Abstract
Oligodendroglioma is characterized by unique clinical, pathological, and genetic features. Recurrent losses of chromosomes 1p and 19q are strongly associated with this brain cancer but knowledge of the identity and function of the genes affected by these alterations is limited. We performed exome sequencing on a discovery set of 16 oligodendrogliomas with 1p/19q co-deletion to identify new molecular features at base-pair resolution. As anticipated, there was a high rate of IDH mutations: all cases had mutations in either IDH1 (14/16) or IDH2 (2/16). In addition, we discovered somatic mutations and insertions/deletions in the CIC gene on chromosome 19q13.2 in 13/16 tumours. These discovery set mutations were validated by deep sequencing of 13 additional tumours, which revealed 7 others with CIC mutations, thus bringing the overall mutation rate in oligodendrogliomas in this study to 20/29 (69%). In contrast, deep sequencing of astrocytomas and oligoastrocytomas without 1p/19q loss revealed that CIC alterations were otherwise rare (1/60; 2%). Of the 21 non-synonymous somatic mutations in 20 CIC-mutant oligodendrogliomas, 9 were in exon 5 within an annotated DNA interacting domain and 3 were in exon 20 within an annotated protein interacting domain. The remaining 9 were found in other exons and frequently included truncations. CIC mutations were highly associated with oligodendroglioma histology, 1p/19q co-deletion and IDH1/2 mutation (p<0.001). Although we observed no differences in the clinical outcomes of CIC mutant versus wild-type tumors, in a background of 1p/19q co-deletion, hemizygous CIC mutations are likely important. We hypothesize that the mutant CIC on the single retained 19q allele is linked to the pathogenesis of oligodendrogliomas with IDH mutation. Our detailed study of genetic aberrations in oligodendroglioma suggests a functional interaction between CIC mutation, IDH1/2 mutation and 1p/19q co-deletion.
Keywords: Glioma, Oligodendroglioma, Next Generation Sequencing, Capicua, IDH1
INTRODUCTION
Oligodendroglioma, a glioma subtype marked by unique clinical, pathological, and genetic characteristics, is composed of neoplastic cellular elements that resemble oligodendrocytes. Unlike other gliomas such as astrocytomas and ependymomas, oligodendrogliomas are chemosensitive and often progress in a slow and predictable manner [1]. Oligodendrogliomas display a classical appearance of cells with round, regular nuclei associated with clearing of the cytoplasm and in close proximity to fine branching vasculature [2]. Most importantly, oligodendrogliomas are strongly linked to co-deletions of the short arm of chromosome 1 (1p) and the long arm of chromosome 19 (19q). These recurrent genetic alterations in turn have been strongly linked to favourable clinical behaviour and classic oligodendroglioma histology such that 1p/19q co-deletion is considered a defining feature [2–4].
Recent developments in brain tumour genomics have highlighted the uniqueness of oligodendrogliomas [5–7]. In particular, it has been shown that the proneural gene expression signature is enriched in oligodendrogliomas, particularly those with 1p/19q co-deletion [5, 8]. In addition to displaying the proneural signature, 1p/19q codeleted oligodendrogliomas are associated with a constellation of positive prognostic markers including methylation of the MGMT promoter, IDH1 mutations, and the recently-described CpG island methylator phenotype (G-CIMP) [9]. Although these markers are also present in glioblastomas that arise from low- grade astrocytomas, an important divergence in the molecular pathogenesis of low-grade oligodendrogliomas and astrocytomas is 1p/19q co-deletion in the former and TP53 mutations in the latter [10]. The mutual exclusivity of these events underscores the distinct molecular characteristics of oligodendrogliomas.
Despite these advances, our understanding of the genetic underpinnings of oligodendrogliomas remains unclear. Deletions of 1p and 19q, which result in loss of heterozygosity in those regions, may unmask mutations leading to the oligodendroglioma phenotype. This could arise strictly within 1p and 19q or could be the result of global genomic or epigenomic changes. However efforts to find candidate tumour-associated genes in 1p and 19q, and more globally, have met with limited success [11–14]. Until recently, our progress in understanding oligodendrogliomas also has been hampered by a dearth of robust model systems with which to conduct functional studies [15].
Next generation sequencing technology and advancements in bioinformatics have transformed the field of cancer genomics. Together, these developments afford rapid, cost-effective “deep- sequencing” of cancer genomes, thus enabling genome-wide searches for cancer-associated mutations [16–23]. Within the last month, two groups have independently reported their findings from next-generation sequencing of oligodendrogliomas with markedly different results. One group sequenced all exons, microRNA, splice sites, and promoter regions on 1p and 19q in 7 oligodendrogliomas and found no recurrent alterations [14]. The other sequenced the exomes of 7 oligodendrogliomas, and performed directed sequencing in an additional 27 oligodendrogliomas, and reported recurrent mutations in CIC in 53% of their cases and FUBP1 in 15% [24].
Here, we report the results of our concurrent effort to identify somatic mutations potentially contributing to 1p/19q codeleted oligodendroglioma through a combination of exome, transcriptome, and whole genome shotgun sequencing. We found frequent mutations in the CIC gene, noting that these cluster in the DNA-interacting HMG domain and the protein- protein interacting GRO-L domain. 13/16 tumours in our discovery set harbored protein altering somatic mutations, nonsense mutations, or insertions/deletions (INDELs) of CIC. Subsequent deep amplicon sequencing of CIC from 73 gliomas with matched normal tissues identified a further 8 somatic CIC mutations and INDELs, and 7/8 of these were found in 1p/19q codeleted oligodendrogliomas with IDH1 mutation. Our study highlights a unique relationship between recurrent chromosomal aberrations and mutations in CIC and suggests that mutations in CIC are key events in the development of oligodendrogliomas.
