Aurora A and Aurora B have nonredundant functions during mitosis in chromosome segregation and anaphase microtubule dynamics.
Abstract
We established a conditional deletion of Aurora A kinase (AurA) in Cdk1 analogue-sensitive DT40 cells to analyze AurA knockout phenotypes after Cdk1 activation. In the absence of AurA, cells form bipolar spindles but fail to properly align their chromosomes and exit mitosis with segregation errors. The resulting daughter cells exhibit a variety of phenotypes and are highly aneuploid. Aurora B kinase (AurB)–inhibited cells show a similar chromosome alignment problem and cytokinesis defects, resulting in binucleate daughter cells. Conversely, cells lacking AurA and AurB activity exit mitosis without anaphase, forming polyploid daughter cells with a single nucleus. Strikingly, inhibition of both AurA and AurB results in a failure to depolymerize spindle microtubules (MTs) in anaphase after Cdk1 inactivation. These results suggest an essential combined function of AurA and AurB in chromosome segregation and anaphase MT dynamics.
Introduction
Accurate chromosome segregation is a prerequisite for the maintenance of genome integrity. To achieve this, chromosomes are captured during prometaphase and transported to the spindle equator. Once all chromosomes are aligned and correctly attached, the sister chromatids lose their cohesin links and are pulled apart to the opposite poles of the spindle. Several S/T kinases, including Cdk1, Polo-like kinase 1 (Plk1), Aurora A kinase (AurA), Aurora B kinase (AurB), and monopolar spindle 1 (Mps1), control mitotic progression (Hochegger et al., 2008; Takaki et al., 2008; Taylor and Peters, 2008). Cdk1 activation is essential for mitotic entry, whereas its inhibition is fundamental for anaphase onset (Sullivan and Morgan, 2007). Plk1-inhibited cells arrest in prometaphase with defects in spindle assembly and chromosome alignment (Lénárt et al., 2007). AurB inhibition allows mitotic progression with missegregated chromosomes and a cytokinesis defect (Piekorz, 2010). AurA has been intensively studied (Barr and Gergely, 2007), but its functions in mitosis remain unclear. In fly embryos and Xenopus laevis egg extracts, the hallmark phenotypes of AurA mutants are monopolar spindles (Glover et al., 1995; Sardon et al., 2008). In contrast, in studies using somatic cells, AurA deficiency can elicit a range of apparently contradictory phenotypes. Some studies showed a prolonged G2 arrest (Marumoto et al., 2002; Hirota et al., 2003), whereas others describe a predominant defect in mitosis with chromosome misalignment, cytokinesis failure, centrosome fragmentation, and multipolar or monopolar spindles (Kunitoku et al., 2003; Marumoto et al., 2003; De Luca et al., 2008; Cowley et al., 2009; Sloane et al., 2010). Interestingly, the AurA inhibitor MLN8054 induces chromosome misalignment and delays, but does not block, mitotic progression (Hoar et al., 2007; Scutt et al., 2009). The reported differences highlighted in these studies might be caused by incomplete AurA depletion or inhibition as well as differential effects of kinase inhibition versus protein removal. Thus, AurA functions in mitosis remain elusive. In this study, we develop a chemical genetic strategy to analyze AurA requirements in mitosis and define its genetic interactions with other mitotic kinases, including Cdk1, Plk1, Mps1, and AurB.
Results and discussion
Characterization of AurA conditional knockout cells
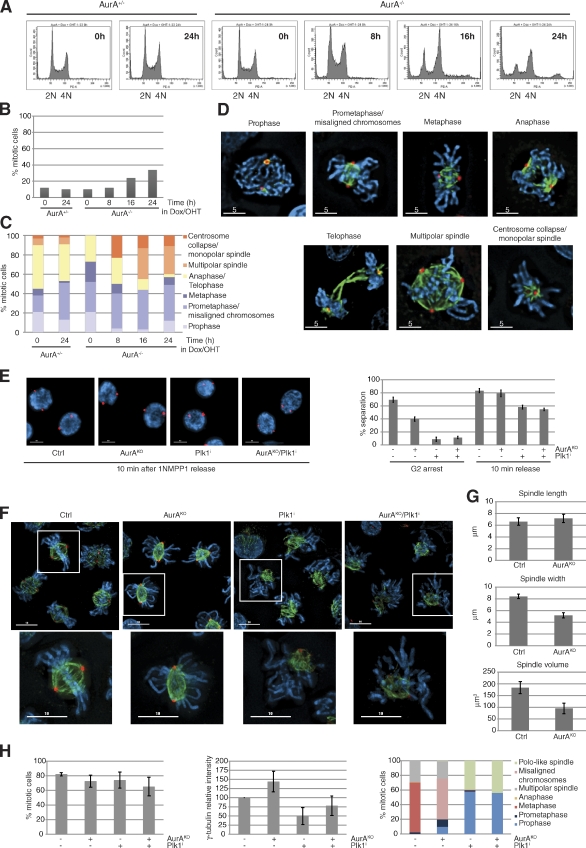
We established a conditional AurA deletion in DT40 cdk1as cells (Hochegger et al., 2007) by simultaneous transcriptional down-regulation and gene deletion (for a detailed description of the knockout strategy see Fig. S1, A and B). Upon AurA depletion, the cells stopped proliferating within 24 h (Fig. S1, C and D), accumulated with 4N DNA content, and initiated endoreplication (Fig. 1 A). However, we only detected a two- to threefold increase in mitotic index in the AurA-depleted cells (Fig. 1 B). These mitotic cells displayed a variety of phenotypes, including monopolar and multipolar spindles (Fig. 1, C and D). Precise dynamics of Cdk1 activation and mitotic entry and exit cannot be determined in these experiments because cells enter mitosis with variable levels of total AurA and remain arrested in mitosis for various periods of time. To circumvent this problem, we used reversible inhibition of analogue-sensitive Cdk1 (cdk1as) by 1NMPP1 to block cells in late G2 phase, with or without AurA, and then release them into mitosis (Fig. S1, E–G). Similar to previous results using Plk1 inhibitors (Smith et al., 2011), we found that centrosome separation could be triggered by Cdk1 independently of AurA and even after double inactivation of AurA and Plk1 (Fig. 1 E). After 1NMPP1 release, most control cells had reached metaphase within 30 min (Fig. 1 F). Both control and AurAKO cells showed a comparable subpopulation of multipolar spindles as a result of centrosome amplification in the 1NMPP1 arrest (Hochegger et al., 2007). However, most AurAKO cells displayed bipolar spindles but failed to align the chromosomes at the metaphase plate (Fig. 1 H, right). Recruitment of γ-tubulin to the centrosome and pole to pole distances were comparable between controls and AurAKO cells (Fig. 1, G and H), but spindle volume and width were significantly smaller (Fig. 1 G), suggesting differences in mitotic microtubule (MT) dynamics in the AurAKO cells. In these cells, the balance between MT depolymerization and polymerization may be altered compared with the control. AurA has been reported to positively control the MT-stabilizing protein complex TACC3–chTOG, protecting MTs from mitotic centromere-associated kinesin (MCAK)–induced MT destabilization (Barros et al., 2005). Moreover, AurA has been shown to inhibit another MT depolymerase, Kif2A, at the poles and to suppress MCAK activity in combination with AurB (Jang et al., 2009; Tanenbaum et al., 2011). We next analyzed the interdependence between Cdk1, AurA, AurB, and Plk1. We found that AurA is required for Plk1 activation in G2 (Fig. S2 A), consistent with previous studies (Macůrek et al., 2008; Seki et al., 2008), but not in M phase. Indeed, even double inactivation of AurA and AurB did not interfere with mitotic Plk1 activation (Fig. S2, A and D) and bipolar spindle formation (Fig. S2 B). In summary, we found that the activity of Plk1, AurA, and AurB was increased after Cdk1 activation but that these kinases appeared to act independently of each other in mitosis (Fig. S2, A–D). Moreover, simultaneous depletion of AurA and Plk1 inhibition showed similar phenotypes to Plk1 inhibition (Fig. 1, F and H), suggesting that the Plk1 phenotype is epistatic to AurA as previously suggested (Scutt et al., 2009).
