Abstract
Obesity is a well-established risk factor for endometrial cancer, the most common gynecologic malignancy. Recent genome-wide association studies (GWAS) have identified multiple genetic markers for obesity. The authors evaluated the association of obesity-related single nucleotide polymorphisms (SNPs) with endometrial cancer using GWAS data from their recently completed study, the Shanghai Endometrial Cancer Genetics Study, which comprised 832 endometrial cancer cases and 2,049 controls (1996–2005). Thirty-five SNPs previously associated with obesity or body mass index (BMI; weight (kg)/height (m)2) at a minimum significance level of ≤5 × 10−7 in the US National Human Genome Research Institute's GWAS catalog (http://genome.gov/gwastudies) and representing 26 unique loci were evaluated by either direct genotyping or imputation. The authors found that for 22 of the 26 unique loci tested (84.6%), the BMI-associated risk variants were present at a higher frequency in cases than in population controls (P = 0.0003). Multiple regression analysis showed that 9 of 35 BMI-associated variants, representing 7 loci, were significantly associated (P ≤ 0.05) with the risk of endometrial cancer; for all but 1 SNP, the direction of association was consistent with that found for BMI. For consistent SNPs, the allelic odds ratios ranged from 1.15 to 1.29. These 7 loci are in the SEC16B/RASAL, TMEM18, MSRA, SOX6, MTCH2, FTO, and MC4R genes. The associations persisted after adjustment for BMI, suggesting that genetic markers of obesity provide value in addition to BMI in predicting endometrial cancer risk.
Keywords: body mass index, endometrial neoplasms, genetics, genome-wide association study, obesity, risk factors
Endometrial cancer is one of the most common gynecologic malignancies in the United States and many other countries. Although endometrial cancer is relatively uncommon among Chinese women, its incidence has been increasing at an alarming rate. For example, incidence of endometrial cancer among Chinese women in urban Shanghai has increased 90% over the last 2 decades, from 4.0 per 100,000 in 1987 (1) to 7.62 per 100,000 in 2007 (2). Obesity, typically defined as a body mass index (BMI; weight (kg)/height (m)2) of 30 or higher, is a well established risk factor for endometrial cancer, as summarized by the World Cancer Research Fund (3). Obese persons have a 4.5–6.25 times’ higher risk of endometrial cancer compared with nonobese persons (4, 5). The increase in obesity prevalence worldwide over the past few decades (6) may have contributed to the increased incidence of endometrial cancer. In China, the obesity rate reached 7.1% in 2004, a 97% increase compared with the rate in 1992 (7). The contribution of obesity to endometrial cancer risk among Chinese women has been demonstrated previously (8–10).
Although environmental factors are major determinants of obesity risk, genetic susceptibility to obesity is also well recognized. The genetic architecture of BMI and obesity has recently been investigated through the use of genome-wide association studies (GWAS) (11–13). To date, 18 studies have identified 96 single nucleotide polymorphisms (SNPs), representing approximately 75 loci, associated with high BMI and/or obesity. While some loci (e.g., FTO, MC4R) have been verified in multiple studies, others have been reported only in discovery samples. Findings have been replicated in other studies for only 13 loci.
The heritability of BMI is estimated to be 0.3–0.7 (14–17), which is comparable to that of endometrial cancer (approximately 0.5). The strong relation between BMI and endometrial cancer and the availability of GWAS-identified risk variants for obesity motivated us to evaluate the association of BMI-related GWAS markers with endometrial cancer. To our knowledge, this is the first study to systematically test the hypothesis that endometrial cancer and measures of obesity share a common genetic architecture. We accomplished this using GWAS data from our recently completed study, the Shanghai Endometrial Cancer Genetics Study. We further evaluated whether associations between these variants and endometrial cancer were mediated by BMI.
MATERIALS AND METHODS
Study population
In the Shanghai Endometrial Cancer Genetics Study, we recruited incident endometrial cancer cases from the Shanghai Endometrial Cancer Study and controls from the Shanghai Breast Cancer Study. Both the Shanghai Endometrial Cancer Study and the Shanghai Breast Cancer Study were population-based case-control studies. The study design has been described in detail elsewhere (18). Briefly, through Shanghai's population-based tumor registry, we identified 1,449 female residents of Shanghai aged 30–69 years who were newly diagnosed with endometrial cancer (and had no prior history of cancer) between 1997 and 2003. Of these 1,449 women, 1,199 (83%) were successfully recruited. DNA samples from 839 cases who donated a blood sample for the study were genotyped using the Affymetrix 6.0 platform (Affymetrix, Santa Clara, California). Controls came from the Shanghai Breast Cancer Study, which was carried out during approximately the same time period (1996–2005) using a study protocol identical to that of the Shanghai Endometrial Cancer Study (19). Controls without a history of cancer were randomly selected from the general population using the Shanghai Resident Registry in 2 phases. The overall response rate for controls was 91% for phase I and 74% for phase II. DNA samples from 2,049 controls were genotyped using the Affymetrix 6.0 platform. Women with a prior hysterectomy were excluded from this study.
Participants completed a detailed in-person interview at the time of enrollment and provided a blood or buccal cell sample. Trained interviewers, who were retired medical professionals, conducted in-person interviews using a structured questionnaire and took anthropometric measurements, including height, weight, and waist and hip circumferences, according to a standard protocol. Indicators of energy balance (i.e., exercise participation and total energy intake) were derived from the questionnaire data (20). This study was approved by the relevant committees for the use of human subjects at all participating institutions, and all study participants provided written informed consent.
Candidate SNP selection
Our SNP selection scheme is shown in Figure 1. The US National Human Genome Resource Institute (NHGRI) GWAS catalog (21) was used to identify SNPs previously associated with high BMI and/or obesity. We selected the traits obesity, BMI, weight, waist circumference, and adiposity (or “anthropometric” or “quantitative” traits that included these) to identify SNPs. By using this approach, we found 96 SNPs previously associated with the above traits by GWAS. We used the following criteria to select SNPs: 1) a P value less than or equal to 5 × 10−7 in the initial GWAS (n = 41) and 2) SNPs for which the obesity-associated allele was known or could be determined (n = 38). For the remaining 3 SNPs, the obesity-increasing risk allele (e.g., A or G) was not reported in the original publication.
Figure 1.