MATERIALS AND METHODS
Sample acquisition and preparation
Fresh frozen tumour tissue, matched normal blood, and formalin-fixed paraffin embedded (FFPE) tissue from patients undergoing surgery for oligodendroglioma, oligoastrocytoma, and astrocytoma were obtained from the Calgary Brain Tumour Bank. H&E-stained sections of the samples were examined for sample quality. Tumour samples with estimated >60% tumour cell content and <20% necrosis were used for nucleic acid extraction. DNA was extracted from fresh frozen tissue, FFPE tissue, and frozen blood buffy coat using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA). RNA was extracted from tumour tissue using Trizol (Invitrogen, Burlington, ON) and was evaluated for quality using an Agilent 2100 Bioanalyzer (Agilent, Mississauga, ON). RNA samples with a RIN ≥8 were used for transcriptional analyses. Additional tumour and matched normal specimens were provided by the Cancer Genetics Laboratory at BCCA and from the Massachusetts General Hospital Brain Tumour Repository. All samples were collected, de-identified, and used in accordance with approval by the respective local research ethics boards.
Clinicopathologic information
Demographic, diagnostic, treatment, and follow-up information from Calgary cases was retrieved from medical records. 1p and 19q status was determined previously by routine PCR microsatellite analysis or FISH. IDH1 and IDH2 genotyping was performed as described previously [25]. Progression-free survival (PFS) was calculated from the date of diagnosis to the date of recurrence or the date last seen without recurrence. Overall survival (OS) was calculated to the date of death or date last seen. Comparisons of mutation status with tumour histology or grade were performed using Fisher’s exact test. Stratified analyses were performed using Mantel-Haenszel statistic. PFS and OS according to mutation status were examined using Kaplan-Meier survival curves with statistical testing by log-rank test. P-values were 2-tailed and a level of <0.05 was considered significant. SAS version 9.2 (SAS Institute, Cary, NC) was used for statistical analyses.
Sequencing of matched tumour and blood specimens
Whole genome shotgun sequencing (WGSS)
WGSS was performed on the SOLiD 4.0 platform as previously described [26]. Paired end reads were aligned to the reference human genome (NCBI Build 36.1, hg18) using diBayes. Single nucleotide variant (SNV) detection was performed using the Samtools package (0.1.5) [27] and candidate somatic mutations were identified by comparison of tumour and matched normal DNA sequence data. Candidate somatic mutations were disqualified if one or more reads from the matched normal sample presented evidence for the SNV.
Exomic and transcriptomic sequencing
Agilent SureSelect target enrichment kit was used for exome library construction as per the manufacturer’s protocol (Agilent, Mississauga, ON). Library construction for RNA Seq was performed as described [16, 28]. Sequencing was performed on the Illumina GAIIx platform. RNA-Seq reads were processed as previously described [16, 19], and paired end reads were aligned to the reference human genome assembly using BWA (0.5.7) [29]. SNV detection was performed using SNVMix2 with somatic non-synonymous SNVs identified by referring to the matched normal pileup [29] with filtering to include SNVs such that the combined probability of either heterozygous or homozygous SNV > 0.99 [30].
CIC deep amplicon sequencing
Deep sequencing of the entire CIC gene (chr19: 47,467,725-47,480,658; hg18), including the unannotated exon 0 identified in RNA-Seq, was performed using the Illumina HiSeq2000 platform (Supplemental Data for detailed methods).
Validation of mutations
All somatic SNVs identified in WGSS and exome sequencing were examined and confirmed manually using IGV Browser 2.0 (Broad Institute, Cambridge, MA, USA). PCR and Sanger sequencing were used to validate all CIC mutations and INDELs in tumour and matched normal samples (Supplemental Data). Lastly, exons 5 and 20 were Sanger sequenced in an extended validation set of 111 gliomas.
RESULTS
Next-generation sequencing of 1p/19q codeleted oligodendrogliomas identifies known and novel somatic mutations
To survey the spectrum of somatic aberrations in the coding sequence present in oligodendrogliomas, we conducted Illumina exome sequencing on a discovery set of sixteen pairs of 1p/19q codeleted oligodendrogliomas and matched normal bloods. Sequencing was performed to >100X average redundancy of the on-target region from the Agilent SureSelect kit (v1.0). After identification of somatic SNVs, non-synonymous mutations were identified and grouped by gene (Table 1). This analysis revealed 3212 candidate somatic non-synonymous mutations in 2696 genes. Of the 3212 mutations, 340 SNV’s were manually verified in IGV.
Table 1. Recurrent mutations and recurrently mutated genes seen in three or more libraries.
List of genes with recurrent or more than one non-synonymous change. Sixteen tumour and matched normal exomes were sequenced and SNV calls were annotated. Novel non-synonymous mutations were identified by filtering against the normal pileup and dbSNP129. All mutations were manually curated using the IGV browser.
