Abstract
Background
Communities of bacteria, termed biofilms, develop on biotic and abiotic surfaces including medical devices and surgical suture. Biofilm-associated bacteria are typically recalcitrant to antibiotic therapy, and the effects of antibiotics on microbial biofilms are not clearly understood. There is emerging evidence that under specific conditions, aminoglycosides may actually promote biofilm development. Experiments were designed to study the effects of gentamicin on suture-associated Staphylococcus aureus biofilms.
Materials and methods
S. aureus biofilms were formed after 24 hr incubation of bacteria with silk suture. Susceptibility of planktonic S. aureus (from broth culture) to gentamicin was compared to the susceptibility of cells from mechanically dispersed S. aureus biofilms. Subinhibitory and inhibitory concentrations of gentamicin were subsequently incubated with intact suture-associated biofilms. S. aureus viability and metabolic capacity were assessed, and biofilm biomass was quantified with crystal violet (binds negatively charged surface molecules) and with the nucleic acid stain Syto 9. Scanning electron microscopy was used to assess the effect of gentamicin on the ultrastructure of suture-associated S. aureus biofilms.
Results
Planktonic cells and S. aureus cells from mechanically dispersed biofilms had similar susceptibility to gentamicin. However, after incubation of high concentrations of gentamicin with intact biofilms, high numbers of S. aureus remained both viable and metabolically active; biofilm biomass was increased and biofilm ultrastructure showed staphylococcal cells within copious amounts of extracellular material.
Conclusion
Gentamicin does not effectively kill S. aureus within intact suture-associated biofilms, and gentamicin also promotes the biomass of S. aureus biofilms.
Keywords: Staphylococcus aureus, suture, biofilm, biomass, electron microscopy, gentamicin
INTRODUCTION
It is now widely accepted that the majority of clinical infections involve microbial growth as a biofilm, including infections associated with indwelling devices such as catheters, joint prostheses, heart valves, stents, and suture materials [1–3]. A biofilm is a community of microbes growing on a biotic or abiotic surface and surrounded by a complex extracellular polymeric substance composed of proteins, glycoproteins, glycolipids, polysaccharides, and extracellular DNA [4]. Biofilm infections are important in postsurgical patients and >20% of nosocomial infections are surgical site infections (SSIs) [5]. Although there are no definitive data on the incidence of SSIs associated with surgical suture, it is reasonable to assume that a substantial proportion of SSIs involve suture materials. Using an in vitro model of Staphylococcus aureus contamination of surgical suture, we recently reported that the resulting bacterial growth resembles a biofilm [6]. Unfortunately, bacteria within a biofilm are typically more antibiotic resistant than planktonic (free living) microbes [3, 7–9].
There are no universally accepted methods for studying the antibiotic susceptibility of bacteria within a biofilm. Most antibiotic susceptibility studies involving biofilms assess bacterial killing within preformed biofilms. This is an important distinction because clinical microbiology laboratories report the antibiotic susceptibility of comparatively low numbers of actively growing planktonic cells [10], not the ability of an antibiotic to kill dense clusters of bacteria living within a biofilm community. Because biofilms are notoriously recalcitrant to antibiotic therapy, it has seemed logical to assume that biofilm-associated bacteria have increased intrinsic antibiotic resistance compared to their planktonic counterparts. However, growing evidence indicates this is not the case, and it is now thought that the antibiotic resistance of chronic biofilm infections is due to existence of a small percentage of “persister cells” that are stochastically generated and highly tolerant to antibiotics [11, 12]. To further complicate this situation, there is evidence that subinhibitory concentrations of some antibiotics actually promote biofilm formation, perhaps by facilitating production of the extracellular polymeric substance. For example, Hoffman et al. [13] reported that subinhibitory concentrations of the aminoglycoside tobramycin induced biofilm formation by Pseudomonas aeruginosa and Escherichia coli, and went on to suggest that biofilm formation may be a defensive reaction to the presence of an antimicrobial agent.
