrupted, which enhances the infection. That these effects are not a function of vMIA itself was demonstrated by replacing the N-terminal amphipathic α-helix of viperin with a mitochondrial targeting sequence, either from vMIA or from the mammalian mitochondrial protein Tom70. Directly targeted viperin had the same effects on ATP generation and the actin cytoskeleton with no requirement for vMIA. The Fe-S binding motif of the radical SAM domain was necessary for the effects, although it was not required for the interaction with TFP (Seo et al., 2011).
Figure 4.
Viperin modulates cellular metabolism during HCMV infection. Viperin is relocalized from the ER to mitochondria by binding to the HCMV vMIA protein. Viperin interacts with trifunctional protein (TFP), responsible for fatty acid β-oxidation, at the inner membrane as well as the outer membrane of mitochondria, reducing total cellular ATP generation. A major consequence is that the actin cytoskeleton is disrupted, which enhances viral infection.
How does viperin work?
Four host proteins have been proposed to interact with viperin: FPPS, TFP, IRAK1 and TRAF6. The first two are involved in metabolism, and the last two in signaling. Functions have been ascribed to all of these interactions but it is surprising that viperin is so promiscuous in its choice of binding partners. It is the product of an ancient gene, and acquisition of multiple functions by a protein over evolutionary time may not be unreasonable, but it does give one pause and certainly suggests that the ultimate answers to the function of viperin will be far from simple.
Studies of the protective functions of viperin have to date focused on its effects on viral infections. However, viperin induction by non-viral microbial products such as LPS suggests that viperin might be involved in host responses to bacteria or protozoan parasites. IFN-γ, most commonly associated with antibacterial responses, stimulates viperin expression in macrophages in vitro and the high expression of viperin in macrophages and neutrophils during LCMV infection was also seen in mice injected with LPS (Hinson et al., 2010). Why this is so remains unclear, but the phagocytic and bacteriocidal capacities of these cell types are well established.
Localization to the ER and lipid droplets by the N-terminal amphipathic α-helix seems likely to be functionally important. Lipid droplet association may be particularly important for suppression of flaviviruses, as described above. The amphipathic helix itself has inhibitory effects on the transport of soluble molecules from the ER, perhaps because an increase in membrane curvature reduces the inclusion of soluble cargo in transport vesicles, but it remains unclear what intact viperin does at this site. The variety of experiments suggesting a role for Fe-S binding by mutation of the CxxxCxxC motif indicates that a free radical-based mechanism is likely to underlie most of the observed effects of viperin. The Radical SAM family of enzymes catalyzes a diverse range of reactions, and viperin could, for example, activate glycyl or alkane radicals that modify cellular metabolites or proteins. However, while recombinant viperin can bind Fe-S clusters in vitro, we do not find Fe association with a non-membrane-associated viperin construct that lacks the amphipathic α-helix when it is expressed in either insect cells or yeast (unpublished results). It may be that Fe-S cluster binding is regulated in a manner that requires membrane association. It may even require that viperin interacts with or enters mitochondria, which are the intracellular source of Fe-S clusters (Lill and Muhlenhoff, 2008). The N-terminal sequence of mouse and particularly human viperin has a high probability of being a mitochondrial targeting sequence according to the MitoProt program that identifies such sequences. However, a construct that incorporates this sequence N-terminal to the dsRed protein does not localize to mitochondria (Hinson and Cresswell, 2009a, b). This suggests that any interaction with mitochondria would require either a regulated post-translational modification or an interaction with another host protein, analogous to the vMIA interaction seen in HCMV infection.
Viruses rely on the metabolism of the infected cells to provide the energy and building blocks required for viral replication, and can modify the metabolic state of the infected cell. For example, HCMV infection causes dramatic alterations in intermediary metabolism, similar to those found in tumor cells. The infection upregulates flux through central carbon metabolism, including glycolysis, and increases efflux to fatty acid biosynthesis, a function presumably selected for because the virus requires membrane for the viral envelope (Munger et al., 2008). Similarly, HCV increases host catabolic and biosynthetic activities early in infection, while the virus induces a compensatory metabolic shift to maintain energy homeostasis and cell viability during the progression of infection. Interestingly, the HADHB subunit of TFP was predicted to be one of key regulators of HCV-associated metabolic reprogramming (Diamond et al., 2010), raising the possibility that the viperin interaction with TFP may be involved in this case too.
The effects on cellular metabolism of viperin targeted to mitochondria almost certainly reflect a normal function of IFN-induced viperin. It seems highly unlikely that HCMV `invented' the process; it is much more probable that the virus has co-opted, even exaggerated, a natural function. The inhibition of fatty acid β-oxidation induced by mitochondrial viperin and the resulting reduction of cellular ATP levels may be inhibitory for viruses other than HCMV, and it will be interesting to examine cells infected with different viruses to determine if a fraction of the induced viperin is targeted to mitochondria. Viperin localization to mitochondria might be also related to mitochondrial dynamics. Mitochondrial fusion and fragmentation can regulate mitochondrial antiviral signaling, including MAVS-mediated activation of the RLR pathway (Castanier et al., 2010; Yasukawa et al., 2009). In addition, it has been suggested that the HCMV protein vMIA mediates mitochondrial fragmentation is a strategy to interfere with antiviral signaling pathways (Castanier et al., 2010). Studies to assess the relationship between viperin and mitochondrial dynamics may provide additional insights into the function of viperin during viral infections.