| Gene | Chr | Position | Sample(s) | Nucleotide (cDNA) |
Amino Acid |
|---|---|---|---|---|---|
| IDH1 | 2 | 208821357 | HS1893, HS2040, HS2042, HS2805, HS2806, HS2807, HS2808, HS2809, HS2810, HS2811, HS2812, HS2813, HS2815, HS3124 | c. 395 C > T | p.R132H |
| CIC | 19 | 47483558 | HS2040 | c.604 C > T | p.R202W |
| CIC | 19 | 47483100 | HS2814 | c.142 G > A | p.G107E |
| CIC | 19 | 47488171 | HS2813 | c.2980 C > T | p.Q994* |
| CIC | 19 | 47483598 | HS2815 | c.644 G > A | p.R215Q |
| CIC | 19 | 47483690 | HS2810 | c.736 A > G | p.K246E |
| CIC | 19 | 47490900 | HS2045 | c.4544 G > T | p.R1515L |
| CIC | 19 | 47490900 | HS3124 | c.4544 G > A | p.R1515H |
| CIC | 19 | 47483580 | HS2805 | c.626 T > G | p.I209S |
| CIC | 19 | 47483653–47483655 | HS2807 | DEL:CCT | |
| CIC | 19 | 47486863–47486864 | HS2806 | DEL:AA | |
| CIC | 19 | 47490858-47490858 | HS2814 | INS:A | |
| NOTCH1 | 9 | 138528889 | HS2805, HS2811 | c.2101 T > G | p.T701P |
| NOTCH1 | 9 | 138532470 | HS2811 | c.1195 T > G | p.T399P |
| NOTCH1 | 9 | 138522244 | HS3124 | c.3494 C > T | p.G1165D |
| NOTCH1 | 9 | 138532105–138532107 | HS2814 | DEL:TTG | |
| IDH2 | 15 | 88432842 | HS2045, HS2814 | c.515 C > T | p.R172K |
| PCDH8 | 13 | 52318741 | HS2805, HS2811 | c.1832 T > G | p.H611P |
Three genes were mutated in three or more cases (IDH1, CIC, NOTCH1). The canonical mutation resulting in p.Arg132His in IDH1 was seen in 14 cases and the remaining 2 tumours were mutated in IDH2 resulting in p.Arg172His. Out of a total set of 340 IGV validated non-synonymous mutations, 54 more mutations were seen in the same gene in more than one case – these included alterations of SOX11, DACH1, PTPRZ1 and ATM. Two post-treatment tumours displayed significantly elevated rates of somatic mutation approximately 9- fold greater than the rest of the tumour exomes (Supplemental Data). Similar to a subgroup of recurrent glioblastoma, somatic mutations in the mismatch repair gene MSH6 were present in both of these cases [31, 32]. TP53 mutations were present only in these two hypermutator tumours.
Although somatic coding mutations of FUBP1 were recently reported in a subset of oligodendrogliomas [24], we did not find any non-synonymous somatic mutations in this gene in the 16 oligodendroglioma cases, despite having 5 fold redundant coverage or better of all the FUBP1 exonic regions except in the first and last exons. We did, however, identify two FUBP1 candidate somatic splice site variants at chr1:78205319 and chr1:78193604.
CIC mutations are common in oligodendrogliomas but rare in other cancers
After IDH1, the gene next most frequently mutated in our discovery cohort was CIC (Source:UniProtKB/Swiss-Prot;Acc:Q96RK0), located on chromosome 19q within the commonly deleted region [33]. In our exome libraries, we found 13 of 16 tumours mutated in CIC, one of which carried two distinct mutations. To validate the exome findings and to determine the frequency of CIC mutations in oligodendrogliomas, we performed CIC-focused deep sequencing on the original 16 tumour/normal pairs in the discovery set as well tumour/normal pairs from 13 additional known 1p/19q codeleted tumours. All mutations identified in the 13/16 discovery samples by exome sequencing were confirmed by deep amplicon sequencing. Seven of the 13 additional cases were also identified as having CIC mutations, yielding a CIC mutation rate in oligodendrogliomas of 20/29 (69%).
In contrast, deep sequencing of 60 tumour/normal pairs from a variety of astrocytomas and mixed oligoastrocytomas without 1p/19q co-deletion revealed only 1 mutant tumour (2%). This set contained 4 cases with 1p loss only and 9 cases with 19q loss only. Interestingly, the single somatic non-coding mutation was found in a tumour with 19q loss only.
To further determine whether CIC mutations were commonly found in other cancer types, including those outside the brain, we searched for CIC alterations in the Genome Sciences Centre (GSC) database containing SNVs from next-generation sequencing of 2315 cancer and normal libraries [34]. Whereas CIC mutations were present in 13/16 (81%) of our oligodendroglioma exome libraries, the GSC database search revealed that CIC alterations in general are rare -- 0% of normal libraries and only 7.3% of libraries from other non-oligodendroglial cancers.
Preferential distribution of CIC mutations in oligodendrogliomas
Of the 21 mutations detected in the 20 CIC-mutant oligodendrogliomas (one carrying two distinct mutations), 9 were clustered in exon 5 within an annotated DNA interacting domain; 3 were in exon 20 within an annotated protein interacting domain, and the remaining 9 were in other exons (Figure 1). We also detected recurrent somatic mutations resulting in p.Arg201Trp and p.Arg215Trp changes and a recurrent out-of-frame deletion at chr19:47483653delTCC in exon 5. Another recurrent somatic mutation in codon 1515 of exon 20, leading to change of Arg>His and Arg>Leu, was also identified. When the rare CIC mutations were identified in the GSC human variant database, exons 5 and 20 mutations were represented in 8.6% of non-oligodendroglial tumours but disproportionally in 69% of oligodendrogliomas. Similarly, a survey of the Catalogue of Somatic Mutations in Cancer (COSMIC v54) database showed 10 reported mutations in CIC, none occurring in exons 5 or 20.
Figure 1.
Mutations are identified along a schematic of the human CIC gene with the highly conserved DNA- interacting HMG domain (exon 5) and protein-protein interacting GRO-L homology domain (exon 20) identified. Somatic mutations are identified with arrows and amino acid changes. Recurrent mutations are outlined. The vast majority of mutations from 1p/19q co-deleted oligodendroglioma (IDH1/2 mutated) are concentrated within exons 5 and 20.
To assess the significance of the clustering of somatic mutations in CIC exons 5 and 20, we performed multiple sequence alignments using CLUSTALW [35]. This analysis revealed that CIC exon 5 encoded a conserved annotated DNA-binding region, while exon 20 encoded a conserved domain lacking annotation in humans (Supplemental Data). Side-by-side alignments of human and fruit fly CIC protein sequences revealed that the second conserved region mapped to a protein-protein interaction region, annotated in the fly as the “GRO-L interaction region” (not shown).