S. aureus remains one of the most frequent etiologic agents of SSIs (as well as bloodstream infections), and the morbidity and mortality associated with these infections is high in postsurgical and critically ill patients [14, 15]. We used an in vitro model to test the hypothesis that subinhibitory and/or inhibitory concentrations of gentamicin alter biomass, metabolic activity, and ultrastructure of S. aureus suture-associated biofilms. Though gentamicin is typically a third-line antibiotic in the treatment of clinical S. aureus infections, we developed a model using gentamicin because aminoglycosides have been reported to induce biofilm formation with certain gram-negative bacteria [13], and this model would test the hypothesis that antibiotics may promote biofilm growth in an S. aureus biofilm. Although planktonic and mechanically dispersed biofilm cells had similar intrinsic susceptibility to gentamicin, a stable population of bacteria within intact biofilms remained viable after incubation with high gentamicin concentrations; gentamicin also increased the biofilm biomass and gentamicin-treated biofilms had copious extracellular material that might potentially inhibit antibiotic binding to pertinent bacterial cell targets.
MATERIALS AND METHODS
Susceptibility of planktonic and biofilm-associated S. aureus to gentamicin
S. aureus RN6390 and ATCC 25923 are wild type strains known to produce biofilms [6,16, 17], and gentamicin sulfate was obtained from Sigma-Aldrich, Inc., St. Louis, MO. Using S. aureus inocula (described below) of 5 × 105/ml for both planktonic cells and mechanically dispersed biofilm cells, macrodilution susceptibility testing for the minimum inhibitory concentration (MIC) and the minimum bactericidal concentration (MBC) followed the Clinical and Laboratory Standards Institute (CLSI) guidelines [10], and results represent at least two replicate experiments. Clinical microbiology laboratories in the USA adhere to CLSI guidelines for antimicrobial susceptibility testing. The MIC is defined as the lowest concentration that inhibits visible growth in a broth culture, and the MBC is the minimum concentration that results in 99.9% killing of the bacterial cells in the inoculum. To prepare planktonic cells, static cultures were incubated overnight at 37°C in tryptic soy broth (TSB), washed, and resuspended to the appropriate concentration using densitometry with results confirmed by quantitative plate culture. Suture-associated biofilms were cultivated as described [6]. Briefly, each well of a six-well microtiter plate contained four 1-cm segments of black braided 3-0 silk suture (Ethicon, Inc., Somerville, NJ) suspended in 3 ml of biofilm growth medium, namely 66% TSB supplemented with 0.2% glucose [17]. Each well was inoculated with ~105 S. aureus cells and incubated for 24 hr at 37°C with gentle rotation. Suture was then gently rinsed, transferred to 2 ml sterile phosphate buffered saline (PBS), sonicated at ~50 joules at 100% amplitude for 5 sec using a sonicator at 20kHz (Sonics and Materials, Newtown, CT); the resulting cell suspension was adjusted by densitometry with results confirmed by quantitative culture. Sonication had no noticeable effect on bacterial viability, and microscopy confirmed that planktonic and biofilm inocula were similarly dispersed single-cell suspensions.
To assess the antibiotic susceptibility of S. aureus cells within intact biofilms, S. aureus was incubated for 24 hr with suture segments as described above. Biofilm-laden sutures were transferred to wells containing fresh medium supplemented with varying concentrations of gentamicin that ranged from subinhibitory concentrations to concentrations up to 100 times the MIC/MBC, and incubated overnight at 37°C under static conditions. Control wells contained no antibiotic. Biofilms were assayed for the effect of gentamicin on the numbers of viable bacteria in biofilm sonicates, as well as on biofilm biomass, metabolic capacity, and ultrastructure.
Biofilm biomass, metabolic capacity, and ultrastructure
Biofilm biomass was measured with crystal violet and with Syto 9 as described [18] with minor modifications. Crystal violet is a basic dye that binds negatively charged surface molecules, including live and dead bacteria and matrix polysaccharides; green fluorescent Syto 9 is a nucleic acid stain that passively diffuses through bacterial membranes and binds DNA within live and dead cells as well as DNA in the extracellular matrix [19]. Biofilm-laden sutures were rinsed with PBS, fixed in 99% methanol for 15 min, air-dried, incubated 20 min with 0.5% crystal violet (Fisher Chemical, Pittsburgh, PA), washed, and then incubated for 20 to 30 min in 33% acetic acid to release the crystal violet, with absorbance read at 590 nm. A 5 mM stock solution of Syto 9 (Invitrogen, Carlsbad, CA) was diluted 1:50,000 in PBS, added to rinsed biofilms, and incubated 45 min in the dark, with fluorescence read at 528±10 nm.