Figure 1.
AurA-depleted cells exhibit substantial chromosome alignment defects in early mitosis. (A) FACS profiles of AurA+/− and AurA−/− cells incubated with doxycyclin (Dox)/4-hydroxytamoxifen (OHT). (B) Mitotic index (n ≥ 237 cells for each condition, three independent experiments). (C) Cells from B classified into different mitotic stages (n ≥ 29 cells for each condition). (D) Representative pictures of mitotic cells counted in C. (E) Centrosome separation analysis. Cells were synchronized in G2 with or without AurA according to the procedure described in Fig. S1 E, with or without the Plk1 inhibitor BI2536. Cells with separated centrosomes were scored in G2 and 10 min after 1NMPP1 washout. Representative pictures and quantifications are shown (n = 100 cells for each condition, three independent experiments). (F) Images of cells immunostained 30 min after 1NMPP1 release. The boxes in the larger images are further magnified below. (G) Spindle analysis. Pictures from F were processed by 3D rendering to determine the spindle pole distance, width, and volume (n ≥ 75 cells for each condition, three independent experiments). (H) Quantitative data of results in F. The mitotic index (left), γ-tubulin intensity measurements (middle), and quantification of mitotic phenotypes (right) are shown (n ≥ 75 cells for each condition, three independent experiments). γ-Tubulin (red), α-tubulin (green), and DNA (blue) are shown. Bars: (D) 5 µm; (E and F) 10 µm. Error bars indicate means ± SD. Ctrl, control; PE-A, phycoerythrin area.
AurAKO cells exit mitosis and are proficient in spindle checkpoint signaling
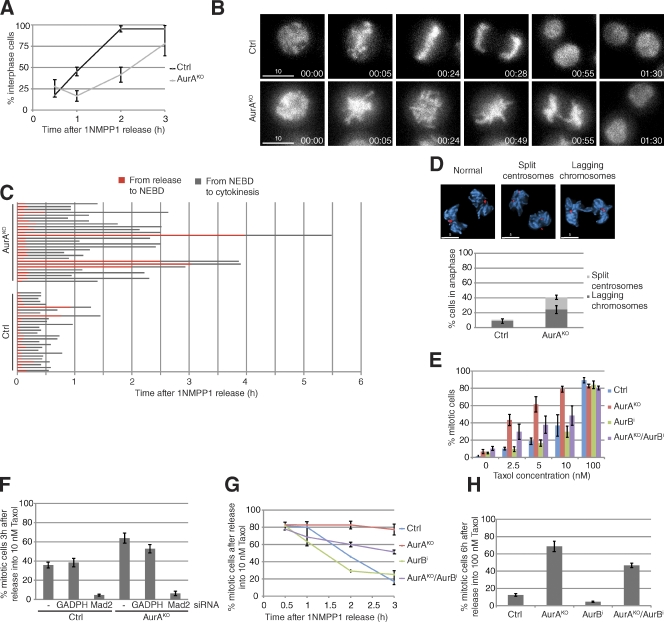
Despite the spindle and chromosome alignment defects, AurAKO cells were not arrested but only delayed in exiting mitosis (Fig. 2 A). Live-cell imaging of histone H2B-GFP–expressing AurAKO cells showed that these cells managed to initiate anaphase and chromosome segregation (Fig. 2, B and C). However, we observed a threefold increase in lagging chromosomes in AurAKO cells, suggestive of merotelic kinetochore attachments (Fig. 2 D; Cimini et al., 2001). AurAKO cells could exit from mitosis as a result of problems in spindle assembly checkpoint (SAC) maintenance or might satisfy the SAC because of sufficient kinetochore attachments to MTs. We analyzed SAC proficiency in AurAKO cells by releasing them into mitosis in the presence of increasing doses of taxol. Surprisingly, AurAKO cells were not only SAC proficient but were also highly sensitive to low doses of taxol when compared with controls (Fig. 2, E–G). This taxol sensitivity was strictly SAC dependent and abrogated after codepletion of the SAC regulator Mad2 (Fig. 2 F). SAC effectors, such as the AurB–INCENP complex and BubR1, were localized at the kinetochores in AurAKO cells (Fig. S2 E), and effective SAC execution in these cells was partially dependent on AurB activity (Fig. 2, E, G, and H). These data suggest that AurAKO cells exit mitosis as a result of recovery from the SAC rather than mitotic slippage.
Figure 2.
Mitotic progression in AurAKO cells. (A) Mitotic exit. Cells were released from 1NMPP1 and stained with DAPI at the indicated time points. The cells with decondensed chromosomes were scored as interphase cells (n ≥ 52 cells for each condition, three independent experiments). (B) Time-lapse sequences of H2B-GFP–expressing cells after 1NMPP1 release (n = 28 cells for each condition). Times are given in minutes. (C) Quantitative analysis of B. The time spent in the indicated phases was determined from live-cell images. (D) Anaphase defects. γ-Tubulin (red) and DNA (blue; n = 50 cells for each condition, three independent experiments) are shown. (E–H) SAC proficiency. Taxol was added after 1NMPP1 release at the indicated concentrations (E) or at 10 nM (F and G) or 100 nM (H). Cells were collected, and mitotic index was determined at 3 h (E and F), 6 h (H), or at the indicated time points (G). In F, cells were transfected with either glyceraldehyde 3-phosphate dehydrogenase (GAPDH) siRNA or Mad2 siRNA 24 h before the experiment (n = 100 cells for each condition, three independent experiments). Bars: (B) 10 µm; (D) 5 µm. Error bars indicate means ± SD. Ctrl, control; NEBD, nuclear envelope breakdown.