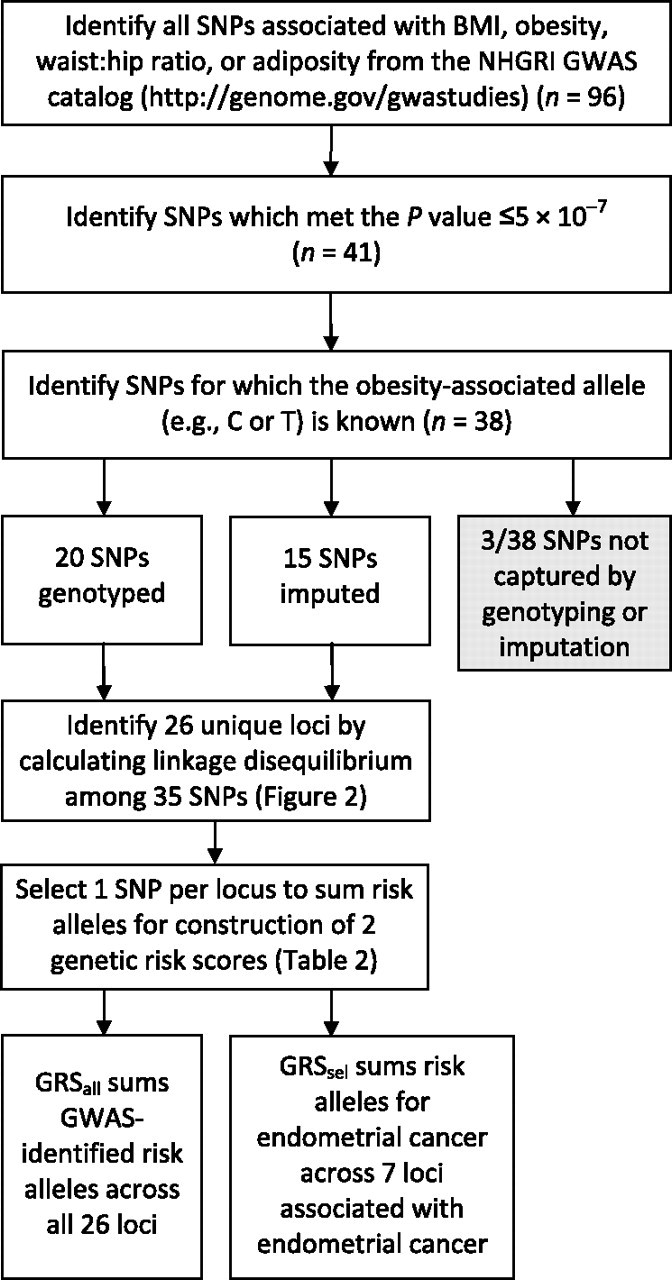
Scheme used for selection of single nucleotide polymorphisms (SNPs) and identification of loci used for genetic risk score (GRS) calculation, Shanghai Endometrial Cancer Genetics Study, Shanghai, China, 2010. (BMI, body mass index; GWAS, genome-wide association study; NHGRI, National Human Genome Research Institute).
Genotyping, quality control, and imputation
Genotyping was performed using the Affymetrix 6.0 array, which includes 906,602 SNPs. The Birdseed algorithm (version 2; http://www.broad.mit.edu/mpg/birdsuite/) was used to call genotypes. Quality control procedures included removal of SNPs with minor allele frequencies less than 0.01, Hardy-Weinberg P values less than 0.00001, and samples with more than 5% of genotypes missing. In addition, we included a number of other quality control measures to verify the integrity of our genotyping. Three sets of SNPs on the Affymetrix SNP Array 6.0 were previously genotyped using different platforms, including: 1) 669 SNPs genotyped by the Affymetrix Targeted Genotyping System, 17 SNPs genotyped by TaqMan, and 56 SNPs genotyped by Sequenom (Sequenom, Inc., San Diego, California). These SNP sets served for cross-platform sample verification. The mean concordance rates were 99.4%, 98.2%, and 98.8% for the Affymetrix Targeted Genotyping System, TaqMan, and Sequenom, respectively, when compared with the Affymetrix SNP Array 6.0. Additionally, we included 1 negative control (water) and 3 positive quality control samples (NA15510, NA10851, and NA18505) purchased from the Coriell Cell Repositories (Coriell Institute, Camden, New Jersey; http://ccr.coriell.org/) in each of the 96-well plates genotyped to assess batch-to-batch validation. The average concordance rate between the quality control samples was 99.8% (median, 100%). Of the 38 BMI-associated variants selected for this study, 20 were directly genotyped with high quality, while the remainder were imputed as described below.
We used the hidden Markov model as implemented in MACH 1.0 (http://www.sph.umich.edu/csg/abecasis/MaCH/) to impute the genotype for variants of interest that were not directly genotyped. Of the remaining 18 selected SNPs that were not directly genotyped, 15 showed high imputation quality, as measured by an Rsq value (an estimate of the squared correlation between imputed and true genotypes) greater than 0.3. Our average Rsq was 0.95. Thus, through either genotyping or imputation, we were able to evaluate the majority (35/38; 92.1%) of selected variants. To determine the number of unique loci these 35 SNPs represented, we used the SNAP (SNP Annotation and Proxy Search) server (Broad Institute, Cambridge, Massachusetts; http://www.broadinstitute.org/mpg/snap/ldsearch.php) to identify variants with r2 > 0.3 (in CEU or CHB + JPT populations). HapMap Phase II data were used as the reference, since these data contain a greater selection of SNPs. Any 2 SNPs with r2 > 0.3 in the HapMap sample were considered to be in sufficient linkage disequilibrium to represent the same locus (i.e., they were not sufficiently independent to warrant artificially weighting the locus). In total, the 35 selected SNPs of interest were found to represent 26 distinct loci (Figure 1).
Statistical analysis
Statistical analysis was carried out using SAS, version 9.2 (SAS Institute, Inc., Cary, North Carolina). Unconditional logistic regression was used to calculate odds ratios and 95% confidence intervals for associations between genotypes and endometrial cancer risk, with adjustment for age, income, and education. Income and education were categorized into 4 and 5 strata, respectively, while age was a continuous variable.
We derived 2 genetic risk scores (GRSs) by summing the number of risk alleles. For imputed SNPs, we used dosage data to capture the probability of a given genotype. For variants that were directly genotyped or found by linkage disequilibrium to represent nonindependent loci, only 1 SNP per locus was included in the GRS calculations. The choice of which SNPs to include per locus was determined first by data source, selecting genotyped variants over imputed variants, and second by the significance of P values for BMI- or obesity-related traits as reported in the NHGRI GWAS catalog. GRSall was defined by summing the risk alleles, according to the original BMI GWAS report, of 26 SNPs that represented all BMI-selected loci. GRSsel was defined by summing the 7 selected variants that were significantly associated with endometrial cancer (P ≤ 0.05) in our study. The SNPs selected were a subset of those used for the calculation of GRSall, selecting 1 SNP per locus and summing the alleles associated with endometrial cancer. GRSs were analyzed both continuously and by categorizing them into quartiles based on the distributions among controls. Relations between GRS and BMI were evaluated by linear regression, whereas relations between GRS and endometrial cancer were evaluated by logistic regression. We evaluated joint effects and tested for the presence of multiplicative interactions of GRSsel with 3 major indicators of energy balance: BMI, exercise participation, and total energy intake.
The probability that an endometrial cancer risk allele and a BMI risk allele have the same direction of effect or the probability of observing a larger number of significant associations (P ≤ 0.05) than would be expected by chance is a function of the binomial distribution (12). We conducted a global test for the hypothesis that the risk alleles for BMI- or obesity-related SNPs would have a higher frequency among endometrial cancer cases than among controls based on the probability of deviation from the binomial distribution, as well as a test evaluating whether the number of significant associations with endometrial cancer (P ≤ 0.05) was greater than expected for the number of loci tested.
All statistical tests were based on 2-sided probabilities, with significance determined by P ≤ 0.05.
RESULTS
Associations of selected demographic characteristics and known risk factors with endometrial cancer are shown in Table 1. As expected, cases and controls differed in regards to age, income, education, BMI, waist:hip ratio, and other traditional risk factors for endometrial cancer.
Table 1.