The BioTimer assay [19] was used to assess the effect of gentamicin on the metabolic capacity of S. aureus in undisturbed biofilms. Briefly, Mueller-Hinton broth supplemented with 1% glucose and 0.0025% phenol red was added to washed biofilms and incubated at 37°C without shaking. The time for a color switch from red to yellow was observed every 30 min for 7 hr. A standard curve was generated using known numbers of planktonic bacteria and results from biofilm cultures were reported as planktonic-equivalent colony forming units (CFUs). Some samples switched from red to yellow after the 7 hr observation period, while others never switched and remained red for 24 hr; for mathematical purposes, these samples were assumed to have switched color at 15 hr, corresponding to a time when 10 CFU of planktonic S. aureus (lower limit of assay detection) has been reported to cause the color switch [19].
To observe biofilm ultrastructure, each of the two S. aureus strains was incubated with silk suture for 24 hr as described above, and then transferred to fresh medium containing 0, 1, 10, or 100 µg/ml gentamicin. After overnight incubation with antibiotic, sutures were processed for scanning electron microscopy (SEM) as described [6, 20]. Samples were viewed with a Hitachi S-4700 field emission scanning electron microscope operated at 2.5 kV. Each biofilm treatment was processed in duplicate and each sample was examined at least 45 min.
Statistical analysis
Bacterial numbers were converted to log10 prior to statistical analysis, and significance was set at P<0.05. Differences between gentamicin concentrations were analyzed by one-way analysis of variance followed by Fisher’s test for significant difference.
RESULTS
Gentamicin susceptibility of planktonic and biofilm-associated S. aureus
The MIC/MBCs of gentamicin were similar for both S. aureus strains, and similar MIC/MBCs were obtained using planktonic cells or cells mechanically dispersed from intact biofilms (Table 1). According to CLSI breakpoints [10], both S. aureus strains were susceptible to gentamicin, and the antibiotic was bactericidal based on similar MIC and MBC values, i.e., similar values for inhibition of visible growth and bacterial killing.
TABLE 1.