AurAKO cells exhibit defects in chromosome biorientation and display high levels of aneuploidy
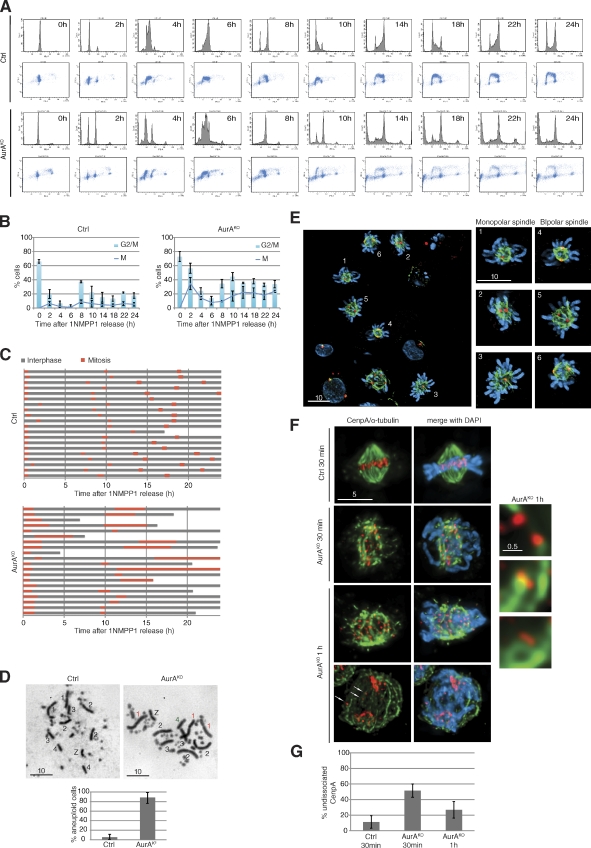
After mitotic exit, AurAKO cells passed through S phase and accumulated with a 4N DNA content at 8 h (Fig. 3 A). During the following 16 h, we noted an increase in the sub-G1 population (∼30% of the total cells). Most cells (∼50%) were blocked in either G2 or M phase (Fig. 3 B). When tracked by live-cell imaging (Fig. 3 C), AurAKO cells showed a variety of phenotypes after exiting the first mitosis. Some cells died in interphase, and some underwent one more cell division, arrested, or died in the next mitosis but never completed more than one additional cell division. AurAKO karyotype analysis after the first division (10 h after release) showed that 80% of the AurAKO cells displayed aberrant numbers of chromosomes 1–4 (Fig. 3 D and Table S1), suggesting high levels of aneuploidy. These changes in chromosome number are a likely cause for the observed spectrum of phenotypes in the next cell cycle. Consistently, we found a variety of defects in mitotic cells 10 h after release (Fig. 3 E). To gain mechanistic insight in the causes of aneuploidy, we investigated centromere biorientation in mitosis by monitoring CenpA on MT kinetochore fibers (Fig. 3 F). In control cells 30 min after 1NMPP1 release, most centromeres were biorientated. In contrast, AurAKO cells displayed a fivefold increase of syntelic and/or monotelic kinetochores as judged by undissociated CenpA. This was only partly corrected in the following 30 min (Fig. 3 G). We also found an increase from 10 to 25% in lagging kinetochores in AurAKO anaphase cells (Fig. 3 F, bottom, arrows).
Figure 3.
Phenotypes of AurAKO cells after mitotic exit. (A) Cell cycle profiles. Cells were synchronized as in Fig. S1 E and, after release in mitosis, were harvested at the indicated time points and colabeled with BrdU and PI. (B) Mitotic index. Mitotic cells were scored from cells in A (n ≥ 100 cells for each condition, three independent experiments). Results were combined to G2/M fraction determined from A (three independent experiments). (C) Phase succession. Time-lapse microscopy was performed for 24 h after 1NMPP1 release. Interphase and mitosis time lengths were scored (n = 20 cells for each condition). (D) Metaphase spreads 10 h after 1NMPP1 release. Chromosomes 1, 2, 3, 4, and Z are shown with extra chromosomes in red and missing chromosomes in green. Chart indicates quantification of aneuploidy (n ≥ 20 cells for each condition, three independent experiments). (E) Representative images of AurAKO cells at 10 h after 1NMPP1 release. γ-Tubulin (red), α-tubulin (green), and DNA (blue) are shown. (F) Kinetochore/MT attachment in mitotic AurAKO cells. HA-SNAP-CenpA–expressing cells were collected in mitosis at indicated time points after 1NMPP1 removal and cold treated before fixation. The right images show a close-up of kinetochore/MT attachment in AurAKO. HA-SNAP-CenpA (red) and α-tubulin (green) are shown. Arrows indicate missegregated centromeres in anaphase. (G) Quantitative analysis of undissociated CenpA observed in F (n = 20 cells for each condition). Bars: (E) 10 µm; (F, left) 5 µm; (F, right) 0.5 µm. Error bars indicate means ± SD. Ctrl, control.
These findings demonstrate that AurA is not required to build a bipolar spindle in chicken DT40 cells, and its deletion delays but does not halt mitotic progression. Cells lacking AurA exhibit reduced spindle volume and misaligned, maloriented chromosomes, resulting in massive aneuploidy. It is surprising that a kinase located at the centrosome has such a strong impact on subsequent chromosome alignment. One possible explanation for this phenotype in AurAKO cells could be a dysfunctional centrosomal regulation of CenpE by AurA (Kim et al., 2010). The strong centromere biorientation defect could also explain the increased taxol sensitivity of AurAKO cells. The correction of syntelic and monotelic centromeres is likely to involve localized MT rearrangements that are suppressed by taxol, resulting in increased unattached kinetochores. Collectively, our results present a concise genetic analysis of AurA and its interactions with other mitotic kinases that are summarized in Table 1.
Table 1.
Summary of kinases used in this study
| Kinases | Required for activation of | Main mitotic functions | Effects of deficiency |
| Cdk1 | AurA, Plk1, and AurB | G2/M transition | G2 arrest |
| Plk1 | N/A | Spindle formation | Prophase/prometaphase arrest |
| AurA | Plk1 in G2 | Spindle MT stability and chromosome alignment | Mitotic delay and aneuploidy |
| AurB | N/A | Chromosome alignment, SAC, and cytokinesis | Cytokinesis failure and binucleate cells |
| AurA/AurB | N/A | Spindle MT stability, chromosome segregation, and anaphase spindle disassembly | No chromosome disjunction and polyploidy |
N/A, not available.
