Formation of a Protocadherin-19–N-cadherin complex facilitates adhesion by Protocadherin-19 while suppressing homophilic interactions by N-cadherin.
Abstract
During embryonic morphogenesis, adhesion molecules are required for selective cell–cell interactions. The classical cadherins mediate homophilic calcium-dependent cell adhesion and are founding members of the large and diverse cadherin superfamily. The protocadherins are the largest subgroup within this superfamily, yet their participation in calcium-dependent cell adhesion is uncertain. In this paper, we demonstrate a novel mechanism of adhesion, mediated by a complex of Protocadherin-19 (Pcdh19) and N-cadherin (Ncad). Although Pcdh19 alone is only weakly adhesive, the Pcdh19–Ncad complex exhibited robust adhesion in bead aggregation assays, and Pcdh19 appeared to play the dominant role. Adhesion by the Pcdh19–Ncad complex was unaffected by mutations that disrupt Ncad homophilic binding but was inhibited by a mutation in Pcdh19. In addition, the complex exhibited homophilic specificity, as beads coated with Pcdh19–Ncad did not intermix with Ncad- or Pcdh17–Ncad-coated beads. We propose a model in which association of a protocadherin with Ncad acts as a switch, converting between distinct binding specificities.
Introduction
Differential cell adhesion is required for the development of multicellular organisms. The expression of distinct subsets of cell adhesion molecules can confer cellular identity, which is essential for the formation of tissues as well as their fine-grained architecture (Steinberg, 1963; Takeichi, 1988, 1990; Steinberg, 1996, 2007; Redies, 2000). The classical cadherins are a major class of adhesion molecules that were originally identified on the basis of their ability to mediate calcium-dependent cell adhesion (Takeichi, 1977; Urushihara and Takeichi, 1980; Yoshida and Takeichi, 1982). The classical cadherins are founding members of a superfamily of cell surface glycoproteins defined by the presence of multiple repeats of an ∼110–amino acid cadherin domain (Nollet et al., 2000; Hulpiau and van Roy, 2009; Hulpiau and van Roy, 2011).
The mechanism of cadherin adhesion has been studied extensively, and it is widely believed that the EC1 domains of partner cadherins are responsible for homophilic adhesion (Shapiro et al., 1995; Boggon et al., 2002; Harrison et al., 2010, 2011; Vendome et al., 2011). Reciprocal exchange of EC1 A β-strands results in the strand dimer, which is stabilized by insertion of a conserved Trp2 residue into a hydrophobic pocket on the partner cadherin, and mutations of Trp2 disrupt cadherin adhesion. Recently, a second adhesive interaction has been identified that involves the A β-strands as well as other contacts near the EC1–EC2 boundary (Harrison et al., 2010; Vendome et al., 2011). It has been proposed that this X dimer represents an intermediate state leading to the formation of the strand dimer (Harrison et al., 2010). Mutations designed to inhibit X dimer formation also impair cadherin adhesion.
The protocadherins are the largest group within the cadherin superfamily, yet their role in cell adhesion remains unclear. Cell-based assays have provided evidence for homophilic interactions by protocadherins, yet these interactions are generally much weaker than those of cadherins (Yoshida, 2003; Frank et al., 2005; Triana-Baltzer and Blank, 2006; Schreiner and Weiner, 2010; Tai et al., 2010). Direct tests using expressed ectodomains (ECs) have not supported a role for protocadherins as adhesion molecules (Chen and Gumbiner, 2006; Morishita et al., 2006; Biswas et al., 2010). In Xenopus laevis, it was shown that the observed cell-sorting activity of paraxial protocadherin is a result of antagonizing C-cadherin adhesion (Chen and Gumbiner, 2006) rather than directly through adhesion. Similarly, forced expression of Pcdh8/arcadlin in rat hippocampal neurons results in the internalization of synaptic N-cadherin (Yasuda et al., 2007). Thus, some protocadherins may play indirect roles in adhesion rather than acting as bona fide cell adhesion molecules.
We have previously shown that Protocadherin-19 (Pcdh19) forms a cis-complex with N-cadherin (Ncad) and that these molecules collaborate to control cell movements during morphogenesis of the anterior neural tube in zebrafish (Biswas et al., 2010). Here, we show that the Pcdh19–Ncad complex is adhesive and that the homophilic interaction is likely mediated by Pcdh19 not Ncad. Within the complex, Ncad appears unable to mediate homophilic interactions. Our results suggest a new mechanism of homophilic cell adhesion mediated by protocadherins with Ncad acting as a required cis-cofactor.