Gentamicin susceptibility of S. aureus RN6390 and ATCC 25923 cultivated as planktonic cells and as mechanically dispersed biofilm cells, with results representing at least two replicate experiments performed according to CLSI guidelines [10]
|
S. aureus strain |
MIC (µg/ml) | MBC (µg/ml) | ||
|---|---|---|---|---|
| planktonic | biofilm | planktonic | biofilm | |
| RN 6390 | 1 | 1–2 | 1–2 | 1–2 |
| ATCC 25923 | 1 | 1 | 2–4 | 2–4 |
Viability and metabolic capacity of gentamicin-treated biofilms
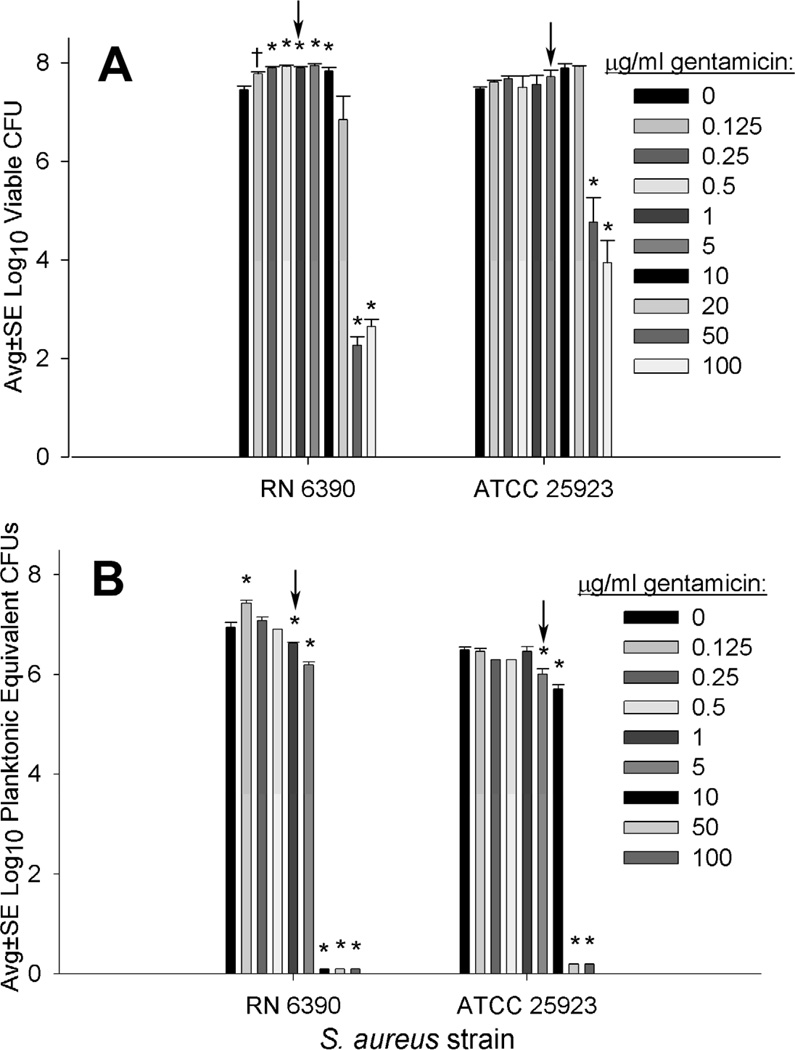
As noted in table 1, ≤4 µg/ml of gentamicin effectively killed planktonic S. aureus as well as biofilm-associated S. aureus cells that were mechanically dispersed before exposure to the antibiotic (Table 1). However, up to 20 µg/ml of gentamicin did not effectively kill S. aureus when gentamicin was incubated with intact biofilms (Fig. 1A), and biofilm-associated S. aureus remained metabolically active after incubation with high concentrations of gentamicin above the MIC/MBC for planktonic and mechanically dispersed biofilm cells (Fig. 1B). Interestingly, compared to incubation with 0 µg/ml gentamicin, the RN 6390 strain had significantly increased numbers of viable biofilm-associated cells following overnight incubation with 0.125 to 10 µg/ml gentamicin (Fig. 1A), and this strain had significantly increased metabolic capacity in biofilms incubated with 0.125 µg/ml gentamicin compared to those incubated with 0 µg/ml gentamicin.
FIG 1.
Viability (A) and metabolic capacity (B) of intact biofilms formed by incubating silk suture with S. aureus RN6390 or ATCC 25923 for 24 hr, followed by overnight incubation with varying concentrations of gentamicin. A, Viable CFUs from sonicated biofilms (n≥5); B, Metabolic capacity of undisturbed biofilms measured as planktonic equivalent CFUs (n≥8). Arrows highlight approximate MIC/MBC of planktonic cells as well as cells from mechanically dispersed biofilms. Significant difference compared to 0 µg/ml at P<0.01 (*) and P<0.05 (†).
Biomass of gentamicin-treated biofilms
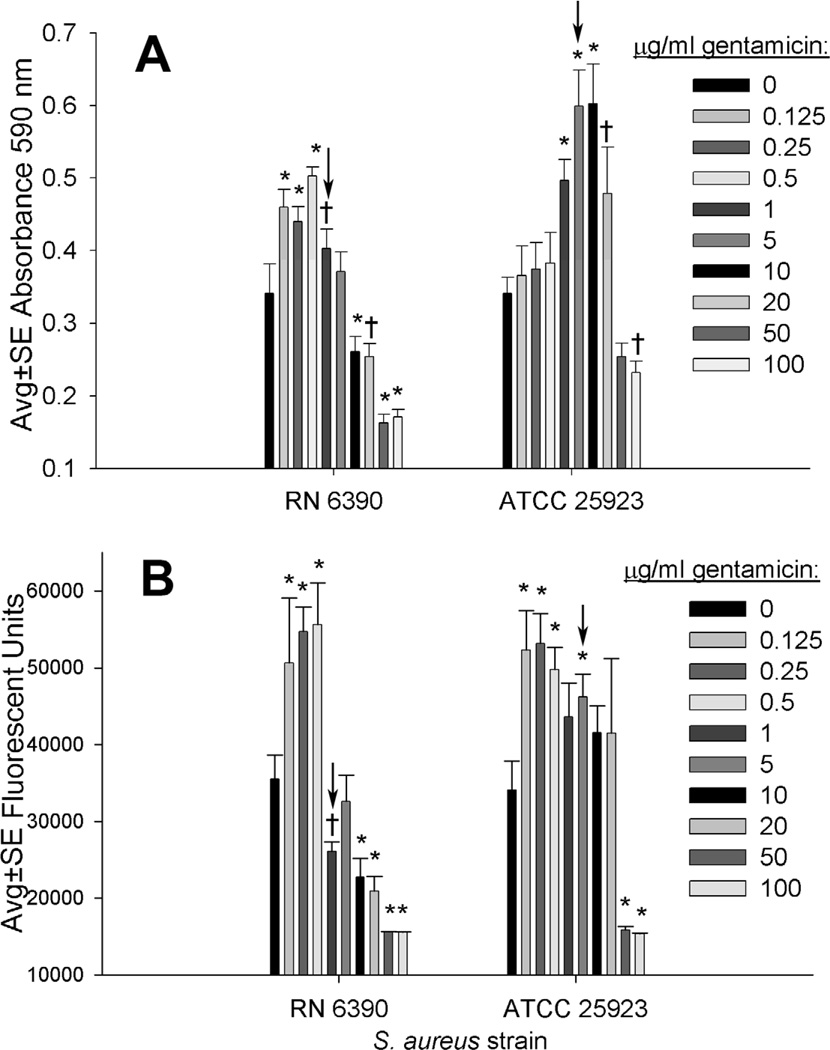
As measured with crystal violet, subinhibitory concentrations of gentamicin (<1 µg/ml) were associated with increased S. aureus RN 6390 biofilm biomass, while higher concentrations (5 to 20 µg/ml) were optimally effective in stimulating biomass with the ATCC 25923 strain (Fig. 2A). As measured with the nucleic acid stain Syto 9, the nucleic acid in RN 6390 biofilms was increased after incubation with subinhibitory concentrations of gentamicin (<1 µg/ml), while the nucleic acid in ATCC 25923 biofilms was increased after incubation with up to 5 µg/ml gentamicin (Fig. 2B).