AurA and AurB cooperate to coordinate chromosome segregation
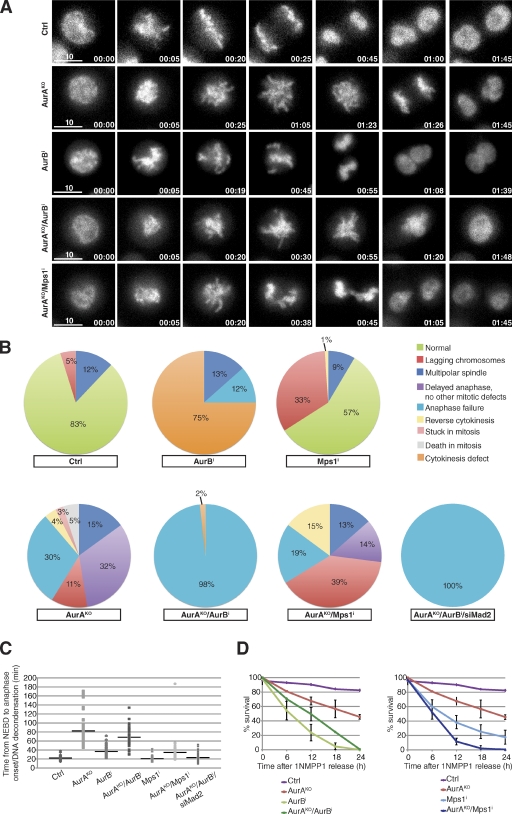
We undertook a detailed time-lapse microscopy analysis using histone H2B-GFP–expressing AurAKO cells to investigate the effects of double inhibition of AurA, AurB, and the SAC kinase Mps1 (Fig. 4). It took on average 82 min to complete mitosis in the absence of AurA compared with 22 min in control cells (Fig. 4, A–C; and Videos 1 and 2). Inhibition of AurB caused a shorter delay (mean length of mitosis was 40 min) and a distinct cytokinesis failure (Video 3). Strikingly, absence of AurB activity in AurAKO cells caused complete inhibition of chromosome segregation (Fig. 4, A and B; and Video 4). In these cells, mitosis was shortened (∼68 min), and chromosomes decondensed without segregation. This failure to undergo anaphase could be a result of the SAC function of AurB. However, inhibition of another SAC kinase, Mps1 (Fig. 4, B and C; and Video 5), or codepletion of Mad2 in AurAKO cells (not depicted) did not interfere with anaphase onset but strongly reduced the time to complete mitosis and increased the number of lagging chromosomes. Likewise, checkpoint inactivation by codepletion of Mad2 shortened M-phase duration in AurAKO/AurB-inhibited cells without altering the chromosome segregation defect (Fig. 4, B and C; and Video 6). We found that inactivation of both Mps1 and AurA was the most effective way to kill cells after mitotic exit (Fig. 4 D). These synergistic effects between AurA inactivation and Mps1 inhibitors or low dose taxol treatments could represent therapeutic opportunities for a combinatorial treatment of cancer cells.
Figure 4.
Chromosome segregation failure in AurAKO/AurB-inhibited cells. (A) Time-lapse microscopy of H2B-GFP–expressing cells after 1NMPP1 release. Shown are still images of cells with the indicated treatments. AurB was inhibited with 60 nM AZD1152-HQPA and Mps1 with 500 nM reversine. Times are given in minutes. Bars, 10 µm. (B) Quantitative analysis of mitotic defects observed in A (n ≥ 57 cells for each condition). In addition, AurAKO and AurB-inhibited cells were transfected with Mad2 siRNA (siMad2) 24 h before the experiment. (C) Duration of mitosis (n ≥ 48 cells for each condition). (D) Cell survival analysis of cells released from 1NMPP1 and treated as indicated. Cell survival was followed for 24 h by long-term live-cell imaging and analyzed every 6 h (n ≥ 20 cells for each video, three different videos). The horizontal lines represent the mean values for each data series. Error bars indicate means ± SD. Ctrl, control; NEBD, nuclear envelope breakdown.
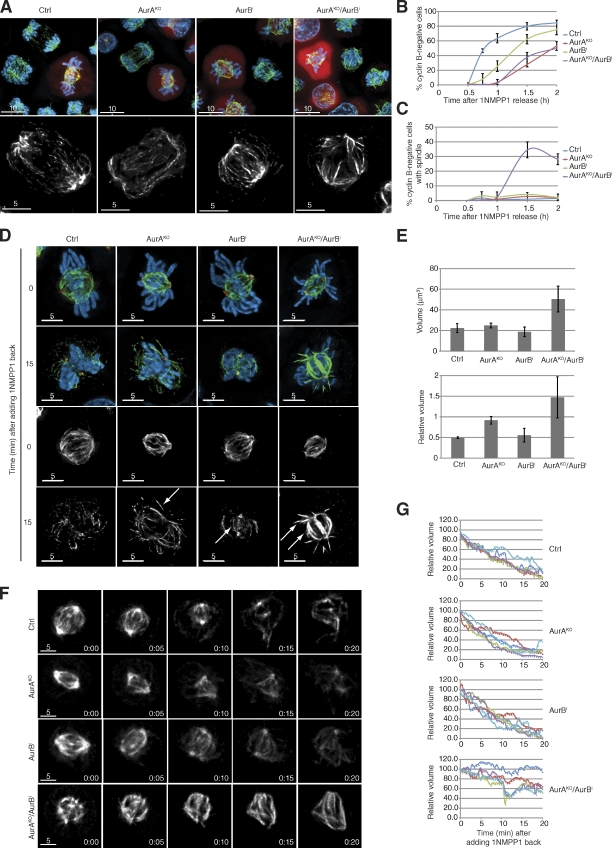
AurA and AurB control MT depolymerization at anaphase onset
To investigate the combined functions of AurA and B in anaphase, we tested the dynamics of cyclin B destruction (Fig. 5, A and B). AurAKO/AurB-inhibited cells triggered cyclin B proteolysis with the same kinetics as AurAKO cells. However, we noted the persistence of long and apparently stable MT fibers in cells lacking both AurA and AurB, whereas all control cells showed the typical anaphase spindle contraction (Fig. 5, A and C). One of the current models of anaphase force generation implies that chromosome motion is induced by MT destabilization (Maiato and Lince-Faria, 2010; Rath and Sharp, 2011). A likely trigger for this is the anaphase-promoting complex/cyclosome-mediated inactivation of Cdk1, but the precise control mechanism of this process is not understood. To further investigate these anaphase spindle dynamics, we took advantage of the cdk1as mutation to mimic mitotic exit by 1NMPP1-mediated inactivation of Cdk1 in metaphase cells. This was sufficient to trigger rapid dephosphorylation of Cdk1 substrates in control and Aurora kinase single and double inactivated cells (Fig. S2 F). We also observed a rapid spindle MT depolymerization after Cdk1 inactivation in control cells and AurB-inhibited cells (Fig. 5 D). Astral MTs appeared to be partially stable in AurAKO cells (Fig. 5 D, arrows), but the entire spindle was significantly stabilized in AurAKO/AurB-inhibited cells (Fig. 5, D and E). We also found a strong spindle stabilization effect in AurAKO/AurB-inhibited cells using 4D live-cell imaging after Cdk1 inhibition compared with controls (Fig. 5, F and G; and Videos 7, 8, 9, and 10). Collectively, these results strongly suggest a role of AurA and AurB in controlling spindle disassembly after Cdk1 inactivation at anaphase onset.
Figure 5.