Demographic and Other Characteristics Associated With Endometrial Cancer in the Shanghai Endometrial Cancer Genetics Study, Shanghai, China, 1996–2005
| Controls (n = 2,049) |
Cases (n = 832) |
Odds Ratioa | 95% Confidence Interval | |||
| % | Mean (SD) | % | Mean (SD) | |||
| Age, years | ||||||
| <40 | 9.4 | 4.1 | 1 | Reference | ||
| 40–49 | 47.4 | 28.0 | 1.60 | 1.07, 2.39 | ||
| 50–59 | 26.8 | 35.7 | 3.22 | 2.16, 4.81 | ||
| ≥60 | 16.4 | 32.2 | 5.33 | 3.50, 8.12 | ||
| Income, yuan/year | ||||||
| <10,000 | 32.2 | 12.5 | 1 | Reference | ||
| 10,000–19,999 | 37.2 | 44.0 | 3.29 | 2.56, 4.24 | ||
| 20,000–29,999 | 18.5 | 20.3 | 3.66 | 2.73, 4.90 | ||
| ≥30,000 | 12.2 | 23.2 | 6.76 | 4.98, 9.17 | ||
| Education | ||||||
| None | 5.2 | 9.3 | 1 | Reference | ||
| Elementary school | 8.5 | 14.9 | 1.11 | 0.76, 1.64 | ||
| Middle school | 42.4 | 38.9 | 1.15 | 0.80, 1.65 | ||
| High school | 34.4 | 24.2 | 0.82 | 0.56, 1.20 | ||
| College/post-high-school | 9.5 | 12.7 | 0.88 | 0.58, 1.34 | ||
| Waist:hip ratio | ||||||
| <0.77 | 23.9 | 8.4 | 1 | Reference | ||
| 0.77–<0.81 | 28.0 | 19.1 | 1.91 | 1.39, 2.62 | ||
| 0.81–<0.84 | 22.1 | 19.7 | 2.23 | 1.61, 3.08 | ||
| ≥0.84 | 26.0 | 52.8 | 4.89 | 3.63, 6.59 | ||
| Body mass indexb | ||||||
| <21.7 | 33.4 | 14.0 | 1 | Reference | ||
| 21.7–24.5 | 33.4 | 28.3 | 1.68 | 1.30, 2.18 | ||
| >24.5 | 33.3 | 57.7 | 3.13 | 2.44, 4.01 | ||
| Physical activity participation | ||||||
| Median or more | 15.3 | 13.4 | 1 | Reference | ||
| Less than median | 14.9 | 15.6 | 1.52 | 1.10, 2.10 | ||
| None | 69.7 | 71.0 | 2.03 | 1.56, 2.65 | ||
| Energy intake, kcal | ||||||
| <1,460 | 25.0 | 23.3 | 1 | Reference | ||
| 1,460–<1,701 | 25.0 | 23.8 | 0.95 | 0.74, 1.22 | ||
| 1,701–<1,983 | 24.9 | 23.2 | 0.95 | 0.74, 1.22 | ||
| ≥1,983 | 25.0 | 29.7 | 1.22 | 0.96, 1.55 | ||
| Age at menarche, years | ||||||
| <13 | 8.4 | 11.5 | 1 | Reference | ||
| 13–<15 | 39.7 | 40.0 | 0.68 | 0.50, 0.93 | ||
| 15–<16 | 19.1 | 20.0 | 0.66 | 0.47, 0.92 | ||
| ≥16 | 32.8 | 28.5 | 0.50 | 0.37, 0.69 | ||
| Age at menopause, years | ||||||
| <47 | 24.0 | 12.7 | 1 | Reference | ||
| 47–<49 | 20.2 | 16.7 | 1.34 | 0.88, 2.04 | ||
| 49–<51 | 26.8 | 30.9 | 1.72 | 1.17, 2.52 | ||
| ≥51 | 29.0 | 39.8 | 1.81 | 1.25, 2.62 | ||
| Reproductive years | 31.1 (5.2) | 34.6 (4.9) | 1.68 | 1.09, 1.14 | ||
| No. of pregnancies | 2.54 (1.33) | 2.64 (1.57) | 0.82 | 0.76, 0.88 | ||
| Family history of cancer | 26.7 | 34.5 | 1.38 | 1.14, 1.66 | ||
| Family history of endometrial or colorectal cancer | 2.1 | 5.2 | 2.68 | 1.68, 4.28 | ||
Abbreviation: SD, standard deviation.
Odds ratios were adjusted for age, income, and education.
Weight (kg)/height (m)2.
Table 2 presents the associations of GWAS-identified obesity-related SNPs with BMI in our study population. Ten variants in 4 gene regions (SEC16B, TRHR, FTO, and MC4R) were significantly associated with BMI (Figure 2). The probability of observing 4 of 26 loci with significant associations with BMI (at P ≤ 0.05) was significantly greater than expected by chance (P = 0.039). Further, 26 of 35 SNPs associated with BMI traits by GWAS showed a consistent direction of association (as indicated by positive beta coefficients) with BMI in our study population (i.e., the BMI risk allele was the same as in the GWAS from which it was selected). For loci with more than 1 SNP identified in the NHGRI GWAS catalog, we selected 1 SNP to represent each locus (see Materials and Methods), resulting in a total of 26 unique loci. On this per-locus basis, 19 of 26 loci had consistent directions of association for BMI in both prior GWAS and our study population (binomial sign test: P = 0.01). For all 10 SNPs in the 4 loci (2, 13, 21, and 25) that were significantly associated with BMI in our study (P ≤ 0.05), the directions of association were consistent with prior GWAS.
Table 2.
Obesity-related Single Nucleotide Polymorphisms Identified in Genome-wide Association Studies and Their Associations With Body Mass Index, Shanghai Endometrial Cancer Genetics Study, Shanghai, China, 1996–2005
| Locus | dbSNP Identification No. | Chr | Position | Nearby Gene(s) | Initial GWAS Report |
SECGS (n = 2,371 Persons) |
|||||||
| Trait | PMID | Reference P Valuea | GWAS Risk Alleleb | βc | 95% Confidence Interval | P Valued | Typing | GRSalle | |||||
| 1 | rs2568958 | 1 | 72537704 | NEGR1 | BMI | 19079260 | 1.0 × 10−11 | A | −0.20 | −0.54, 0.15 | 0.2583 | I | |
| 1 | rs2815752 | 1 | 72585028 | NEGR1 | BMI | 19079261 | 6.0 × 10−08 | A | −0.21 | −0.55, 0.14 | 0.2381 | T | Yes |
| 2 | rs10913469 | 1 | 176180142 | SEC16B, RASAL2 | BMI | 19079260 | 6.0 × 10−08 | C | 0.39 | 0.15, 0.63 | 0.0014 | I | Yes |
| 3 | rs2605100 | 1 | 217710847 | LYPLAL1 | Adiposity | 19557161 | 3.0 × 10−08 | G | −0.11 | −0.34, 0.12 | 0.3471 | T | Yes |
| 4 | rs6429082 | 1 | 233666752 | TBCE | Adiposity | 19557161 | 3.0 × 10−07 | C | −0.01 | −0.20, 0.18 | 0.9197 | T | Yes |
| 5 | rs6548238 | 2 | 624905 | TMEM18 | BMI | 19079261 | 1.0 × 10−18 | C | 0.18 | −0.12, 0.49 | 0.2425 | T | Yes |
| 5 | rs7561317 | 2 | 634953 | TMEM18 | BMI | 19079260 | 4.0 × 10−17 | G | 0.19 | −0.12, 0.50 | 0.2386 | I | |
| 6 | rs1260326 | 2 | 27584444 | GCKR | WC | 18454146 | 4.0 × 10−08 | T | 0.08 | −0.13, 0.29 | 0.4611 | I | Yes |
| 7 | rs7647305 | 3 | 187316984 | SFRS10, ETV5, DGKG | BMI | 19079260 | 7.0 × 10−11 | C | −0.16 | −0.60, 0.28 | 0.4745 | I | Yes |
| 8 | rs10938397 | 4 | 44877284 | GNPDA2 | BMI | 19079261 | 3.0 × 10−16 | G | 0.03 | −0.17, 0.23 | 0.7801 | I | Yes |
| 9 | rs12517906 | 5 | 180103425 | MGAT1 | Weight | 19851299 | 7.0 × 10−08 | T | −0.04 | −0.35, 0.28 | 0.8215 | T | Yes |