Validation of CIC exon 5 and 20 mutations in an expanded number of gliomas
Focused PCR and Sanger sequencing of CIC exons 5 and 20 was then performed on an extension set of 111 primary glioma specimens including 68 oligodendrogliomas (WHO II and III), 11 mixed oligoastrocytomas (WHO II and III), 32 astrocytomas (WHO II–IV), and 4 normal brain specimens. The association of CIC mutations with 1p/19q codeleted oligodendrogliomas was seen again in this cohort. In this extension set, 15 mutations were detected of which 14 (93%) were found in IDH1 mutated, 1p/19q codeleted oligodendrogliomas. (See Supplemental Data for detailed list of samples and mutations).
Association of CIC mutations with oligodendroglioma histology, IDH1/2 mutations, and 1p/19q status
Of the 204 tumour samples studied (16 discovery set exome and CIC deep sequenced, 77 CIC deep sequenced, 111 CIC ex5/20 Sanger sequenced), 191 were informative for CIC, IDH1/2, and 1p/19q. CIC mutations were closely associated with classic oligodendroglioma histology (p<0.001), and therefore also associated with other oligodendroglioma-related mutations in IDH1/2 mutations (p<0.001), and 1p/19q loss (p<0.001). No exon 5 CIC mutations were noted in tumours that were both IDH1/2 wild-type and 1p/19q intact. The single instance of a CIC mutation outside of classic oligodendrogliomas occurred in exon 20 in a case of WHO IV astrocytoma/gliomatosis cerebri, raising the possibility that exon 20 mutations may be less specific for oligodendrogliomas than other mutations. No CIC mutations were identified in 4 normal brain tissue samples. Thus, CIC mutations appear to be a specific to prototypical oligodendrogliomas wherein we frequently observed co-alteration of IDH, 1p/19q, and CIC abnormalities.
Clinicopathologic correlation of oligodendrogliomas with and without mutations in CIC
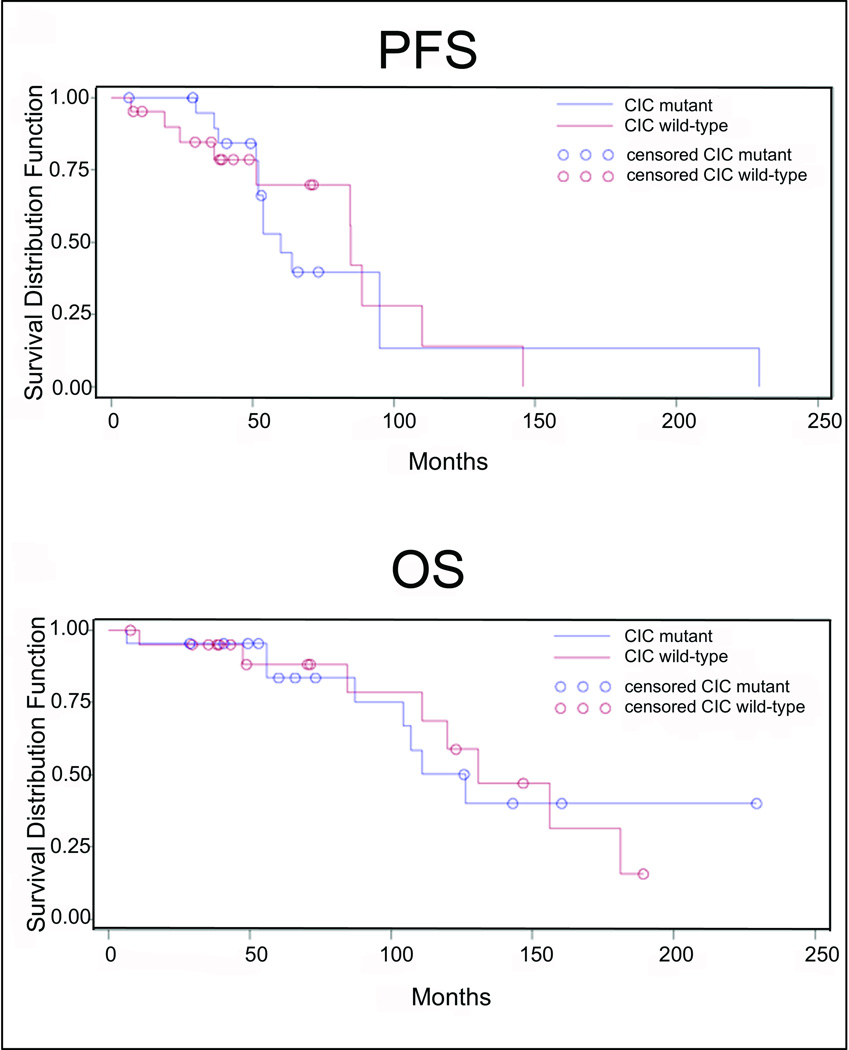
To determine whether the presence of CIC mutation has prognostic significance, we examined relationships with tumour grade, progression-free survival, and overall survival. Among the 84 oligodendrogliomas with 1p/19q co-deletion (out of 99 oligodendrogliomas total), CIC mutations were detected in 15/44 (34.1%) of low-grade tumours and 19/40 (47.5%) of high-grade tumours (p=0.27). Controlling for 1p/19q status in a stratified analysis, there was a trend toward an association between the presence of a CIC mutation and high-grade histology (OR 2.05, 95% CI 0.86–4.66, p=0.10). Clinical follow-up was available for 43 patients with oligodendrogliomas of known CIC status (23 wild-type, 20 mutant). In this cohort, CIC mutation was not associated with recurrence (13/22 wild type vs. 10/21 mutant, p=0.55), progression-free survival (p=0.89), or overall survival (p=0.99) (Figure 2).
Figure 2.
Kaplan-Meier analyses show no differences in progression-free survival (PFS) or overall survival (OS) in CIC-mutant versus wild-type oligodendroglioma patients.