FIG 2.
Biomass of biofilms formed by incubating silk suture with S. aureus RN6390 or ATCC 25923 for 24 hr, followed by overnight incubation with varying concentrations of gentamicin, with biomass measured (n≥5) as absorbance of crystal violet (A) and as fluorescence of Syto 9 (B). Arrows highlight approximate MIC/MBC of planktonic cells as well as cells from mechanically dispersed biofilms. Significant difference compared to 0 µg/ml at P<0.01 (*) and P<0.05 (†).
Ultrastructure of gentamicin-treated biofilms
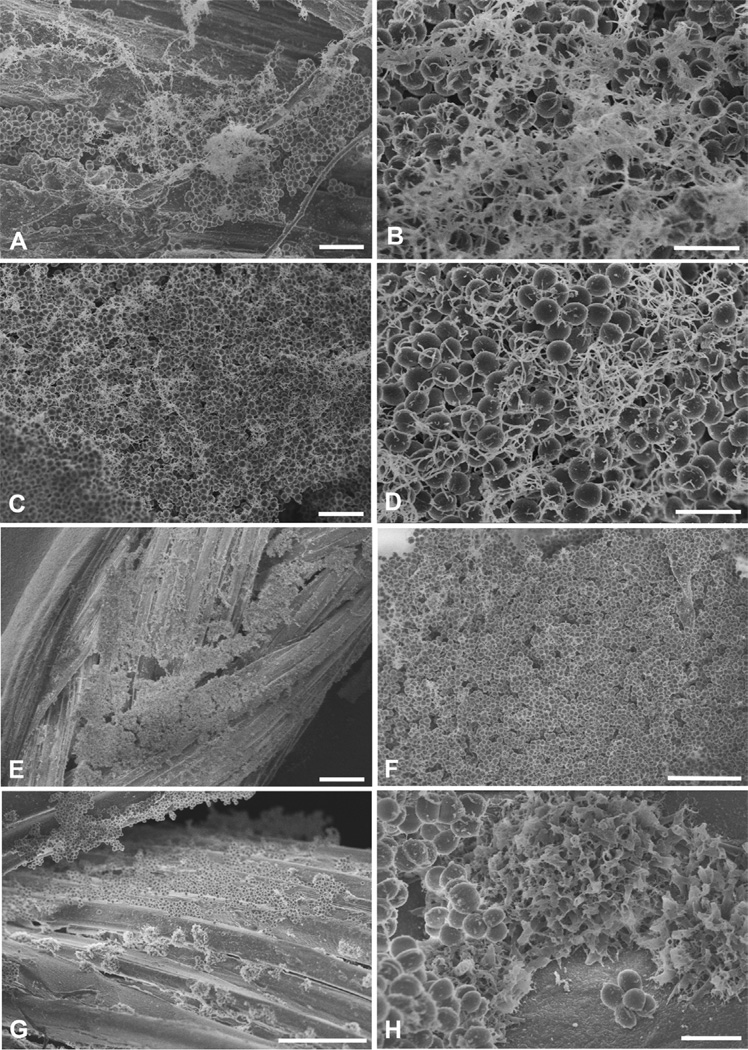
To assess the effect of gentamicin on biofilm ultrastructure, intact 24 hr suture-associated biofilms were incubated overnight with 0, 1, 10, and 100 µg/ml gentamicin, and then observed with SEM. Similar results were obtained with the two strains of S. aureus, and representative images from S. aureus ATCC 25923 biofilms are presented in figure 3. Control biofilms without gentamicin contained large clusters of cocci both on and between individual strands of braided silk suture; these cocci were enmeshed in extracellular material that often appeared fibrillar although some of this material had a more consolidated appearance (Fig 3A-B). In general, the amount of extracellular material appeared increased in biofilms treated with gentamicin. However, SEM is not a quantitative technique and observing samples at high resolution can introduce sampling error. Nonetheless, general observations can be made. Like control biofilms, robust biofilms were noted in samples incubated with 1 and 10 µg/ml gentamicin (Fig. 3C-F), and this observation was consistent with the recovery of high numbers of viable bacteria from biofilms treated with 1 and 10 µg/ml gentamicin (Fig. 1). In biofilms treated with 10 to 100 µg/ml gentamicin, not only were intact bacteria easily seen, but discreet clumps of material were noted along the silk strands (Fig. 3G) and these clumps appeared as “ruffled” extracellular matrix material at higher magnifications (Fig. 3H).
FIG 3.