| 10 | rs2844479 | 6 | 31680935 | AIF1, NCR3 | Weight | 19079260 | 2.0 × 10−08 | A | 0.15 | −0.04, 0.34 | 0.1302 | I | Yes |
| 11 | rs987237 | 6 | 50911009 | TFAP2B | Adiposity | 19557161 | 2.0 × 10−11 | G | 0.10 | −0.15, 0.34 | 0.4404 | T | Yes |
| 12 | rs545854 | 8 | 9897490 | MSRA | Adiposity | 19557161 | 9.0 × 10−09 | C | 0.10 | −0.08, 0.28 | 0.2867 | T | Yes |
| 13 | rs7832552 | 8 | 110184852 | TRHR | Lean body mass | 19268274 | 4.0 × 10−10 | T | 0.20 | 0.01, 0.38 | 0.0377 | T | Yes |
| 14 | rs297325 | 11 | 16346170 | SOX6 | Obesity | 19714249 | 4.0 × 10−07 | C | −0.06 | −0.25, 0.13 | 0.5366 | T | Yes |
| 15 | rs925946 | 11 | 27623778 | BDNF | BMI | 19079260 | 9.0 × 10−10 | T | 0.09 | −0.39, 0.57 | 0.7054 | I | Yes |
| 16 | rs6265 | 11 | 27636492 | BDNF | BMI | 19079260 | 5.0 × 10−10 | C | 0.14 | −0.04, 0.32 | 0.1352 | T | Yes |
| 17 | rs10838738 | 11 | 47619625 | MTCH2 | BMI | 19079261 | 5.0 × 10−09 | G | 0.04 | −0.15, 0.23 | 0.6806 | I | Yes |
| 18 | rs7138803 | 12 | 48533735 | BCDIN3D, FAIM2 | BMI | 19079260 | 1.0 × 10−07 | A | −0.02 | −0.22, 0.18 | 0.8464 | T | Yes |
| 19 | rs7498665 | 16 | 28790742 | SH2B1, ATP2A1 | BMI | 19079261 | 5.0 × 10−11 | G | 0.02 | −0.26, 0.31 | 0.8845 | I | Yes |
| 20 | rs6499640 | 16 | 52327178 | FTO | BMI | 19079260 | 4.0 × 10−13 | A | −0.01 | −0.25, 0.24 | 0.9676 | I | Yes |
| 21 | rs1421085 | 16 | 52358455 | FTO | Obesity | 19151714 | 1.0 × 10−28 | C | 0.41 | 0.13, 0.69 | 0.0036 | T | |
| 21 | rs1558902 | 16 | 52361075 | FTO | WC | 19557197 | 5.0 × 10−19 | A | 0.40 | 0.13, 0.68 | 0.0041 | I | |
| 21 | rs1121980 | 16 | 52366748 | FTO | BMI | 18159244 | 1.0 × 10−07 | A | 0.31 | 0.07, 0.55 | 0.0107 | T | |
| 21 | rs8050136 | 16 | 52373776 | FTO | BMI/weight | 19079260 | 1.0 × 10−47 | A | 0.42 | 0.15, 0.69 | 0.0027 | T | Yes |
| 21 | rs9939609 | 16 | 52378028 | FTO | BMI | 17434869 | 2.0 × 10−20 | A | 0.42 | 0.14, 0.69 | 0.0029 | T | |
| 21 | rs9941349 | 16 | 52382989 | FTO | Obesity (extreme) | 19553259 | 6.0 × 10−12 | T | 0.32 | 0.08, 0.56 | 0.0101 | T | |
| 22 | rs1424233 | 16 | 78240252 | MAF | Obesity | 19151714 | 4.0 × 10−13 | T | 0.03 | −0.17, 0.22 | 0.7973 | I | Yes |
| 23 | rs1805081 | 18 | 19394430 | NPC1 | Obesity | 19151714 | 3.0 × 10−07 | T | 0.09 | −0.15, 0.33 | 0.4453 | I | Yes |
| 24 | rs1840440 | 18 | 21511184 | NR | Weight | 19851299 | 3.0 × 10−07 | C | 0.10 | −0.08, 0.28 | 0.2934 | T | Yes |
| 25 | rs17782313 | 18 | 56002077 | MC4R | BMI | 18454148 | 3.0 × 10−15 | C | 0.33 | 0.11, 0.55 | 0.0039 | T | Yes |
| 25 | rs12970134 | 18 | 56035730 | MC4R | WC | 18454146 | 2.0 × 10−09 | A | 0.25 | 0.02, 0.48 | 0.0300 | T | |
| 26 | rs29941 | 19 | 39001372 | KCTD15, CHST8 | BMI | 19079260 | 7.0 × 10−12 | G | 0.10 | −0.12, 0.32 | 0.3751 | I | |
| 26 | rs11084753 | 19 | 39013977 | KCTD15 | BMI | 19079261 | 2.0 × 10−08 | G | 0.09 | −0.1, 0.28 | 0.3712 | T | Yes |
Abbreviations: BMI, body mass index; Chr, chromosome; dbSNP, Database of Single Nucleotide Polymorphisms; GRS, genetic risk score; GWAS, genome-wide association study; I, imputed; PMID, PubMed identification number; SECGS, Shanghai Endometrial Cancer Genetics Study; T, typed; WC, waist circumference.
P value for the trait in the referenced PMID.
BMI- or obesity-increasing risk allele identified in the associated PMID and GWAS.
Mean difference in BMI (weight (kg)/height (m)2) per allele, adjusted for age, income, and education. Positive β’s indicate that the BMI risk allele was the same between the GWAS and the SECGS.
P value for association with BMI in the SECGS.
One single nucleotide polymorphism was selected per locus to be used in the calculation of GRSall (see Materials and Methods).
Figure 2.
Associations of selected genetic loci with body mass index and/or endometrial cancer, Shanghai Endometrial Cancer Genetics Study, Shanghai, China, 2010. The chromosome number is indicated on the left side of chromosomes 1–11 and on the right side of chromosomes 12–22. Locus numbers corresponding to the text and Tables 2 and 3 are shown above their chromosome positions. Black circles indicate loci associated with body mass index; black triangles indicate loci associated with endometrial cancer. All associations marked were significant at P ≤ 0.05.
With regard to endometrial cancer, for 22 of the 26 loci (84.6%), the obesity risk allele showed a higher frequency among cases than among controls (P = 0.0003). In addition, 9 of 35 SNPs (25.7%) were significantly associated with endometrial cancer (P ≤ 0.05) (Table 3). For 8 of these 9 SNPs, the directions of association with endometrial cancer were consistent with reported associations for obesity (i.e., the obesity-related allele was the same as the one that increased the risk of endometrial cancer). These 9 SNPs represent 7 of the 26 loci evaluated. The probability of observing this number of significant results at P ≤ 0.05, based on the binomial distribution under the null hypothesis, was 0.0002. Two of the 4 loci associated with BMI in our population were also associated with endometrial cancer (locus 2, SEC16B/RASAL2 and locus 25, MC4R). Since these 2 loci were associated with both BMI and endometrial cancer, we examined the extent to which the inclusion of BMI in the regression analysis changed the resultant odds ratio for the SNPs at these loci. No significant differences were observed. In the only gene spanning more than 1 locus, FTO, 1 locus was associated with BMI (locus 21), and an independent locus within this gene (locus 20) was associated with endometrial cancer.