MT depolymerization defect in AurAKO and AurB-inhibited cells. (A) Mitotic exit. Representative images of mitotic cells before and after cyclin B degradation. Cyclin B (red), α-tubulin (green), and DNA (blue) are shown. (B and C) Quantitative analysis of cyclin B–negative cells (B) and stable spindles in cyclin B–negative cells (C; n ≥ 100 cells for each condition, three independent experiments). (D and E) Analysis of MT depolymerization. 30 min after 1NMPP1 release, MT depolymerization was induced by adding back 1NMPP1. Cells were treated as indicated and collected before 1NMPP1 addition (0) and 15 min later (15). Representative pictures are shown in D. γ-Tubulin (red), α-tubulin (green or gray), and DNA (blue) are shown. Intensity scale of the FITC channel was kept the same between 0 and 15 min for all images. Arrows indicate stable MT populations. In E, quantitative data from D are shown. We measured the volume of polar MTs by 3D rendering. Quantifications of absolute volume at 15 min after 1NMPP1 treatment and relative volume referenced to spindles before 1NMPP1 addition are shown (n = 10 cells for each condition, three independent experiments). (F) Time-lapse microscopy of mCherry–α-tubulin–expressing cells after adding 1NMPP1 back. Shown are still images of cells with the indicated treatments. Intensity scales were kept constant in all images for each time course. (G) Quantitative data from F. The spindle volume was measured over time by 3D rendering. The volume at 0 min was used as a reference, and relative volumes were scored over time. Each curve represents a single cell (n = 5 cells for each condition). Bars: (A, top) 10 µm; (A [bottom], D, and F) 5 µm. Error bars indicate means ± SD. Ctrl, control.
Recent studies suggest that anaphase A requires both MT destabilization at the spindle poles (the Flux model) and at the kinetochores (the Pacman model; Rogers et al., 2004; Manning et al., 2007; Zhang et al., 2007). Given that AurA and AurB are localized to the centrosomes and the kinetochores, respectively, it is tempting to speculate that these kinases could regulate Flux and Pacman activities after Cdk1 inactivation. The precise mechanism of Pacman–Flux is unknown, but a combined action of MT-severing enzymes and MT depolymerases has been proposed (Rath and Sharp, 2011). Kinesin-13 MT depolymerases, such as Kif2A, Kif2B, and MCAK, are logical targets of the Aurora kinases in this pathway. However, Aurora kinases have been shown to inhibit rather than activate these enzymes (Lan et al., 2004; Zhang et al., 2008; Jang et al., 2009; Knowlton et al., 2009; Tanenbaum et al., 2011). Aurora kinases may thus be responsible for both spindle stability in metaphase and spindle disassembly in anaphase. Another potential group of targets in this pathway are MT-severing enzymes of the class II AAA ATPase family, such as Katanin, Spastin, and Fidgetin (Roll-Mecak and McNally, 2010). These proteins are localized to both centrosomes and kinetochores and have been implicated in the Pacman–Flux pathway in Drosophila melanogaster cells (Zhang et al., 2007). However, their regulation by mitotic kinases has not been closely evaluated. This anaphase role for Aurora kinases appears to be conserved because budding yeast Ipl1 has also been reported to contribute to spindle disassembly during mitotic exit (Buvelot et al., 2003; Zimniak et al., 2009; Woodruff et al., 2010). Based on these results, further work will be necessary to unravel the control of anaphase A spindle dynamics.
Materials and methods
Generation of conditional AurA knockout in DT40 cells
A conditional deletion of AurA was established in DT40 cdk1as cells (Hochegger et al., 2007). Using the gene-targeting construct shown in Fig. S1 A, we replaced exons 1–4 of the first allele with the puromycin resistance gene floxed by loxP sites. The cdk1as DT40 cell line stably expressed tamoxifen-inducible Cre recombinase; therefore, the puromycin resistance gene was removed by 4-hydroxytamoxifen–induced Cre recombinase activity. We next introduced human Flag-AurA cDNA (a gift from T. Hirota, The Cancer Institute, Japanese Foundation for Cancer Research, Tokyo, Japan) cloned in a pTre2-loxP-ires-luciferase plasmid (Wakasugi et al., 2007) to express the cDNA from a tetracycline-repressible cytomegalovirus promoter. For repressible expression of AurA in the obtained heterozygotes, the construct encoding the tetracycline-controlled transactivator and the human Flag-AurA pTre2 vector were cotransfected with a puromycin resistance gene expression construct. A stably expressing cell line was isolated, and the second AurA allele was deleted. This targeting strategy was confirmed by Southern blotting using EcoRV digestion (Fig. S1 B). In the AurA+/− cell line, a specific probe recognized a 7-kb band for the wild-type allele and a 6-kb band for the targeted allele after puromycin depletion. The second wild-type allele was targeted with the histidinol resistance gene, and a new 9.4-kb band was detected by Southern blotting in the AurAKO cell line. Note that in our construct, human Flag-AurA cDNA was flanked by loxP sites; therefore, the depletion of the gene can be controlled by both 4-hydroxytamoxifen and doxycyclin.
Cell culture, synchronization, and inhibitor treatments
Chicken DT40 mutant cells were cultured in RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum, 5% chicken serum, 2 mM l-glutamine, 0.1% β-mercaptoethanol, and antibiotics. These cells were incubated at 39°C in a humidified cell culture chamber with 5% CO2. G2-synchronized AurAKO cells were treated as indicated in Fig. S1 E. In brief, cells were incubated with 1 µg/ml doxycyclin and 0.5 µM 4-hydroxytamoxifen for 10–12 h followed by 6–8 h with the addition of 10 µM 1NMPP1 (Smith et al., 2011). To inhibit Plk1, 100 nM BI2536 (Axon Medchem) was added to the media for 6 h. To inhibit AurB, 60 nM AZD1152-HQPA (Selleck Chemicals, LLC) was added in the last 2 h. Cells were synchronized in mitosis by washing out 1NMPP1. 500 nM reversine (Cambridge Bioscience) was added to the media to inhibit Mps1. To deplete Mad2, 8 × 105 cells were transfected with 300 pmol MAD2 siRNA (QIAGEN) with DNA sequence 5′-TACCACGATTCACAAAGTAAA-3′ using the Neon system (Invitrogen). An siRNA-targeting glyceraldehyde 3-phosphate dehydrogenase was used as a negative control. 1,400-V, 10-ms, and 3-pulse electroporation conditions were used.
Immunostaining
For mitotic index, cells (∼5 × 104 per 0.1 ml) were spun onto slides at 1,000 rpm for 3 min then fixed with 3.7% formaldehyde (Sigma-Aldrich) in PBS for 10 min. After several PBS washes, cells were stained and mounted with DAPI solution (ProLong Gold; Invitrogen). Cells with condensed DNA were scored as mitotic.