SEMs of biofilms formed by incubating silk suture with S. aureus ATCC 25923 for 24 hr, followed by overnight incubation with varying concentrations of gentamicin. A and B, Control biofilms after incubation with 0 µg/ml gentamicin, showing clusters of cocci some of which are covered with fibrillar elements; C, D, E, and F, Robust biofilms remain evident after overnight incubation with 1 µg/ml (C and D) and 10 µg/ml (E and F) gentamicin; G, Biofilm on silk suture after incubation with 100 µg/ml gentamicin, showing clusters of normal-appearing cocci (upper photo) and clusters of ruffled matrix (lower photo) seen at higher magnification in H. Scale bars in µm: A=5, B=2, C=5, D=2, E=50, F=10, G=20, H=2.
DISCUSSION
Bacterial biofilms have increased antibiotic resistance, but the mechanisms responsible for this resistance remain unclear. To confound this situation, there is substantial evidence that planktonic and dispersed biofilm bacteria have similar intrinsic antimicrobial susceptibility [7, 8, 11, 21], a finding consistent with data from the present study where planktonic and dispersed suture-associated biofilm cells of S. aureus had similarly low MIC/MBCs to gentamicin (<4µg/ml, Table 1). If mechanically dispersed cells from an intact S. aureus biofilm do not have increased intrinsic resistance to gentamicin, why are intact biofilms recalcitrant to antibiotic therapy (Fig 1)? A partial answer is related to the fact that the data in Table 1 were generated according to CLSI guidelines [10] that define effective MBC killing as a 99.9% (≥3 log10) reduction in an inoculum of relatively low numbers of viable bacteria, e.g., 5 × 105/ml in the present study. In contrast, the inoculum in intact biofilms is typically much higher, and was ~107/ml in the present study (Fig. 1). Although effective bacterial killing (≥3 log10) was obtained with a high concentration (50 µg/ml) of gentamicin, 102 to 105 biofilm-associated S. aureus remained viable. Thus, compared to planktonic bacteria, as well as dispersed biofilm bacteria, not only were S. aureus cells within an intact biofilm more resistant to gentamicin, the CLSI guideline for effective bacterial killing (99.9% killing) may not be relevant for intact biofilms. It has been suggested that persister cells are largely responsible for the recalcitrance of biofilms to all known antibiotics [11], and the 102 to 105 viable S. aureus recovered from biofilms treated with 50 and 100 µg/ml gentamicin may represent this persister population.