Table 3.
Association Between Obesity-related Single Nucleotide Polymorphisms Identified in Genome-wide Association Studies and Endometrial Cancer Risk, Shanghai Endometrial Cancer Genetics Study, Shanghai, China, 1996–2005
| Locus | dbSNP Identification No. | Nearby Gene(s) | GWAS Risk Allele Frequencya | GWAS Risk Alleleb | Other Allele | Odds Ratioc | 95% Confidence Intervalc | SECGS P Valued | Typing | GRSsele |
| 1 | rs2568958 | NEGR1 | 0.925 | A | G | 1.08 | 0.86, 1.36 | 0.5200 | I | |
| 1 | rs2815752 | NEGR1 | 0.923 | A | G | 1.06 | 0.84, 1.34 | 0.6176 | T | |
| 2 | rs10913469 | SEC16B, RASAL2 | 0.216 | C | T | 1.21 | 1.04, 1.41 | 0.0145 | I | Yes |
| 3 | rs2605100 | LYPLAL1 | 0.794 | G | A | 1.06 | 0.91, 1.23 | 0.4741 | T | |
| 4 | rs6429082 | TBCE | 0.666 | C | T | 0.94 | 0.82, 1.06 | 0.3082 | T | |
| 5 | rs6548238 | TMEM18 | 0.902 | C | T | 1.28 | 1.04, 1.59 | 0.0215 | T | Yes |
| 5 | rs7561317 | TMEM18 | 0.909 | G | A | 1.25 | 1.01, 1.56 | 0.0395 | I | |
| 6 | rs1260326 | GCKR | 0.528 | T | C | 1.08 | 0.95, 1.24 | 0.2430 | I | |
| 7 | rs7647305 | SFRS10, ETV5, DGKG | 0.946 | C | T | 1.05 | 0.78, 1.41 | 0.7540 | I | |
| 8 | rs10938397 | GNPDA2 | 0.298 | G | A | 1.01 | 0.89, 1.16 | 0.8540 | I | |
| 9 | rs12517906 | MGAT1 | 0.095 | T | C | 0.93 | 0.75, 1.16 | 0.5241 | T | |
| 10 | rs2844479 | AIF1, NCR3 | 0.563 | A | C | 0.97 | 0.85, 1.10 | 0.6405 | I | |
| 11 | rs987237 | TFAP2B | 0.159 | G | A | 1.04 | 0.88, 1.23 | 0.6421 | T | |
| 12 | rs545854 | MSRA | 0.583 | C | G | 1.19 | 1.05, 1.35 | 0.0068 | T | Yes |
| 13 | rs7832552 | TRHR | 0.478 | T | C | 1.01 | 0.89, 1.15 | 0.8313 | T | |
| 14 | rs297325 | SOX6 | 0.336 | C | T | 1.15 | 1.01, 1.30 | 0.0348 | T | Yes |
| 15 | rs925946 | BDNF | 0.044 | T | G | 1.09 | 0.80, 1.48 | 0.5839 | I | |
| 16 | rs6265 | BDNF | 0.510 | C | T | 1.06 | 0.93, 1.19 | 0.3820 | T | |
| 17 | rs10838738 | MTCH2 | 0.328 | G | A | 0.84 | 0.73, 0.96 | 0.0094 | I | Yes |
| 18 | rs7138803 | BCDIN3D, FAIM2 | 0.283 | A | G | 1.01 | 0.88, 1.16 | 0.9249 | T | |
| 19 | rs7498665 | SH2B1, ATP2A1 | 0.115 | G | A | 1.06 | 0.88, 1.27 | 0.5643 | I | |
| 20 | rs6499640 | FTO | 0.164 | A | G | 1.26 | 1.08, 1.48 | 0.0045 | I | Yes |
| 21 | rs1421085 | FTO | 0.122 | C | T | 1.01 | 0.84, 1.22 | 0.9238 | T | |
| 21 | rs1558902 | FTO | 0.117 | A | T | 1.05 | 0.87, 1.26 | 0.6130 | I | |
| 21 | rs1121980 | FTO | 0.171 | A | G | 1.05 | 0.89, 1.23 | 0.5700 | T | |
| 21 | rs8050136 | FTO | 0.123 | A | C | 1.08 | 0.90, 1.30 | 0.4085 | T | |
| 21 | rs9939609 | FTO | 0.123 | A | T | 1.07 | 0.89, 1.29 | 0.4731 | T | |
| 21 | rs9941349 | FTO | 0.172 | T | C | 1.05 | 0.89, 1.23 | 0.5840 | T | |
| 22 | rs1424233 | MAF | 0.673 | T | C | 1.07 | 0.94, 1.22 | 0.3306 | I | |
| 23 | rs1805081 | NPC1 | 0.772 | T | C | 1.10 | 0.95, 1.29 | 0.2135 | I | |
| 24 | rs1840440 | NR | 0.490 | C | T | 1.08 | 0.95, 1.22 | 0.2324 | T | |
| 25 | rs17782313 | MC4R | 0.216 | C | T | 1.29 | 1.11, 1.50 | 0.0007 | T | Yes |
| 25 | rs12970134 | MC4R | 0.207 | A | G | 1.19 | 1.03, 1.38 | 0.0208 | T | |
| 26 | rs29941 | KCTD15, CHST8 | 0.231 | G | A | 0.96 | 0.83, 1.11 | 0.5607 | I | |
| 26 | rs11084753 | KCTD15 | 0.356 | G | A | 1.00 | 0.88, 1.14 | 0.9783 | T |
Abbreviations: BMI, body mass index; dbSNP, Database of Single Nucleotide Polymorphisms; GRS, genetic risk score; GWAS, genome-wide association study; I, imputed; SECGS, Shanghai Endometrial Cancer Genetics Study; T, typed.
Frequency (proportion) of the GWAS risk allele in cases and controls.
BMI- or obesity-increasing risk allele identified in a prior GWAS (see Table 2 for study identification).
Odds ratio and 95% confidence interval for association with endometrial cancer, adjusted for age, income, and education. Positive values indicate that the BMI risk allele was the same between the GWAS and the SECGS.
P value for association with endometrial cancer in cases and controls.
“Yes” indicates that the single nucleotide polymorphism was used in the calculation of GRSsel (see Materials and Methods).
GRSall, based on 26 SNPs in unique loci, and GRSsel, based on 7 SNPs significantly associated with endometrial cancer in unique loci, were evaluated. Risk alleles were defined by previously reported BMI GWAS results for all variants included in GRSall. For GRSsel, risk alleles were those associated with endometrial cancer. In 6 of 7 loci, the endometrial cancer risk allele was identical to the BMI risk allele in the source GWAS from which it was selected. Both GRSall and GRSsel were positively correlated with BMI in our study (Table 4), with beta coefficients of 0.07 and 0.04, respectively. Further, both types of GRS had stronger associations with BMI among controls than among cases, whether analyzed continuously or in quartiles based on distributions among controls.
Table 4.