For immunofluorescence, cells were spun onto slides at 1,000 rpm for 3 min and then fixed with 3.7% formaldehyde in PBS for 10 min. Cells were permeabilized in PBS–0.1% NP-40 for 10 min. Cells were then blocked in 1% BSA for 30 min and probed with primary antibodies for 45 min. Slides were rinsed in PBS and probed with Alexa Fluor secondary antibodies (Invitrogen) for 45 min. Slides were then rinsed in PBS, and coverslips were mounted using ProLong Gold mounting solution containing DAPI. γ-Tubulin rabbit polyclonal and α-tubulin mouse monoclonal antibodies were purchased from Abcam. Anti–chicken cyclin B2 rabbit polyclonal antibody (Gallant and Nigg, 1994) was a gift from E. Nigg (Biozentrum, University of Basel, Basel, Switzerland). Phosphorylated T232 AurB antibody was as previously described (Tyler et al., 2007), and BubR1 and INCENP rabbit polyclonal antibodies were gifts from W. Earnshaw (Wellcome Trust Centre for Cell Biology, University of Edinburgh, Edinburgh, Scotland, UK).
Images were acquired on microscope (DeltaVision) equipped with a UPLS Apochromat NA 1.40, 100× oil immersion objective (Olympus), standard filter sets (excitation 360/40, 490/20, and 555/28; emission 457/50, 528/38, and 617/40), and a camera (CoolSNAP HQ2; Photometrics). Z series of 0.3-µm stacks were acquired using softWoRx software (version 4.0.0; Applied Precision), and deconvolution was performed using Huygens Professional Deconvolution Software (version 3.5; Scientific Volume Imaging). Maximum intensity projections were obtained in softWoRx software (version 3.7.1) and exported as Photoshop (Adobe) files.
For quantitative data on spindle morphology, DeltaVision files were imported into Imaris software (version 6.3.0; Bitplane) for 3D rendering measurements using the volume rendering algorithm for α-tubulin signals. Measurements were then exported to Excel (Microsoft) and plotted. γ-Tubulin intensity on centrosomes was determined using ImageJ (version 1.44i; National Institutes of Health) with the plugin Time Series Analyzer (version 2.0; National Institutes of Health). DeltaVision files were imported into ImageJ. A rectangle (6 × 6 pixels) delimiting the centrosome was defined, and γ-tubulin intensities through all sections were summed. The same measurement was performed outside the centrosome to determine the background. Measurements were then exported to Excel software. For each experimental condition, a mean was calculated from γ-tubulin intensity values obtained after background subtraction. Relative intensities were determined as a ratio relative to intensities measured in control cells.
Live-cell imaging
AurAKO cells were stably transfected with histone H2B-GFP (a gift from S. Geley, Medical University Innsbruck, Innsbruck, Austria). Note that these cells already express GFP (Hochegger et al., 2007), so that both the entire cell as well as DNA can be visualized. Time-lapse microscopy was performed on coverglass chambers (Thermo Fisher Scientific) in CO2-independent medium (Invitrogen) in an environmental chamber heated to 39°C using a personal microscope (DeltaVision) and a UPLS Apochromat NA 1.40, 100× oil immersion objective with EGFP filters (excitation 470/40; emission 520/40). One single plane was taken at 1-min intervals for 4 h using 10% neutral density filters and a 50× gain on the EM charge-coupled device camera. The 3D time series was exported into QuickTime (Apple) format for presentation as supplementary videos. For long-term live-cell imaging, the same microscope camera set up was used with a 60× 1.4 NA lens and 2 × 2 binning. Cells were grown in RPMI 1640 growth medium supplemented with 20% FCS in an environmental chamber (Okolab H201-MEC-SLIM-Universal Micro; Warner Instruments) with 5% CO2 supply.
MT depolymerization in live cells was investigated in AurAKO cells transfected with mCherry–α-tubulin (a gift from S. Geley). 8 × 105 cells were transfected using Neon system with 4 µg mCherry–α-tubulin expression vector. The plasmid was purified using Endofree Plasmid Maxi kit (QIAGEN). After 1NMPP1 add back, z series of 0.3-µm stacks were acquired at 30-s intervals for 20 min. Maximum intensity projections were obtained in softWoRx software and exported into QuickTime format for presentation as supplementary videos. To obtain volume data, the 3D time series was imported into Imaris software. Volume rendering was quantified by filtering all volumes >5,000 µm3 and >2,500 maximum intensity (scoring absolute intensity values). The volume thus represents the total number of polymerized spindle MTs per cell. Relative volumes were determined as a ratio relative to volume measured at 0 min.
FACS analysis
Cells were spun down, washed once with PBS, and then fixed using 70% ethanol. Cells were centrifuged at 1,500 rpm for 3 min, rinsed twice with PBS containing 3% BSA (BSA/PBS), and resuspended in BSA/PBS solution containing 150 µg/ml RNase A and 5 µg/ml propidium iodide (PI). Cells were then analyzed for DNA content using a flow cytometer (FACSCanto; BD) and FACSDiva software (BD) to plot PI area versus cell counts.
To visualize replicating cells, 25 µM BrdU (Sigma-Aldrich) was added to the media for 20 min, and then cells were collected and fixed as described in the previous paragraph. To stain BrdU-labeled DNA, cells were washed once with BSA/PBS and then incubated with 2 M HCl for 30 min. Next, cells were washes three times with BSA/PBS and incubated with BrdU mouse monoclonal antibody (BD) for 1 h. Cells were then rinsed with PBS/BSA and incubated with Alexa Fluor 488 anti–mouse antibody (Invitrogen). PI staining was performed as in the previous paragraph. BrdU staining intensity was plotted against PI staining.
Metaphase spreads
Approximately 5 × 106 cells were pulsed with 0.1 µg/ml colcemid (Sigma-Aldrich) for 2 h, centrifuged at 1,000 rpm for 5 min, and then swollen in 1 ml of 75-mM KCl for 30 min and fixed in 5 ml of freshly prepared fixative (methanol/acetic acid [3:1]). Next, cells were pelleted and resuspended in 5 ml of fixative for at least 30 min. Cells were pelleted again then resuspended in 100–200 µl of fixative. The cell suspension was dropped onto a slide glass prewetted with ethanol and directly flame dried. The slides were air dried overnight followed by staining with 3% Giemsa diluted in 50 mM phosphate buffer, pH 6.4. Images were acquired on a microscope (Axioplan 2; Carl Zeiss) equipped with a camera (ORCA-ER; Hamamatsu Photonics). Images were acquired with a Plan Apochromat NA 1.40, 100× oil immersion objective (Carl Zeiss) using Simple PCI software (version 5.3.0; Compix, Inc. Imaging Systems).