Validation of CIC mutations in oligodendroglioma cell lines
The initiation and maintenance of different types of brain tumours has been linked to the presence of “tumour initiating cells” (TICs) [36, 37]. These cells have the ability to undergo asymmetric and symmetric division to propagate and to generate various types of neoplastic progeny. TIC lines propagated under stem cell promoting conditions have become valuable reagents in the study of cancer biology as they more reliably maintain phenotypic and genotypic characteristics of their parent cancers. Recently, two oligodendroglioma-derived TIC lines (BT54 and BT88) were established that display the oligodendrogliomatous phenotype and co-deletions of 1p and 19q [15]. To determine whether these cell lines were similar at the DNA sequence level to our patient cohorts and to assist in the use of the lines for functional studies of oligodendroglioma, we sequenced the genomes of the two TIC lines using SOLiD technology to an average of 30X haploid coverage (Supplemental Data). We confirmed that line BT54 carried the mutation chr19:47483801G>C affecting the CIC exon 6 splice acceptor site and line BT88 has a missense mutation leading to p.ARg1515His, both of which were identified in the respective parental tumour tissues from which the lines had been derived.
Expression of mutant CIC transcripts and protein
Analysis of RNA Seq data confirmed that the CIC mutations identified at the DNA level in the exomes resulted in expression of mutation-bearing CIC transcripts. Additional gene expression analyses of 16 oligodendrogliomas, including 4 wildtype and 12 with mutated in CIC also showed no significant difference in transcript expression levels between wildtype and mutated CIC (data not shown). The assembly of RNA Seq data using ABySS [38, 39] revealed the presence of a novel alternative transcript. A previously unknown exon (exon 0) was spliced to exon 2. Thus we predict two predominant transcripts of CIC in oligodendrogliomas: a short isoform predicted to encode a protein of 1608 amino acids (160 kDa), and a long isoform, incorporating exon 0 and predicted to encode 2517 amino acids (250 kDa). We confirmed the specificity of our antibody via CIC sequence specific siRNA knockdown which also confirmed the presence of both short and long form proteins in protein blot and immunoprecipitation-MRM assay (Figure 3A, B1–2). Next we confirmed CIC protein expression in lysates from control human embryonic kidney cell line (HEK293), BT54 (somatic mutation affecting splice acceptor in exon 6), and BT88 (p.Arg1515His) were equivalent, indicating that CIC mutant protein is expressed and that both predicted transcripts encode CIC protein (Figure 3C)
Figure 3.
A representative western blot showing high levels of CIC mutant protein expression both in nuclear and cytoplasmic protein fractions from oligodendroglioma cell lines, line 54 (L54) and line 88 (L88). CE- mouse cerebellum, HEK-HEK293 embryonic kidney line.
DISCUSSION
The data presented in this study, generated from systematic sequencing of tumour exomes, genomes, and transcriptomes of oligodendroglioma, demonstrate the power of massively parallel next generation sequencing in uncovering novel genetic changes in cancers. We sequenced a total of 628,096,883,783 bp of tumour and 499,887,339,249 bp of normal DNA from genome, RNA-seq and exome libraries. From this, we confirmed that IDH mutations are a universal feature of oligodendrogliomas. We also found novel potential driver mutations in CIC in a majority of the tumours.
On a broad level, the data show that oligodendrogliomas carry a relatively modest and stable somatic mutational load compared to many other cancers -- much lower than that present in glioblastoma, melanoma, lymphoma, and colorectal cancer (Broad Institute, unpublished). In addition to known positive prognostic factors such as the proneural gene signature and MGMT promoter methylation, the finding that oligodendrogliomas bear mutations relatively infrequently might also explain the relatively benign clinical course of this tumour and its initial chemosensitivity, an hypothesis that has yet to be formally tested. A particularly striking finding from our analysis, however, was the identification of recurrent mutations in CIC and the apparent relationship between mutations in CIC and IDH genes, and the 1p/19q anomaly, which appear to be distinguishing features of oligodendrogliomas.
CIC, found on chromosome 19q13.2, encodes the mammalian homolog of the Drosophila transcriptional repressor Capicua. In Drosophila, Capicua is a transcriptional repressor with crucial roles in development – it represses genes downstream of the RAS/MAPK pathway that signal the differentiation of wing veins, imaginal eye discs, and head and tail polarity development [40–42]. Mammalian Capicua is highly expressed in the brain, particularly in the external granular cells of the developing cerebellum [43], and its over-expression has been identified in medulloblastomas, an aggressive primitive neuroectodermal tumour of the central nervous system [44]. However CIC mutations in medulloblastoma have not been reported. Association of CIC alterations with human disease is restricted to two diseases affecting cells of the neural crest lineage. Spinocerebellar Ataxia I (SCAI), a progressive neurodegenerative disease, and Ewing’s Family Tumours, a cohort of aggressive soft tissue tumours affecting children and young adults, both involve defects of the CIC gene which in turn lead to dysregulated interactions of the CIC protein with downstream signaling partners [45, 46]. With respect to the Ewing’s family of tumours, rare cases of Ewing’s sarcoma have a recurrent chromosomal translocation t(4;19)(q35;q13) that generates a novel CIC –DUX4 fusion protein. The fusion oncoprotein exhibits enhanced transcriptional activation of the ETS family genes ERM/ETV5and ETV1 [45, 47] whose expression levels are also elevated in “traditional” Ewing’s sarcoma with the EWS-FLI1 fusion [48].
It is intriguing that recurrent mutation in CIC, located on chromosome 19q, is found almost exclusively in 1p/19q codeleted oligodendrogliomas with IDH1 mutation. Yet loss of chromosome 1p is more strongly associated with the oligodendrogliomatous phenotype and clinical behaviour than 19q loss [3]. In addition, 19q loss, which is occasionally found in astrocytomas, has traditionally been considered less specific to oligodendroglioma. Thus, it was unexpected that our sequencing did not yield more recurrent candidate coding point mutations in 1p, including mutations in FUBP1 as reported by others [24]. Since our study focused on the detection of recurrent point mutations, other types of recurrent alterations affecting the 1p region have not been excluded. Larger scale sequencing of panels of whole tumor and normal genomes would address this possibility.