Association of Genetic Risk Scores With Body Mass Index and the Risk of Endometrial Cancer, Shanghai Endometrial Cancer Genetics Study, Shanghai, China, 1996–2005
| Linear Regression of GRS and BMIa |
Association of GRS and Endometrial Cancer (n = 2,881) |
||||||||||||||
| Total (n = 2,881) |
Cases (n = 832) |
Controls (n = 2,049) |
OR1b | 95% CI | P Valuec | OR2d | 95% CI | P Valuee | |||||||
| βf | 95% CI | P Valueg | βh | 95% CI | P Valueg | βh | 95% CI | P Valueg | |||||||
| GRSalli | |||||||||||||||
| Q1 | 0 | Reference | 0 | Reference | 0 | Reference | 1 | Reference | 1 | Reference | |||||
| Q2 | 0.12 | −0.24, 0.48 | 0.508 | 0.18 | −0.63, 0.99 | 0.661 | 0.08 | −0.31, 0.47 | 0.676 | 1.25 | 0.99, 1.60 | 0.065 | 1.38 | 1.07, 1.80 | 0.015 |
| Q3 | 0.33 | −0.03, 0.69 | 0.075 | 0.15 | −0.67, 0.98 | 0.715 | 0.41 | 0.02, 0.8 | 0.04 | 1.13 | 0.88, 1.45 | 0.315 | 1.25 | 0.96, 1.63 | 0.105 |
| Q4 | 0.42 | 0.06, 0.77 | 0.022 | 0.48 | −0.31, 1.27 | 0.233 | 0.39 | 0.00, 0.78 | 0.051 | 1.50 | 1.18, 1.92 | 0.001 | 1.51 | 1.17, 1.95 | 0.002 |
| Continuous | 0.07 | 0.02, 0.11 | 0.002 | 0.05 | −0.04, 0.14 | 0.242 | 0.07 | 0.03, 0.12 | 0.002 | 1.05 | 1.02, 1.08 | 0.001 | 1.04 | 1.01, 1.07 | 0.009 |
| GRSselj | |||||||||||||||
| Q1 | 0 | Reference | 0 | Reference | 0 | Reference | 1 | Reference | 1 | Reference | |||||
| Q2 | 0.06 | −0.30, 0.43 | 0.732 | −0.19 | −1.04, 0.65 | 0.653 | 0.08 | −0.31, 0.47 | 0.679 | 1.15 | 0.88, 1.49 | 0.303 | 1.15 | 0.88, 1.51 | 0.300 |
| Q3 | 0.08 | −0.27, 0.43 | 0.651 | −0.14 | −0.93, 0.66 | 0.732 | 0.10 | −0.28, 0.48 | 0.615 | 1.31 | 1.02, 1.67 | 0.035 | 1.32 | 1.02, 1.70 | 0.035 |
| Q4 | 0.24 | −0.11, 0.59 | 0.177 | −0.25 | −1.01, 0.5 | 0.507 | 0.41 | 0.03, 0.79 | 0.035 | 1.96 | 1.54, 2.5 | <0.001 | 1.91 | 1.49, 2.45 | <0.001 |
| Continuous | 0.04 | −0.04, 0.12 | 0.329 | −0.08 | −0.32, −0.69 | 0.531 | 0.13 | 0.01, 0.25 | 0.041 | 1.21 | 1.14, 1.280 | <0.001 | 1.20 | 1.13, 1.27 | <0.001 |
Abbreviations: BMI, body mass index; CI, confidence interval; GRS, genetic risk score; OR, odds ratio; Q, quartile; SNP, single nucleotide polymorphism.
Weight (kg)/height (m)2.
Odds ratio was adjusted for age, income, and education.
P value for logistic regression for endometrial cancer, adjusted for age, income, and education.
Odds ratio was adjusted for age, income, education, and BMI.
P value for logistic regression for endometrial cancer, adjusted for age, income, education, and BMI.
Mean difference in BMI, adjusted for age, income, education, and case-control status.
P value from linear regression, adjusted for age, income, and education.
Mean difference in BMI, adjusted for age, income, and education.
GRSall was calculated using the 26 SNPs indicated in Table 2 for which the BMI-associated allele was known and was adjusted for age, income, and education.
GRSsel was calculated using the 7 SNPs indicated in Table 3 and was adjusted for age, income, and education.
The associations of GRSall and GRSsel with endometrial cancer were evaluated by logistic regression. Both GRSall (odds ratio (OR) = 1.05, 95% confidence interval (CI): 1.02, 1.08) and GRSsel (OR = 1.21, 95% CI: 1.14, 1.28) were significantly associated with endometrial cancer risk. With adjustment for BMI, these associations were somewhat attenuated, but they remained significant (ORGRSall = 1.04, 95% CI: 1.01, 1.07; ORGRSsel = 1.20, 95% CI: 1.13, 1.27). Categorical analysis showed that odds ratios across the 4 quartiles (lowest to highest) were 1.00, 1.25, 1.13, and 1.50 for GRSall and 1.00, 1.15, 1.31, and 1.96 for GRSsel. Additional adjustment for BMI did not appreciably alter these results.
The joint effect of energy balance-related variables (including BMI, energy intake, and exercise participation) and GRSsel on endometrial cancer risk was evaluated (Table 5). Although there were no statistically significant interactions between energy balance-related variables and GRSsel, we found that persons who were in the higher categories of GRSsel and BMI or who did not exercise had higher odds ratios for endometrial cancer compared with persons in the lowest categories. For example, women in the highest quantiles of both GRSsel and BMI were 8 times more likely to have endometrial cancer than controls (OR = 8.03, 95% CI: 4.78, 13.51), in comparison with women in the lowest quantiles.
Table 5.
Joint Associations of Indicators of Energy Balance (Body Mass Index, Exercise, and Energy Intake) and Genetic Risk Score With Risk of Endometrial Cancer, Shanghai Endometrial Cancer Genetics Study, Shanghai, China, 1996–2005
| Indicator and Category | Quartile of GRSsela |
OR for Indicator | 95% CI | P for Interaction | |||||||
| 1 |
2 |
3 |
4 |
||||||||
| OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | ||||
| Body mass indexb | 0.26 | ||||||||||
| T1 | 1 | Reference | 1.72 | 0.91, 3.26 | 1.37 | 0.72, 2.59 | 3.79 | 2.11, 6.81 | 1 | Reference | |
| T2 | 2.19 | 1.23, 3.91 | 3.33 | 1.89, 5.85 | 3.05 | 1.75, 5.32 | 3.41 | 1.98, 5.89 | 1.63 | 1.26, 2.12 | |
| T3 | 4.56 | 2.68, 7.75 | 3.94 | 2.28, 6.80 | 5.99 | 3.53, 10.16 | 8.03 | 4.78, 13.51 | 3.08 | 2.40, 3.96 | |
| GRSsel OR | 1.18 | 0.90, 1.55 | 1.35 | 1.04, 1.74 | 1.94 | 1.51, 2.49 | |||||
| Exercise participation | 0.92 | ||||||||||
| Yes, median or more | 1 | Reference | 0.89 | 0.44, 1.80 | 1.43 | 0.74, 2.76 | 2.17 | 1.14, 4.13 | 1 | Reference | |
| Yes, less than median | 1.77 | 0.91, 3.44 | 2.01 | 1.04, 3.90 | 1.83 | 0.96, 3.53 | 2.44 | 1.30, 4.59 | 1.50 | 1.09, 2.08 | |
| No | 1.89 | 1.11, 3.22 | 2.35 | 1.37, 4.01 | 2.58 | 1.53, 4.37 | 4.00 | 2.37, 6.73 | 1.99 | 1.52, 2.60 | |
| GRSsel OR | 1.17 | 0.90, 1.52 | 1.32 | 1.02, 1.69 | 1.98 | 1.55, 2.53 | |||||
| Total energy intake | 0.67 | ||||||||||
| T1 | 1 | Reference | 1.10 | 0.68, 1.78 | 1.29 | 0.84, 1.99 | 1.76 | 1.15, 2.70 | 1 | Reference | |
| T2 | 0.83 | 0.53, 1.31 | 0.89 | 0.56, 1.41 | 1.04 | 0.66, 1.64 | 2.11 | 1.38, 3.23 | 0.91 | 0.73, 1.14 | |
| T3 | 1.11 | 0.71, 1.72 | 1.41 | 0.91, 2.19 | 1.52 | 0.99, 2.32 | 1.95 | 1.29, 2.93 | 1.16 | 0.94, 1.43 | |
| GRSsel OR | 1.15 | 0.89, 1.50 | 1.31 | 1.02, 1.69 | 1.97 | 1.54, 2.51 | |||||
Abbreviations: CI, confidence interval; GRS, genetic risk score; OR, odds ratio; T, tertile.