SNAP labeling and cold treatment
Conditional AurA knockout cells were stably transfected with HA-SNAP-CenpA (a gift from L. Jansen, Instituto Gulbenkian de Ciência, Oeiras, Portugal). Within the last hour of 1NMPP1 incubation, cells were pulse labeled with 5 µM TMR-Star (New England Biolabs, Inc.) for 30 min and then washed twice with warm medium containing 10 µM 1NMPP1 and doxycyclin/4-hydroxytamoxifen if required. Cells were reincubated for 30 min to remove unreacted substrate. Cells were released into mitosis after 1NMPP1 removal. Next, cells were deposited on concanavalin-coated coverslips and incubated on ice for 10 min at the required time points. Cells were then fixed for 5 min in PHEM (60 mM Pipes, 25 mM Hepes KOH, pH 7.0, 5 mM EGTA, 4 mM MgSO4, 0.5% NP-40, and 3.7% formaldehyde) followed by 5 min in 95% methanol plus 5 mM EGTA. Immunofluorescence was performed, and images were acquired on a microscope (DeltaVision), deconvolved, and analyzed using Imaris software. An undissociated CenpA and a biorientated CenpA were scored in each cell through z stacks using ImageJ software.
Immunoblotting
Western blotting was performed as previously described (Smith et al., 2011). Primary antibodies were purchased from Abcam and were used at the manufacturer’s recommended concentrations. T288 AurA rabbit monoclonal and panphospho-(Ser) Cdk substrate rabbit polyclonal antibody were bought from Cell Signaling Technology, and S10 histone H3 was obtained from Millipore. HRP-conjugated and polyclonal goat anti–rabbit or –mouse antibodies were purchased from Dako.
Kinase assays
AurAKO cells were transfected with Myc-Plk1 expressed in pcDNA5/Flip recombination target vector using the Nucleofector kit (Lonza). 24 h later, cells were treated with 10 µM 1NMPP1 with or without 60 nM AZD1152-HQPA. Cells were released from 1NMPP1 by washout and collected 30 min later. For immunoprecipitation (IP), 106 cells were lysed in 500 µl IP lysis buffer (20 mM Tris, pH 7.5, 137 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA, 0.05% β-mercaptoethanol, and protease and phosphatase inhibitors [Roche]). Cells were sonicated; cell debris was then cleared by centrifugation at 13,000 rpm for 10 min at 4°C, and the supernatants were transferred to fresh tubes. Protein concentrations were determined by the Bradford method, and lysates were equalized for protein concentration using IP buffer. Lysates were incubated with 3–5 µg Myc antibody at 4°C for 1 h with end over end rotation, and 20 µl protein G Dynabeads (Invitrogen) was added to precipitated proteins. Samples were incubated at 4°C for a further 2 h. Beads were then rinsed with 3× 1 ml IP lysis buffer. Beads were then resuspended in SDS-PAGE sample buffer (12.5 mM Tris-HCl, pH 6.8, 1.4% [wt/vol] SDS, 4% [wt/vol] sucrose, 0.002% [wt/vol] bromophenol blue, and 0.4 mM β-mercaptoethanol) or prepared for kinase assays. For Plk1 kinase assays, immunoprecipitates were washed once in 1 ml kinase assay buffer (50 mM MOPS, pH 7.5, 5 mM MgCl2, 0.4 mM EGTA, 2 mM EDTA, and protease/phosphatase inhibitors). Beads were then resuspended in 20 µl kinase assay buffer. To each reaction, 2 µg casein protein (Sigma-Aldrich) was added, and to start reactions, 10 µl of 100-µM ATP including 0.1 MBq γ-[32P]ATP (made up in kinase assay buffer) was added. Reactions were incubated at 37°C for 20 min and were terminated with the addition of 15 µl 5× SDS-PAGE sample buffer and boiling at 95°C for 5 min.
Online supplemental material
Fig. S1 shows the strategy and the efficiency of AurA depletion in DT40 cells. Fig. S2 shows kinase activation in G2 and M phases, the recruitment of SAC proteins, and the efficiency of Cdk1 inhibition in mitotic cells. Table S1 shows chromosome counting from metaphase spreads performed in control and AurAKO cells. Videos 1–6 show control and AurAKO cells progressing in mitosis in the absence (Videos 1 and 2) or in the presence of an AurB inhibitor (Videos 3 and 4) or a Mps1 inhibitor (Videos 5 and 6). Videos 7–10 show MT depolymerization in control and AurAKO in absence (Videos 7 and 8) or in presence of AurB inhibitor (Videos 9 and 10). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201105058/DC1.
Acknowledgments
We would like to thank Bill Earnshaw, Toru Hirota, Stephan Geley, Lars Jansen, and Erich Nigg for contributing reagents.
H. Hochegger and N. Hégarat were funded by a Wellcome Trust Career Development Fellowship (082267/Z/07/Z). E. Smith was funded by a Cancer Research project grant (C28206, A9057).
Footnotes
Abbreviations used in this paper:
- AurA
- Aurora A kinase
- AurB
- Aurora B kinase
- DIC
- differential interference contrast
- IP
- immunoprecipitation
- MCAK
- mitotic centromere-associated kinesin
- Mps1
- monopolar spindle 1
- MT
- microtubule
- PI
- propidium iodide
- Plk1
- Polo-like kinase 1
- SAC
- spindle assembly checkpoint
References
- Barr A.R., Gergely F. 2007. Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120:2987–2996 10.1242/jcs.013136 [DOI] [PubMed] [Google Scholar]
- Barros T.P., Kinoshita K., Hyman A.A., Raff J.W. 2005. Aurora A activates D-TACC–Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J. Cell Biol. 170:1039–1046 10.1083/jcb.200504097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S.Y., Vermaak D., Biggins S. 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160:329–339 10.1083/jcb.200209018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D., Howell B., Maddox P., Khodjakov A., Degrassi F., Salmon E.D. 2001. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 153:517–527 10.1083/jcb.153.3.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley D.O., Rivera-Pérez J.A., Schliekelman M., He Y.J., Oliver T.G., Lu L., O’Quinn R., Salmon E.D., Magnuson T., Van Dyke T. 2009. Aurora-A kinase is essential for bipolar spindle formation and early development. Mol. Cell. Biol. 29:1059–1071 10.1128/MCB.01062-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M., Brunetto L., Asteriti I.A., Giubettini M., Lavia P., Guarguaglini G. 2008. Aurora-A and ch-TOG act in a common pathway in control of spindle pole integrity. Oncogene. 27:6539–6549 10.1038/onc.2008.252 [DOI] [PubMed] [Google Scholar]
- Gallant P., Nigg E.A. 1994. Identification of a novel vertebrate cyclin: cyclin B3 shares properties with both A- and B-type cyclins. EMBO J. 13:595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D.M., Leibowitz M.H., McLean D.A., Parry H. 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 81:95–105 10.1016/0092-8674(95)90374-7 [DOI] [PubMed] [Google Scholar]
- Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., Saya H. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 114:585–598 10.1016/S0092-8674(03)00642-1 [DOI] [PubMed] [Google Scholar]