Gentamicin increased the biomass of biofilms formed with both S. aureus RN 6390 and ATCC 25923, and this increase was detected using both the crystal violet assay (for negatively charged surface molecules) and the nucleic acid stain Syto 9 (Fig. 2). Others have noted that subinhibitory concentrations of some antibiotics may promote biofilm formation, perhaps by facilitating production of the extracellular polymeric substance, i.e., the biofilm matrix. For example, imipenem stimulated Acinetobacter baumannii biofilms cultivated on polystyrene [22], cefotaxime promoted biofilm formation and extracellular matrix production with selected isolates of Salmonella enterica serovar Typhimurium on plastic [23], tobramycin induced biofilm formation in P. aeruginosa and E. coli [13], tetracycline decreased expression of the icaADBC operon in Staphylococcus epidermidis resulting in decreased production of a polysaccharide intracellular adhesin thought to play a role in biofilm formation [24], and imipenem increased alginate production as well as the volume of P. aeruginosa biofilms [25]. Each of these examples used developing biofilms and tested subinhibitory (below the MIC/MBC) drug concentrations, while the present study tested a wide range of gentamicin concentrations (subinhibitory and inhibitory) with preformed biofilms. Consistent with the above findings of others, subinhibitory concentrations of gentamicin increased the biomass of biofilms formed with S. aureus RN 6390, while higher inhibitory concentrations also increased the biomass of the ATCC 25923 strain (Fig. 2). These results provide further evidence that some antibiotics, that now include gentamicin, can promote biofilm biomass. Unique findings from this study were that (a) the ability of gentamicin to induce biofilm biomass varied considerably with the bacterial strain, and (b) in addition to subinhibitory concentrations, inhibitory concentrations of gentamicin also augmented biofilm biomass.
SEMs of sutures incubated with S. aureus revealed robust biofilms localized on and between individual strands of braided silk suture. In this in vitro model, biofilm structure resulted from bacterial growth on a surface without the contribution of host factors. In control samples without gentamicin, biofilm structure generally consisted of clusters of cocci within fibrillar elements. Clusters of normal-appearing staphylococci were also present on gentamicin-treated sutures including those incubated with high drug concentrations, and this observation was consistent with the recovery of high numbers of viable bacteria from biofilms incubated with inhibitory concentrations of gentamicin. Suture-associated biofilms treated with the highest gentamicin concentrations (50 and 100 µg/ml) had a discreet clumps of material along the suture strands that appeared as clumps of ruffled matrix material at higher magnifications. This ruffled material contained hollow depressions suggesting that the circular spaces once held staphylococcal cells, and cocci were occasionally localized within this ruffled matrix. Thus, SEMS of suture-associated biofilms incubated with even high concentrations of gentamicin, revealed large clusters of intact staphylococci and an altered appearance of the extracellular polymeric matrix material.
No single mechanism is likely to explain the increased antimicrobial resistance of intact biofilms [9]. In addition to putative persister cells that are inherently antibiotic resistant, other mechanisms of antibiotic resistance in biofilms have been postulated to include increased expression of resistance genes, reduced bacterial growth rate, decreased antibiotic penetration of biofilm matrix (via biochemical methods or physical walling off), and bacterial expression of specific factors such as multidrug efflux pumps [1, 7, 8]. However, a major difference between mechanically dispersed biofilm cells (with MIC/MBCs comparable to planktonic cells) and bacteria within an intact biofilm is the presence of the extracellular matrix. Mechanically dispersed (sonicated) biofilm cells are likely freed of their extracellular matrix, so this matrix may only be present in an intact biofilm. Additional studies are needed to determine if the biofilm matrix interferes with antibiotic binding to its bacterial target, but if this is so, greater attention should be directed into designing agents that either interfere with matrix development or decrease the amount of extracellular matrix.