Logistic regression odds ratios adjusted for age, income, and education.
Weight (kg)/height (m)2.
DISCUSSION
To our knowledge, this is the first study to have systematically evaluated the shared genetic architecture between measures of obesity and endometrial cancer risk. Of the 35 GWAS-identified SNPs that are associated with high BMI, obesity, weight, or waist:hip ratio, associations with endometrial cancer were found for 9 SNPs (P ≤ 0.05), representing 7 loci (FTO, MC4R, MSRA, MTCH2, SEC16B, SOX6, and TMEM18). The number of endometrial cancer associations among BMI-related variants was significantly higher than would be expected by chance on the basis of the binomial distribution test, indicating that genetic variants associated with BMI are also involved in endometrial cancer. In addition, the direction of association with endometrial cancer for the majority of the variants evaluated (84.6%) was in agreement with the direction of association with BMI found by other GWAS. The binomial distribution test was highly significant, indicating that additional BMI-associated variants may be associated with endometrial cancer, even if they were not found to be significant in the current analysis. Our findings are in agreement with a recent report by Elks et al. (22), in which 4 loci (i.e., FTO, SEC16B, TMEM18, and MTCH2) were also associated with age at menarche, a phenotype that is strongly related to both endometrial cancer risk and BMI. Notably, in our study, adjustment for BMI had little impact on these results, indicating that these SNPs capture more information on endometrial cancer risk than does current BMI alone.
Associations with both endometrial cancer and BMI were found for genetic variants at 2 loci: locus 2, near the SEC16B and RASAL genes, and locus 25, near the MC4R gene. In addition, locus 21 was associated with BMI and locus 20 was associated with endometrial cancer; both of these loci are near the FTO gene. The 2 strongest associations with endometrial cancer in this study were for loci in the FTO gene (locus 20, SNP rs6499640; P = 0.0045) and near the MC4R gene (locus 25, rs17782313; P = 0.0007). These 2 genes have been the subject of multiple reports of associations with BMI (12, 16, 17). FTO, the fat mass and obesity-associated gene, has been detected as a locus affecting BMI in 13 independent GWAS (12, 23). In addition, the FTO gene has been associated with diabetes in 5 independent GWAS (24). FTO has 7 SNPs, of which 6 are within 1 locus by linkage disequilibrium, while the other is independent. The current study included the 6 single-locus SNPs, and the direction of the association with BMI was consistent between our study and previous GWAS. The seventh and independent SNP in the FTO gene, rs6499640, was not associated with BMI in our population but was strongly associated with endometrial cancer. This suggests that FTO may be involved in endometrial cancer via mechanisms other than obesity. MC4R, the melanocortin 4 receptor gene, has been associated with BMI and related body measurements in 8 independent GWAS and across multiple ethnic groups (25, 26). In our study, 2 SNPs at this locus showed an association with endometrial cancer, with the strongest signal coming from rs17782313 (P = 0.0007; OR = 1.29, 95% CI: 1.11, 1.50). MC4R has been linked to both obesity and reproductive dysfunction (26–30). A SNP near SEC16B/RASAL, rs10913469, was also associated with both BMI and endometrial cancer in our study. This locus was also recently associated with early age at menarche (22), a strong risk factor for endometrial cancer (12).
In our study population of Chinese women, other loci that were associated with endometrial cancer but had no statistically significant association with BMI included TMEM18, MSRA, SOX6, and MTCH2. Aside from rs10838738 at MTCH2, all other SNPs showed an association with endometrial cancer that was consistent with the directions of association with BMI reported in previous GWAS.
The GRS provides a measure of the combined genetic effect of obesity-associated loci on endometrial cancer risk. The GRS calculated in this study was associated with both BMI and endometrial cancer. The effect of the GRS on BMI is somewhat paradoxical: The accumulation of risk alleles as measured by GRSall showed that the positive correlation with BMI is driven chiefly by controls but modestly or significantly attenuated among cases. Two explanations seem plausible. First, these genetic variants may increase the risk of obesity initially, but then the disease itself, or its treatment, attenuates the effects among cases. Several common treatments for endometrial cancer may result in weight loss. These include hysterectomy (31), radiation therapy (32), and several common chemotherapeutic agents (33). The second possibility is that the risk alleles comprising the GRS may influence the risk of endometrial cancer through mechanisms other than those related to obesity alone. The persistent significance of associations of the GRS with endometrial cancer risk after adjustment for BMI supports this explanation. It is noteworthy that although no statistically significant interactions were observed between the GRS and factors related to energy balance in our study, we did find that women with a high GRS and high BMI or no exercise participation were at substantially increased risk of developing endometrial cancer. These findings suggest the potential value of using genetic markers to identify populations at high risk of endometrial cancer for targeted prevention.
One limitation of our study is that it relied on the NHGRI GWAS catalog for identification of SNPs for inclusion. Because the threshold for entry of SNPs into the GWAS catalog is higher than that used for most association studies, a number of potentially interesting loci may have been missed. A further limitation is that data were not available for all originally selected SNPs, although the majority were captured (92.1%). In addition, BMI was measured at the time of interview for cases, which may not reflect women's prediagnosis weight due to the effects of either illness or treatment. Further, this study was carried out in a Chinese population with an average BMI of 23.4 in controls and 25.7 in cases. The results may not be directly generalizable to non-Chinese populations. However, almost all of the risk factors for endometrial cancer found in this Chinese population were identical to those found in populations of European ancestry. Because the majority of GWAS-identified SNPs associated with BMI were identified in populations of European ancestry, associations between BMI-associated SNPs and endometrial cancer may be even stronger in those discovery populations. Nevertheless, our study also had many strengths. To our knowledge, this was the first population-based epidemiologic study to comprehensively evaluate GWAS-identified obesity markers in association with endometrial cancer risk. The vast majority of persons in our study population were of a single ethnic group, reducing the potential effects of population stratification. The relatively large sample size and the detailed exposure information allowed us to evaluate the joint effect of obesity-related genetic markers and energy balance measures on endometrial cancer risk.
In summary, our study found strong evidence linking obesity and endometrial cancer at the genetic level. We also found that relations between BMI-associated variants and endometrial cancer risk appear to be independent of BMI, suggesting that genetic markers of obesity have value themselves, in addition to BMI, for defining women who may be at higher risk of endometrial cancer.
Acknowledgments
Author affiliations: Division of Epidemiology, Department of Medicine; Vanderbilt Epidemiology Center; and Vanderbilt-Ingram Cancer Center, School of Medicine, Vanderbilt University, Nashville, Tennessee (Ryan J. Delahanty, Alicia Beeghly-Fadiel, Jirong Long, Qiuyin Cai, Wanqing Wen, Hui Cai, Wei Zheng, Xiao Ou Shu); Department of Epidemiology, Shanghai Cancer Institute, Shanghai Jiao Tong University, Shanghai, China (Yong-Bing Xiang, Wang-Hong Xu, Yu-Tang Gao); and Department of Biostatistics, School of Medicine, Vanderbilt University, Nashville, Tennessee (Jing He).