High levels of cross-species sequence conservation and identification of functional domains encoded by the human CIC gene support possible functional consequences of mutations in CIC. Two highly conserved regions in CIC include the high mobility group (HMG) DNA binding domain in exon 5 and the protein-protein interaction GRO-L domain in exon 20 at the C-terminus of the protein – both these regions contained recurrent mutations in our patient series. We have confirmed that mutant CIC mRNA and mutant CIC protein are expressed. Thus, based on the known structure/function of CIC, it is likely that the oligodendroglioma mutations play a key role in the biology of the disease.
Two- thirds of CIC somatic mutations in oligodendroglioma occurred in exon 5 within the HMG-box domain. A majority of these are clustered around several hotspots in the following order of frequency – p.Arg215 > p.Arg201 > p.Arg202, and the affected residues display a high degree of evolution conservation in all examined vertebrate homologs. Furthermore, 33% of the unique exon 5 mutations were predicted to affect protein function using POLYPHEN2 analysis which included the recurrent p.Arg201Trp and p.Arg215Trp changes. Interestingly, 5/20 exon 5 mutations affected amino acid position 215 and 3 of these caused p.Arg215Trp changes, which corresponds to the Drosophila cicArg505Trp mutation associated with loss of CIC function. These mutations result in aberrant Drosophila eye development [42]. After exon 5, the next most frequently altered CIC region was in the C- terminal portion of exon 20. We found 4 mutations causing amino acid changes at p.Arg1515 to His, Leu, and Cys. All were predicted to be deleterious to protein function by POLYPHEN2 analysis. It is undetermined, however, whether the hemizygous CIC mutations in oligodendroglioma result in loss or gain of protein function. Whatever the consequence, the recurrent nature of CIC mutations in such a high percentage of tumours supports that they are driver mutations central to the genesis and/or identity of the tumours. It is clear that carefully designed functional studies taking into account genomic context (such as 1p/19q loss and IDH mutations) are necessary to delineate the role of CIC in the pathogenesis of oligodendroglioma.
Further investigation of this complex relationship and of the downstream molecular consequences of CIC mutation will necessitate the development of novel biological reagents and appropriate model systems. To this end, the two oligodendroglioma TIC lines analyzed here represent a unique resource to study the relationship between 1p/19q co-deletions and the special biology and clinical behaviour of this tumour. Future studies should also concentrate on sequencing of CIC in larger cohorts of oligodendrogliomas with clinical follow-up data to better distill potential influences on outcome. This will most likely involve international multicenter collaboration similar to a recent study [49].
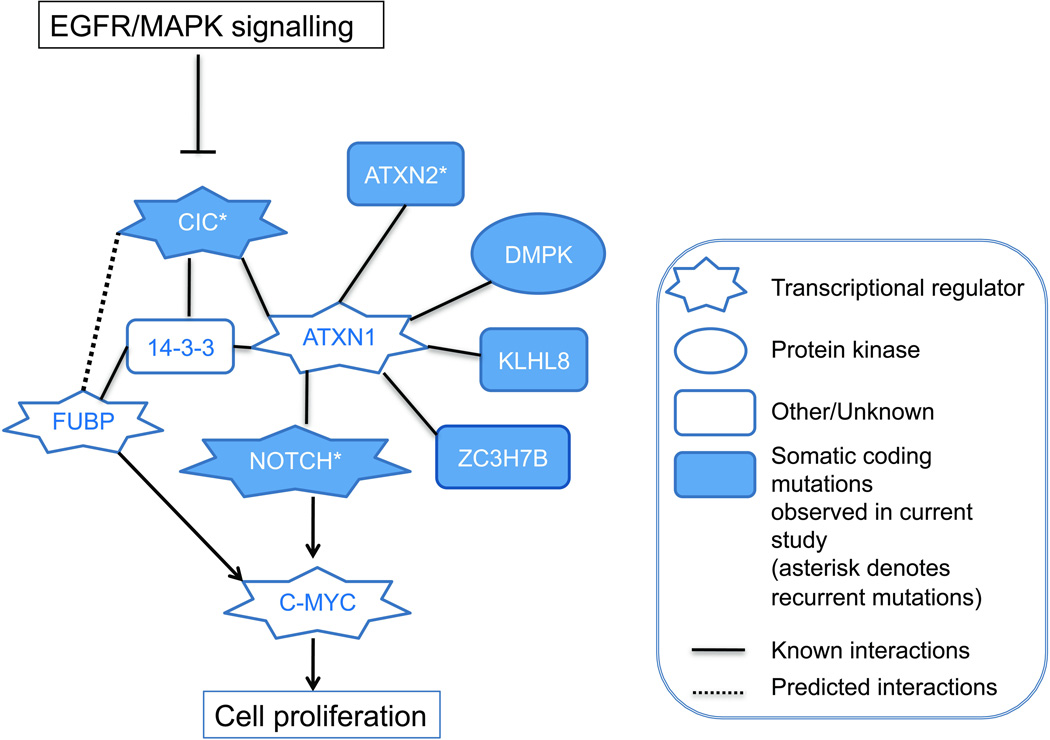
In the minority of oligodendrogliomas where we do not find CIC mutations, it is reasonable to speculate that other mutations in the RAS/MAPK pathway or in CIC interacting proteins may result in a phenocopy of CIC mutants. One potential candidate is ATXN2, which we found mutated in two oligodendrogliomas in our exome dataset. CIC is known to interact with ATXN1 in human disease [46], and ATXN1 is modulated by ATXN2 [50]. Similarly, we and others [24] have found a low frequency of NOTCH mutations in oligodendrogliomas. Recent work suggests direct involvement of ATXN1 in the NOTCH signaling pathway that has critical roles in neural development and tumourigenesis [51] [52]. Lastly, FUBP1 was reported to be mutated in oligodendroglioma [24], and although we did not find non-synonymous coding mutations in our tumours we did detect infrequent somatic splice site variants. The possible relationship between FUBP1 and CIC, however, remains to be elucidated. Our working model is that mutations in CIC (in the majority of tumours) or a number of related genes such as ATXN2, NOTCH1, and FUBP1 (in a minority of tumours) may be functionally equivalent in the context of 1p/19q loss, IDH1/2 mutation, and the genesis of oligodendroglioma (FIGURE 4). In our exome discovery cohort, we noted various patterns of co-occurrence of somatic mutations affecting CIC, NOTCH genes, and ATXN2. However, such a model has yet to be formally tested in a larger cohort of tumours and also in the context of the two predominant patterns of mutations in CIC affecting the two functional domains.