Concluding remarks
The published data indicate that viperin is an important antiviral molecule, and yet its precise mechanism of action remains mysterious. Identification of an activity associated with its membership in the radical SAM family of enzymes would be a great asset in understanding its function. While the ER and lipid droplet associations are clear, mitochondrial localization has so far been observed only in the case of HCMV infection, although it seems likely that viperin must have a mitochondrial function is other circumstances. Mitochondria and the ER are commonly connected in cells by so-called mitochondria-associated membranes (MAM) in a manner that facilitates calcium and phospholipid exchange. It has recently been suggested that MAM is an important site for regulating RIG-I and MAVS signaling (Horner et al., 2011) and it may also be important for viperin-mediated activities.
Viperin knockout mice exist but have failed to show significantly enhanced susceptibility to viral infection until the very recent observation of enhanced WNV infection. The lack of an effect with a particular virus may simply reflect the range of antiviral effectors in addition to viperin that are stimulated by IFNs. The mice have indicated a role for viperin in modulating the T cell response, and cells derived from them have been useful in unraveling the metabolic effects of viperin and its role in type IFN induction in pDCs. However, much more work is required to clarify the multiple functions associated with this enigmatic molecule, which may be more aptly named than originally supposed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Selected reading
- Boehme KW, Singh J, Perry ST, Compton T. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein B. J Virol. 2004;78:1202–1211. doi: 10.1128/JVI.78.3.1202-1211.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudinot P, Riffault S, Salhi S, Carrat C, Sedlik C, Mahmoudi N, Charley B, Benmansour A. Vesicular stomatitis virus and pseudorabies virus induce a vig1/cig5 homologue in mouse dendritic cells via different pathways. J Gen Virol. 2000;81:2675–2682. doi: 10.1099/0022-1317-81-11-2675. [DOI] [PubMed] [Google Scholar]
- Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanier C, Garcin D, Vazquez A, Arnoult D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010;11:133–138. doi: 10.1038/embor.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YL, Chang TH, Liao CL, Lin YL. The cellular antiviral protein viperin is attenuated by proteasome-mediated protein degradation in Japanese encephalitis virus-infected cells. J Virol. 2008;82:10455–10464. doi: 10.1128/JVI.00438-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci U S A. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, et al. 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFilippis VR, Robinson B, Keck TM, Hansen SG, Nelson JA, Fruh KJ. Interferon regulatory factor 3 is necessary for induction of antiviral genes during human cytomegalovirus infection. J Virol. 2006;80:1032–1037. doi: 10.1128/JVI.80.2.1032-1037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, et al. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duschene KS, Broderick JB. The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 2010;584:1263–1267. doi: 10.1016/j.febslet.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, Lin R, Hiscott J. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson ER, Cresswell P. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc Natl Acad Sci U S A. 2009a;106:20452–20457. doi: 10.1073/pnas.0911679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson ER, Cresswell P. The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J Biol Chem. 2009b;284:4705–4712. doi: 10.1074/jbc.M807261200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson ER, Joshi NS, Chen JH, Rahner C, Jung YW, Wang X, Kaech SM, Cresswell P. Viperin is highly induced in neutrophils and macrophages during acute and chronic lymphocytic choriomeningitis virus infection. J Immunol. 2010;184:5723–5731. doi: 10.4049/jimmunol.0903752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, Guo JT. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol. 2008;82:1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, et al. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J Virol. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, Burckstummer T, Stefanovic A, Krieger S, Bennett KL, et al. IFIT1 is an antiviral protein that recognizes 5'-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- Qiu LQ, Cresswell P, Chin KC. Viperin is required for optimal Th2 responses and T-cell receptor-mediated activation of NF-kappaB and AP-1. Blood. 2009;113:3520–3529. doi: 10.1182/blood-2008-07-171942. [DOI] [PubMed] [Google Scholar]
- Rivieccio MA, Suh HS, Zhao Y, Zhao ML, Chin KC, Lee SC, Brosnan CF. TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for viperin/cig5. J Immunol. 2006;177:4735–4741. doi: 10.4049/jimmunol.177.7.4735. [DOI] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Satoh T, Yamamoto N, Uematsu S, Takeuchi O, Kawai T, Akira S. Antiviral protein Viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity. 2011;34:352–363. doi: 10.1016/j.immuni.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Seo JY, Yaneva R, Hinson ER, Cresswell P. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science. 2011;332:1093–1097. doi: 10.1126/science.1202007. [DOI] [PubMed] [Google Scholar]
- Shaveta G, Shi J, Chow VT, Song J. Structural characterization reveals that viperin is a radical S-adenosyl-L-methionine (SAM) enzyme. Biochem Biophys Res Commun. 2010;391:1390–1395. doi: 10.1016/j.bbrc.2009.12.070. [DOI] [PubMed] [Google Scholar]
- Stirnweiss A, Ksienzyk A, Klages K, Rand U, Grashoff M, Hauser H, Kroger A. IFN regulatory factor-1 bypasses IFN-mediated antiviral effects through viperin gene induction. J Immunol. 2010;184:5179–5185. doi: 10.4049/jimmunol.0902264. [DOI] [PubMed] [Google Scholar]
- Szretter KJ, Brien JD, Thackray LB, Virgin HW, Cresswell P, Diamond MS. The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J Virol. 2011;85:11557–11566. doi: 10.1128/JVI.05519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata S, Koshiba T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Burke CW, Ryman KD, Klimstra WB. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J Virol. 2007;81:11246–11255. doi: 10.1128/JVI.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Cong JP, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci U S A. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]