- Hoar K., Chakravarty A., Rabino C., Wysong D., Bowman D., Roy N., Ecsedy J.A. 2007. MLN8054, a small-molecule inhibitor of Aurora A, causes spindle pole and chromosome congression defects leading to aneuploidy. Mol. Cell. Biol. 27:4513–4525 10.1128/MCB.02364-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger H., Dejsuphong D., Sonoda E., Saberi A., Rajendra E., Kirk J., Hunt T., Takeda S. 2007. An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. J. Cell Biol. 178:257–268 10.1083/jcb.200702034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger H., Takeda S., Hunt T. 2008. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat. Rev. Mol. Cell Biol. 9:910–916 10.1038/nrm2510 [DOI] [PubMed] [Google Scholar]
- Jang C.Y., Coppinger J.A., Seki A., Yates J.R., III, Fang G. 2009. Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J. Cell Sci. 122:1334–1341 10.1242/jcs.044321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Holland A.J., Lan W., Cleveland D.W. 2010. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 142:444–455 10.1016/j.cell.2010.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton A.L., Vorozhko V.V., Lan W., Gorbsky G.J., Stukenberg P.T. 2009. ICIS and Aurora B coregulate the microtubule depolymerase Kif2a. Curr. Biol. 19:758–763 10.1016/j.cub.2009.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitoku N., Sasayama T., Marumoto T., Zhang D., Honda S., Kobayashi O., Hatakeyama K., Ushio Y., Saya H., Hirota T. 2003. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev. Cell. 5:853–864 10.1016/S1534-5807(03)00364-2 [DOI] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., Stukenberg P.T. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286 [DOI] [PubMed] [Google Scholar]
- Lénárt P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J.J., Hoffmann M., Rettig W.J., Kraut N., Peters J.M. 2007. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17:304–315 10.1016/j.cub.2006.12.046 [DOI] [PubMed] [Google Scholar]
- Macůrek L., Lindqvist A., Lim D., Lampson M.A., Klompmaker R., Freire R., Clouin C., Taylor S.S., Yaffe M.B., Medema R.H. 2008. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 455:119–123 10.1038/nature07185 [DOI] [PubMed] [Google Scholar]
- Maiato H., Lince-Faria M. 2010. The perpetual movements of anaphase. Cell. Mol. Life Sci. 67:2251–2269 10.1007/s00018-010-0327-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A.L., Ganem N.J., Bakhoum S.F., Wagenbach M., Wordeman L., Compton D.A. 2007. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol. Biol. Cell. 18:2970–2979 10.1091/mbc.E07-02-0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T., Hirota T., Morisaki T., Kunitoku N., Zhang D., Ichikawa Y., Sasayama T., Kuninaka S., Mimori T., Tamaki N., et al. 2002. Roles of aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes Cells. 7:1173–1182 10.1046/j.1365-2443.2002.00592.x [DOI] [PubMed] [Google Scholar]
- Marumoto T., Honda S., Hara T., Nitta M., Hirota T., Kohmura E., Saya H. 2003. Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J. Biol. Chem. 278:51786–51795 10.1074/jbc.M306275200 [DOI] [PubMed] [Google Scholar]
- Piekorz R.P. 2010. Dissecting the role of INCENP-Aurora B in spindle assembly checkpoint function, chromosomal alignment and cytokinesis. Cell Cycle. 9:1678–1679 10.4161/cc.9.9.11514 [DOI] [PubMed] [Google Scholar]
- Rath U., Sharp D.J. 2011. The molecular basis of anaphase A in animal cells. Chromosome Res. 19:423–432 10.1007/s10577-011-9199-2 [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rogers S.L., Schwimmer T.A., Ems-McClung S.C., Walczak C.E., Vale R.D., Scholey J.M., Sharp D.J. 2004. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 427:364–370 10.1038/nature02256 [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., McNally F.J. 2010. Microtubule-severing enzymes. Curr. Opin. Cell Biol. 22:96–103 10.1016/j.ceb.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardon T., Peset I., Petrova B., Vernos I. 2008. Dissecting the role of Aurora A during spindle assembly. EMBO J. 27:2567–2579 10.1038/emboj.2008.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutt P.J., Chu M.L., Sloane D.A., Cherry M., Bignell C.R., Williams D.H., Eyers P.A. 2009. Discovery and exploitation of inhibitor-resistant aurora and polo kinase mutants for the analysis of mitotic networks. J. Biol. Chem. 284:15880–15893 10.1074/jbc.M109.005694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki A., Coppinger J.A., Jang C.Y., Yates J.R., Fang G. 2008. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 320:1655–1658 10.1126/science.1157425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloane D.A., Trikic M.Z., Chu M.L., Lamers M.B., Mason C.S., Mueller I., Savory W.J., Williams D.H., Eyers P.A. 2010. Drug-resistant aurora A mutants for cellular target validation of the small molecule kinase inhibitors MLN8054 and MLN8237. ACS Chem. Biol. 5:563–576 10.1021/cb100053q [DOI] [PubMed] [Google Scholar]
- Smith E., Hégarat N., Vesely C., Roseboom I., Larch C., Streicher H., Straatman K., Flynn H., Skehel M., Hirota T., et al. 2011. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J. 30:2233–2245 10.1038/emboj.2011.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Morgan D.O. 2007. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 8:894–903 10.1038/nrm2276 [DOI] [PubMed] [Google Scholar]
- Takaki T., Trenz K., Costanzo V., Petronczki M. 2008. Polo-like kinase 1 reaches beyond mitosis—cytokinesis, DNA damage response, and development. Curr. Opin. Cell Biol. 20:650–660 10.1016/j.ceb.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M.E., Macurek L., van der Vaart B., Galli M., Akhmanova A., Medema R.H. 2011. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr. Biol. 21:1356–1365 10.1016/j.cub.2011.07.017 [DOI] [PubMed] [Google Scholar]
- Taylor S., Peters J.M. 2008. Polo and Aurora kinases: lessons derived from chemical biology. Curr. Opin. Cell Biol. 20:77–84 10.1016/j.ceb.2007.11.008 [DOI] [PubMed] [Google Scholar]
- Tyler R.K., Shpiro N., Marquez R., Eyers P.A. 2007. VX-680 inhibits Aurora A and Aurora B kinase activity in human cells. Cell Cycle. 6:2846–2854 10.4161/cc.6.22.4940 [DOI] [PubMed] [Google Scholar]
- Wakasugi M., Matsuura K., Nagasawa A., Fu D., Shimizu H., Yamamoto K., Takeda S., Matsunaga T. 2007. DDB1 gene disruption causes a severe growth defect and apoptosis in chicken DT40 cells. Biochem. Biophys. Res. Commun. 364:771–777 10.1016/j.bbrc.2007.10.063 [DOI] [PubMed] [Google Scholar]
- Woodruff J.B., Drubin D.G., Barnes G. 2010. Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. J. Cell Biol. 191:795–808 10.1083/jcb.201006028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Rogers G.C., Buster D.W., Sharp D.J. 2007. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 177:231–242 10.1083/jcb.200612011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ems-McClung S.C., Walczak C.E. 2008. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol. Biol. Cell. 19:2752–2765 10.1091/mbc.E08-02-0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T., Stengl K., Mechtler K., Westermann S. 2009. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186:379–391 10.1083/jcb.200901036 [DOI] [PMC free article] [PubMed] [Google Scholar]