Figure 4.
Proposed CIC interaction network in the context of observed mutations
Our discovery that CIC mutations are a core abnormality in the majority of oligodendrogliomas has translational and clinical implications in neuro-oncology. Improved understanding of the role of CIC in a background of IDH1/2 mutations in codeleted oligodendroglioma will significantly enhance our understanding of this disease and may pave the way for the development of more targeted therapies for this common type of glioma.
Supplementary Material
Acknowledgements
We would like to acknowledge Dr. John Kelly for cell lines, Dr. Peter Forsyth and the Calgary Brain Tumour Bank for tissues, Colleen Anderson and Susan Hui for technical assistance, Alberta Innovates Health Solutions (JAC, EES), Brain Tumour Foundation of Canada (JGC), Alberta Cancer Foundation (JGC), and the Clark Smith Family. We also acknowledge Dr. Robert Jenkins for performing mFISH on the brain tumour initiating cell lines BT054 and BT088 and sharing the data. MAM acknowledges the support of the BC Cancer Foundation, Western Economic Diversification, Genome Canada and Genome British Columbia. MAM is also grateful to Dr. Brian Thiessen, BCCA, for early conversations that helped shape the project. SY acknowledges the generous support of BrainCare BC and the BCCA Hershey & Yvette Porte Neuro-Oncology Endowment Fund. We would also like to acknowledge Maria Mendez-Lago, Diane Trinh, Kane Tse, and Thomas Zeng at the BCGSC for invaluable scientific advice and technical support.
Footnotes
Author contributions
JGC and MAM funded and co- directed the project, designed the study. SY planned and executed the validation experiments, performed data analysis. YSB conducted next generation sequencing (NGS) bioinformatic analysis (WGSS, exome, amplicon). JAC identified tumours and performed the histopathology and analysis, provided tumour and blood samples for NGS. MDB collected and identified tumour samples for sequencing. AK and S Young contributed additional tumour and blood samples for validation. DNL contributed additional tumour samples for validation. CC and MP assisted in collection and identification of tumour samples. OM conducted NGS bioinformatic analysis (WTSS, exome, hypermutation, CIC homology). SM conducted NGS bioinformatic analysis (WTSS). SC coordinated and led the amplicon deep sequencing and functional/proteomic validation, assembled the data. GBM coordinated and performed proteomic analysis. AN performed proteomic analysis. AJM coordinated NGS. YZ performed NGS. SJ coordinated NGS bioinformatic analysis. RC conducted NGS bioinformatic analysis (WGSS). KLM conducted NGS bioinformatic analysis (ABySS pipeline). IB, RC, EC, RD, MH, SJ, HL, JQ, NT, RV assisted in NGS bioinformatic analysis. MF and JTW performed deep amplicon sequencing and biochemical analysis. RM coordinated sequencing and NGS bioinformatic analysis. AM conducted biochemical analysis (Mass spectrometry). GR and EEE contributed clinical information and performed survival analysis. SW generated TIC lines from tumours. WW performed statistical analysis. JGC, MAM, SY, YSB, JAC, OM and SC wrote the manuscript.
REFERENCES
- 1.Mason W, Louis DN, Cairncross JG. Chemosensitive gliomas in adults: which ones and why? J Clin Oncol. 1997;15:3423–3426. doi: 10.1200/JCO.1997.15.12.3423. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, et al., editors. WHO Classification of Tumours of the Central Nervous System. Lyon: IARC; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairncross JG, Ueki K, Zlatescu MC, et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 4.McDonald JM, See SJ, Tremont IW, et al. The prognostic impact of histology and 1p/19q status in anaplastic oligodendroglial tumors. Cancer. 2005;104:1468–1477. doi: 10.1002/cncr.21338. [DOI] [PubMed] [Google Scholar]
- 5.Ducray F, Idbaih A, de Reynies A, et al. Anaplastic oligodendrogliomas with 1p/19q codeletion have a proneural gene expression profile. Mol Cancer. 2008;7:41. doi: 10.1186/1476-4598-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gravendeel LA, Kloosterhof NK, Bralten LB, et al. Segregation of non-p.R132H mutations in IDH1 in distinct molecular subtypes of glioma. Hum Mutat. 2010;31:E1186–E1199. doi: 10.1002/humu.21201. [DOI] [PubMed] [Google Scholar]
- 7.van den Bent MJ, Dubbink HJ, Marie Y, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16:1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 8.Cooper LA, Gutman DA, Long Q, et al. The Proneural Molecular Signature Is Enriched in Oligodendrogliomas and Predicts Improved Survival among Diffuse Gliomas. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huse JT, Phillips HS, Brennan CW. Molecular subclassification of diffuse gliomas: Seeing order in the chaos. Glia. 2011;59:1190–1199. doi: 10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
- 10.Okamoto Y, Di Patre PL, Burkhard C, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108:49–56. doi: 10.1007/s00401-004-0861-z. [DOI] [PubMed] [Google Scholar]
- 11.Boulay JL, Miserez AR, Zweifel C, et al. Loss of NOTCH2 positively predicts survival in subgroups of human glial brain tumors. PLoS ONE. 2007;2:e576. doi: 10.1371/journal.pone.0000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrer-Luna R, Mata M, Nunez L, et al. Loss of heterozygosity at 1p–19q induces a global change in oligodendroglial tumor gene expression. J Neurooncol. 2009;95:343–354. doi: 10.1007/s11060-009-9944-y. [DOI] [PubMed] [Google Scholar]