Results and discussion
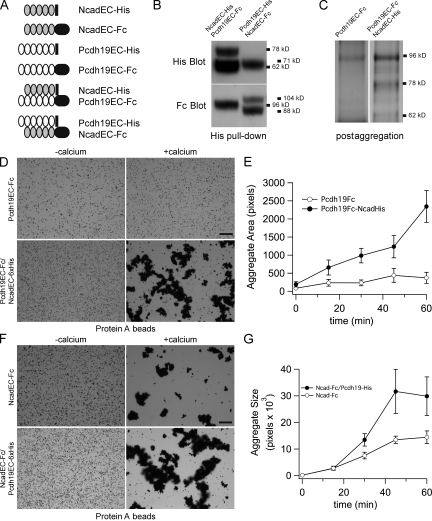
In developing zebrafish embryos, Pcdh19 acts synergistically with Ncad to control convergence cell movements in the anterior neural plate. In addition, Pcdh19 associates directly with Ncad to form a cis-complex, with the interaction likely mediated by their ECs (Biswas et al., 2010). Those observations suggested that the Pcdh19–Ncad cis-complex could comprise a functional unit. To investigate this complex in more detail, we engineered secreted ECs of zebrafish Pcdh19 and Ncad fused to either the Fc region of human IgG or to a 6xHis tag (Fig. 1 A). When HEK293 cells are cotransfected with Pcdh19EC-Fc/NcadEC-6xHis or Pcdh19EC-6xHis/NcadEC-Fc, the secreted proteins can be isolated from culture medium as a stable complex (Fig. 1, B and C). We tested the adhesive properties of expressed Pcdh19 and Ncad EC complexes using standard bead aggregation assays. Evidence from cell-based assays suggests that protocadherins are capable of weak homophilic interactions. However, direct tests using purified protein have generally failed to detect intrinsic adhesive capacity. We previously showed that Pcdh19 exhibits very little calcium-dependent adhesion, which we confirm here (Fig. 1 D). In contrast, protein A beads coated with the complex of Pcdh19EC-Fc–NcadEC-6xHis show calcium-dependent aggregation (Fig. 1, D and E). As NcadEC-6xHis neither binds to the protein A beads nor induces bead aggregation (unpublished data) and Pcdh19EC-Fc alone is not significantly adhesive (Fig. 1 D), the Pcdh19–Ncad complex must be responsible for bead aggregation. Similar results were obtained when Pcdh19EC-Fc and NcadEC-6xHis were purified separately and mixed for the bead aggregation assays (Fig. S1). We also performed the reciprocal experiment, cotransfecting cells with NcadEC-Fc and Pcdh19EC-6xHis (Fig. 1, F and G). Surprisingly, we observed an increase in the size of aggregates formed by the complex of NcadEC-Fc–Pcdh19EC-6xHis compared with those with NcadEC-Fc alone (Fig. 1, F and G). These results demonstrate that Pcdh19 and Ncad comprise an adhesive complex and suggest that the adhesive properties of this complex are distinct from those of either Pcdh19 or Ncad.
Figure 1.
Pcdh19 and Ncad form an adhesive complex. (A) A schematic representation of fusion proteins. Pcdh19 and Ncad ECs were each tagged with either the Fc region of human IgG or with 6xHis. The proposed complexes used in bead aggregation assays are also shown. (B) HEK293 cells were cotransfected with the Fc and 6xHis fusions, which are efficiently produced and secreted into the culture medium. Pcdh19EC and NcadEC form a stable complex, as they can be coisolated using pull-downs of the 6xHis-tagged proteins. (C) Protein A beads were recovered from an aggregation assay, and protein was run on an SDS-PAGE gel and silver stained. In the left lane, aggregation was performed with Pcdh19EC-Fc only, and beads used in the right lane were coated with Pcdh19EC-Fc and NcadEC-6xHis. (D and E) Protein A beads were coated with either Pcdh19EC-Fc or with a complex of Pcdh19EC-Fc and NcadEC-6xHis and allowed to aggregate in the presence of 2 mM CaCl2 or 2 mM EDTA. As previously shown, Pcdh19EC-Fc does not exhibit calcium-dependent homophilic adhesion. However, the complex of Pcdh19EC-Fc and NcadEC-6xHis does mediate bead aggregation. The time course of Pcdh19EC-NcadEC bead aggregation (n = 3) shows robust adhesive activity compared with Pcdh19EC alone (n = 3). (F and G) Although beads coated with NcadEC-Fc (n = 4) aggregate in the presence of calcium, the size of the aggregates is significantly larger for the complex of NcadEC-Fc with Pcdh19EC-6xHis (n = 4). Error bars represent SEM. Bars, 50 µm.