This work was supported by US National Institutes of Health grants R01 CA092585 (to X. O. S.) and R01 CA090899 and R01 CA064277 (to W. Z.).
The authors thank the research staff of the Shanghai Endometrial Cancer Genetics Study. They also thank Drs. Tsogzolmaa Dorjgochoo, Todd Edwards, and Raquel Villegas for consultations, as well as Bethanie Rammer for assistance with manuscript preparation.
The authors take sole responsibility for the content of this article.
Conflict of interest: none declared.
Glossary
Abbreviations
- BMI
body mass index
- CI
confidence interval
- GRS
genetic risk score
- GWAS
genome-wide association study(ies)
- NHGRI
National Human Genome Research Institute
- OR
odds ratio
- SNP
single nucleotide polymorphism
References
- 1.Gao YT, Wei L. Cancer Incidence, Mortality and Survival Rates in Urban Shanghai (1973–2000) Shanghai, China: Second Military Medical University Press; 2007. [Google Scholar]
- 2.Shanghai Municipal Center for Disease Control and Prevention. Shanghai Cancer Report of 2009. Shanghai, China: Shanghai Municipal Center for Disease Control and Prevention; 2009. [Google Scholar]
- 3.American Institute for Cancer Research. Food, Nutrition, Physical Activity, and the Prevention of Cancer: A Global Perspective. (Summary) Washington, DC: American Institute for Cancer Research; 2007. [Google Scholar]
- 4.Schouten LJ, Goldbohm RA. Van Den Brandt PA. Anthropometry, physical activity, and endometrial cancer risk: results from the Netherlands Cohort Study. J Natl Cancer Inst. 2004;96(21):1635–1638. doi: 10.1093/jnci/djh291. [DOI] [PubMed] [Google Scholar]
- 5.Calle EE, Rodriguez C, Walker-Thurmond K, et al. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348(17):1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 6.Finucane MM, Stevens GA, Cowan MJ, et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating Group (Body Mass Index) Lancet. 2011;337(9765):557–567. doi: 10.1016/S0140-6736(10)62037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ministry of Health of the People's Republic of China, Ministry of Science and Technology of the People's Republic of China, National Bureau of Statistics of China. The nutrition and health status of the Chinese people. Chin J Cardiovasc Rev. 2004;2(12):919–922. [Google Scholar]
- 8.Xu WH, Xiang YB, Zheng W, et al. Weight history and risk of endometrial cancer among Chinese women. Int J Epidemiol. 2006;35(1):159–166. doi: 10.1093/ije/dyi223. [DOI] [PubMed] [Google Scholar]
- 9.Xu WH, Matthews CE, Xiang YB, et al. Effect of adiposity and fat distribution on endometrial cancer risk in Shanghai women. Am J Epidemiol. 2005;161(10):939–947. doi: 10.1093/aje/kwi127. [DOI] [PubMed] [Google Scholar]
- 10.Shu XO, Brinton LA, Zheng W, et al. Relation of obesity and body fat distribution to endometrial cancer in Shanghai, China. Cancer Res. 1992;52(14):3865–3870. [PubMed] [Google Scholar]
- 11.Hinney A, Vogel CI, Hebebrand J. From monogenic to polygenic obesity: recent advances. Eur Child Adolesc Psychiatry. 2010;19(3):297–310. doi: 10.1007/s00787-010-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299(11):1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 14.Coady SA, Jaquish CE, Fabsitz RR, et al. Genetic variability of adult body mass index: a longitudinal assessment in Framingham families. Obes Res. 2002;10(7):675–681. doi: 10.1038/oby.2002.91. [DOI] [PubMed] [Google Scholar]
- 15.Bell CG, Walley AJ, Froguel P. The genetics of human obesity. Nat Rev Genet. 2005;6(3):221–234. doi: 10.1038/nrg1556. [DOI] [PubMed] [Google Scholar]
- 16.Farooqi IS, O'Rahilly S. Genetic factors in human obesity. Obes Rev. 2007;8(suppl 1):37–40. doi: 10.1111/j.1467-789X.2007.00315.x. [DOI] [PubMed] [Google Scholar]
- 17.Schousboe K, Willemsen G, Kyvik KO, et al. Sex differences in heritability of BMI: a comparative study of results from twin studies in eight countries. Twin Res. 2003;6(5):409–421. doi: 10.1375/136905203770326411. [DOI] [PubMed] [Google Scholar]
- 18.Xu WH, Zheng W, Xiang YB, et al. Soya food intake and risk of endometrial cancer among Chinese women in Shanghai: population based case-control study. BMJ. 2004;328(7451):1285. doi: 10.1136/bmj.38093.646215.AE. (doi:10.1136/bmj.38093.646215.AE) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41(3):324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu XO, Yang G, Jin F, et al. Validity and reproducibility of the food frequency questionnaire used in the Shanghai Women's Health Study. Eur J Clin Nutr. 2004;58(1):17–23. doi: 10.1038/sj.ejcn.1601738. [DOI] [PubMed] [Google Scholar]
- 21.Hindorff LA, Junkins HA, Hall PN, et al. A Catalog of Published Genome-wide Association Studies. Bethesda, MD: National Human Genome Research Institute; 2010. ( www.genome.gov/gwastudies). (Accessed July 6, 2010) [Google Scholar]
- 22.Elks CE, Perry JR, Sulem P, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42(12):1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 24.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chambers JC, Elliott P, Zabaneh D, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40(6):716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 27.Young EH, Wareham NJ, Farooqi S, et al. The V103I polymorphism of the MC4R gene and obesity: population based studies and meta-analysis of 29 563 individuals. Int J Obes (Lond) 2007;31(9):1437–1441. doi: 10.1038/sj.ijo.0803609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irani BG, Xiang Z, Moore MC, et al. Voluntary exercise delays monogenetic obesity and overcomes reproductive dysfunction of the melanocortin-4 receptor knockout mouse. Biochem Biophys Res Commun. 2005;326(3):638–644. doi: 10.1016/j.bbrc.2004.11.084. [DOI] [PubMed] [Google Scholar]
- 29.Van der Ploeg LH, Martin WJ, Howard AD, et al. A role for the melanocortin 4 receptor in sexual function. Proc Natl Acad Sci U S A. 2002;99(17):11381–11386. doi: 10.1073/pnas.172378699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin WJ, McGowan E, Cashen DE, et al. Activation of melanocortin MC(4) receptors increases erectile activity in rats ex copula. Eur J Pharmacol. 2002;454(1):71–79. doi: 10.1016/s0014-2999(02)02479-2. [DOI] [PubMed] [Google Scholar]
- 31.Weber AM, Walters MD, Schover LR, et al. Functional outcomes and satisfaction after abdominal hysterectomy. Am J Obstet Gynecol. 1999;181(3):530–535. doi: 10.1016/s0002-9378(99)70488-6. [DOI] [PubMed] [Google Scholar]
- 32.Potish RA, Twiggs LB, Adcock LL, et al. Role of whole abdominal radiation therapy in the management of endometrial cancer; prognostic importance of factors indicating peritoneal metastases. Gynecol Oncol. 1985;21(1):80–86. doi: 10.1016/0090-8258(85)90235-5. [DOI] [PubMed] [Google Scholar]
- 33.Decruze SB, Green JA. Hormone therapy in advanced and recurrent endometrial cancer: a systematic review. Int J Gynecol Cancer. 2007;17(5):964–978. doi: 10.1111/j.1525-1438.2007.00897.x. [DOI] [PubMed] [Google Scholar]