ACKNOWLEDGEMENT
This work was supported in part by U.S. National Institutes of Health Grant R01 GM095553 (to CW) and in part by funds from the Department of Surgery, University of Minnesota, Minneapolis (to DH). Parts of this work were carried out in the Institute of Technology Characterization Facility, University of Minnesota, which receives partial support from NSF through the MRSEC program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented at the 6th Annual Academic Surgical Congress (Association for Academic Surgery), Huntington Beach, CA, February 1–3, 2011
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nature Rev Microbiol. 2004;2:95. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 3.Fux CA, Costerton JW, Stewart PS, et al. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Flemming H-C, Neu TR, Wozniak DJ. The EPS matrix: The “house of biofilm cells”. J Bacteriol. 2007;189:7945. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klevens RM, Edwards JR, Richards CL, Jr, et al. Estimating health care-associated infections and deaths in U.S. hospitals 2002. Public Health Reports. 2007;122:160. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry-Stanley MJ, Hess DJ, Barnes AMT, et al. Bacterial contamination of surgical suture resembles a biofilm. Surg Infect. 2010;11:433. doi: 10.1089/sur.2010.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies D. Understanding biofilm resistance to antibacterial agents. Nature Reviews. Drug Discovery. 2003;2:114. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 8.Lewis K. Riddle of biofilm resistance. Antimicrob Agents Chemother. 2001;45:999. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsek MR, Fuqua C. Biofilms 2003: Emerging themes and challenges in studies of surface-associated microbial life. J Bacteriol. 2004;186:4427. doi: 10.1128/JB.186.14.4427-4440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI Guidelines. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 16th edition. Wayne, PA, USA: Approved Standard M100-S16. CLSI; 2006. [Google Scholar]
- 11.Lewis K. Persister cells, dormancy and infectious disease. Nature Rev Microbiol. 2007;5:48. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 12.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman LR, D’Argenio DA, MacCoss MJ, et al. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 14.Kirby JP JE, Mazuski JE. Prevention of surgical site infection. Surg Clinics No Amer. 2009;89:365. doi: 10.1016/j.suc.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 15.National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004;32:470. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- 16.Fux CA, Wilson S, Stoodley P. Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J Bacteriol. 2004;186:4486. doi: 10.1128/JB.186.14.4486-4491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shanks RMQ, Donegan NP, Graber ML, et al. Heparin stimulates Staphylococcus aureus biofilm formation. Infect Immun. 2005;73:4596. doi: 10.1128/IAI.73.8.4596-4606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peeters E, Nelis HJ, Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Meth. 2008;72:157. doi: 10.1016/j.mimet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Pantanella F, Valenti P, Frioni A, et al. BioTimer assay, a new method for counting Staphylococcus spp in biofilm without sample manipulation applied to evaluate antibiotic susceptibility of biofilm. J Microbiol Meth. 2008;75:478. doi: 10.1016/j.mimet.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 20.Erlandsen SL, Kristich CJ, Dunny GM, et al. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J Histochem Cytochem. 2004;52:1427. doi: 10.1369/jhc.4A6428.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spoering AL, Lewis K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol. 2001;183:6746. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nucleo E, Steffanoni L, Migilavacca R, et al. Growth in glucose-based medium and exposure to subinhibitory concentrations of imipenem induce biofilm formation in a multidrug-resistant clinical isolate of Acinetobacter baumannii. BMC Microbiol. 2009;9:270. doi: 10.1186/1471-2180-9-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majtan J, Majtanova L, Xu M, et al. In vitro effect of subinhibitory concentrations of antibiotics on biofilm formation by clinical strains of Salmonella enterica serovar Typhimurium isolated in Slovakia. J Appl Microbiol. 2008;104:1294. doi: 10.1111/j.1365-2672.2007.03653.x. [DOI] [PubMed] [Google Scholar]
- 24.Rachid S, Ohlsen K, Witte W, et al. Effect of subinhibitory concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:3357. doi: 10.1128/aac.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagge N, Schuster M, Hentzer M, et al. 2004. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob Agents Chemother. 2004;48:1175. doi: 10.1128/AAC.48.4.1175-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]