- 13.Benetkiewicz M, Idbaih A, Cousin PY, et al. NOTCH2 is neither rearranged nor mutated in t(1;19) positive oligodendrogliomas. PLoS ONE. 2009;4:e4107. doi: 10.1371/journal.pone.0004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bralten LB, Nouwens S, Kockx C, et al. Absence of common somatic alterations in genes on 1p and 19q in oligodendrogliomas. PLoS ONE. 2011;6:e22000. doi: 10.1371/journal.pone.0022000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly JJ, Blough MD, Stechishin OD, et al. Oligodendroglioma cell lines containing t(1;19)(q10;p10) Neuro Oncol. 2010 doi: 10.1093/neuonc/noq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah SP, Kobel M, Senz J, et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med. 2009;360:2719–2729. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 17.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2009;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2009;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei X, Walia V, Lin JC, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steidl C, Shah SP, Woolcock BW, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471:377–381. doi: 10.1038/nature09754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 Contribute to Human Oligodendroglioma. Science. 2011 doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horbinski C, Kofler J, Kelly LM, et al. Diagnostic use of IDH1/2 mutation analysis in routine clinical testing of formalin-fixed, paraffin-embedded glioma tissues. J Neuropathol Exp Neurol. 2009;68:1319–1325. doi: 10.1097/NEN.0b013e3181c391be. [DOI] [PubMed] [Google Scholar]
- 26.Clark MJ, Homer N, O'Connor BD, et al. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000832. e1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrader KA, Heravi-Moussavi A, Waters PJ, et al. Using next-generation sequencing for the diagnosis of rare disorders: a family with retinitis pigmentosa and skeletal abnormalities. J Pathol. 2011;225:12–18. doi: 10.1002/path.2941. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goya R, Sun MG, Morin RD, et al. SNVMix: predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics. 2010;26:730–736. doi: 10.1093/bioinformatics/btq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter C, Smith R, Cahill DP, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yip S, Miao J, Cahill DP, et al. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15:4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith JS, Alderete B, Minn Y, et al. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999;18:4144–4152. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- 34.Fejes AP, Khodabakhshi AH, Birol I, et al. Human variation database: an open-source database template for genomic discovery. Bioinformatics. 2011;27:1155–1156. doi: 10.1093/bioinformatics/btr100. [DOI] [PubMed] [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh SK, Clarke ID, Terasaki M, et al. Identification of a Cancer Stem Cell in Human Brain. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 37.Taylor MD, Poppleton H, Fuller C, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Simpson JT, Wong K, Jackman SD, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson G, Schein J, Chiu R, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 40.Jimenez G, Guichet A, Ephrussi A, et al. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14:224–231. [PMC free article] [PubMed] [Google Scholar]
- 41.Goff DJ, Nilson LA, Morisato D. Establishment of dorsal-ventral polarity of the Drosophila egg requires capicua action in ovarian follicle cells. Development. 2001;128:4553–4562. doi: 10.1242/dev.128.22.4553. [DOI] [PubMed] [Google Scholar]
- 42.Tseng AS, Tapon N, Kanda H, et al. Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/ras signaling pathway. Curr Biol. 2007;17:728–733. doi: 10.1016/j.cub.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee CJ, Chan WI, Cheung M, et al. CIC, a member of a novel subfamily of the HMG-box superfamily, is transiently expressed in developing granule neurons. Brain Res Mol Brain Res. 2002;106:151–156. doi: 10.1016/s0169-328x(02)00439-4. [DOI] [PubMed] [Google Scholar]
- 44.Lee CJ, Chan WI, Scotting PJ. CIC, a gene involved in cerebellar development and ErbB signaling, is significantly expressed in medulloblastomas. J Neurooncol. 2005;73:101–108. doi: 10.1007/s11060-004-4598-2. [DOI] [PubMed] [Google Scholar]
- 45.Kawamura-Saito M, Yamazaki Y, Kaneko K, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 46.Lam YC, Bowman AB, Jafar-Nejad P, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 47.Yoshimoto M, Graham C, Chilton-MacNeill S, et al. Detailed cytogenetic and array analysis of pediatric primitive sarcomas reveals a recurrent CIC-DUX4 fusion gene event. Cancer Genet Cytogenet. 2009;195:1–11. doi: 10.1016/j.cancergencyto.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 48.Erkizan HV, Uversky VN, Toretsky JA. Oncogenic partnerships: EWS-FLI1 protein interactions initiate key pathways of Ewing's sarcoma. Clin Cancer Res. 2010;16:4077–4083. doi: 10.1158/1078-0432.CCR-09-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lassman AB, Iwamoto FM, Cloughesy TF, et al. International retrospective study of over 1000 adults with anaplastic oligodendroglial tumors. Neuro Oncol. 2011;13:649–659. doi: 10.1093/neuonc/nor040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Ramahi I, Perez AM, Lim J, et al. dAtaxin-2 mediates expanded Ataxin-1-induced neurodegeneration in a Drosophila model of SCA1. PLoS Genet. 2007;3:e234. doi: 10.1371/journal.pgen.0030234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong X, Gui H, Jin F, et al. Ataxin-1 and Brother of ataxin-1 are components of the Notch signalling pathway. EMBO Rep. 2011;12:428–435. doi: 10.1038/embor.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, Kesari S, Rooney C, et al. Inhibition of Notch Signaling Blocks Growth of Glioblastoma Cell Lines and Tumor Neurospheres. Genes Cancer. 2010;1:822–835. doi: 10.1177/1947601910383564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.