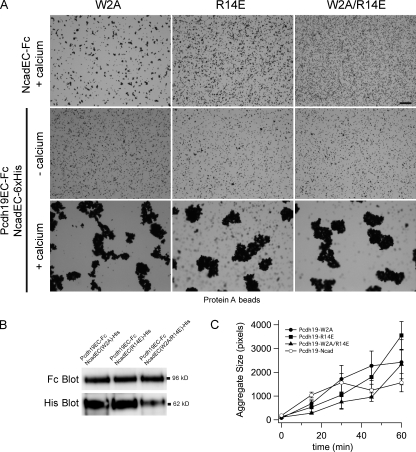
Ncad is one of the founding members of the classical cadherins and mediates calcium-dependent cell adhesion, both in cell- and protein-based assays. Therefore, it is possible that homophilic adhesion of Ncad is responsible for bead aggregation, and Pcdh19 simply tethers Ncad to the beads. To determine the respective roles played by Pcdh19 and Ncad in the adhesive interactions of the Pcdh19–Ncad complex, we generated mutants of zebrafish Ncad that should disrupt formation of the strand dimer (W2A), the X dimer (R14E), or both (W2A/R14E). When used in bead aggregation assays, none of these mutants exhibits significant adhesive activity (Fig. 2 A), which is consistent with a previous study (Harrison et al., 2010). When coexpressed with Pcdh19EC, each of these mutants forms the Pcdh19–Ncad complex (Fig. 2 B), and the complexes of Pcdh19 with each of these Ncad mutants mediate robust bead aggregation, comparable with what is observed with native NcadEC (Fig. 2, A and C). These data argue that Pcdh19–Ncad adhesion is not simply a result of the formation of cadherin homodimers but is mediated by homophilic Pcdh19 interactions.
Figure 2.
Ncad function is not required for Pcdh19–Ncad adhesion. (A) We generated point mutants in NcadEC-Fc that are known to impair adhesion of classical cadherins. The W2A mutation interferes with the formation of the strand dimer, and R14E disrupts the X dimer. Protein A beads coated with the Fc fusions of W2A, R14E, or W2A/R14E mutants do not exhibit aggregation. However, the complexes of Pcdh19EC-Fc with the 6xHis-tagged mutants mediate robust calcium-dependent aggregation. Bar, 50 µm. (B) The formation of the Pcdh19EC-Fc–mutant NcadEC-6xHis complexes was verified by Western blotting after an aggregation experiment. (C) Quantitative characterization of aggregate sizes reveals that complexes formed with the Ncad mutants are as effective as wild-type Ncad in mediating homophilic adhesion (n = 3 for each experiment). Error bars are ±SEM.
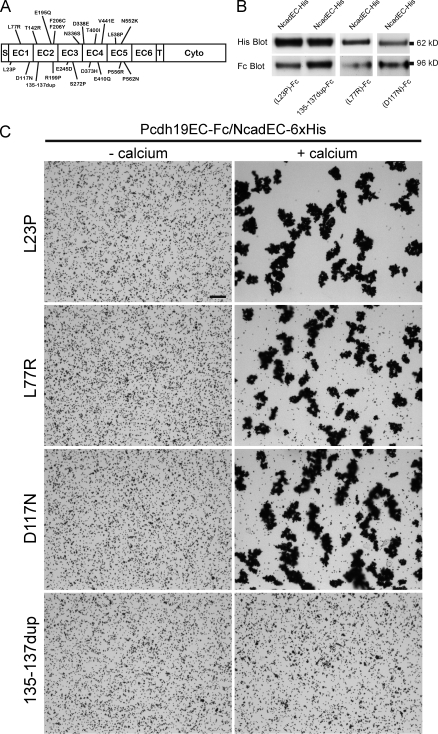
To date, >40 distinct mutations of pcdh19 have been reported in human patients with epilepsy and mental retardation limited to females (EFMR; Fig. 3 A), including ∼20 missense mutations in the Pcdh19 EC (Dibbens et al., 2008; Depienne et al., 2009, 2011; Hynes et al., 2010; Jamal et al., 2010). Nearly all of the mutated residues are conserved between zebrafish and human. We hypothesized that some of these mutations could impair Pcdh19 function through an effect on Pcdh19–Ncad adhesion. We tested several EFMR mutations for their ability to bind Ncad and to mediate bead aggregation (Fig. 3, B and C). Each of the Pcdh19EC mutants associated with NcadEC in pull-downs (Fig. 3 B). Most of the mutations support calcium-dependent bead aggregation when coupled with NcadEC-6xHis (Fig. 3 C). One of the mutants, Pcdh19EC(135–137)dup, exhibited dramatically reduced adhesion (Fig. 3 C), although its association with Ncad was unaffected (Fig. 3 B). This mutation is a duplication of three residues (Ser135-Glu136-Asn137; Ser-Glu-Ala in human) in EC2 (Fig. 3 A), which are predicted to be in a loop adjacent to the EC2–EC3 boundary. Thus, despite the presence of functional NcadEC within the complex, the (135–137)dup mutation in Pcdh19 impairs calcium-dependent adhesion of the Pcdh19–Ncad complex. This result further supports the idea that Pcdh19, rather than Ncad, mediates adhesion within the Pcdh19–Ncad complex. In addition, our data demonstrate for the first time a specific functional defect for one of the mutations identified in EFMR.
Figure 3.
The EFMR mutation 135–137dup disrupts homophilic adhesion by the Pcdh19–Ncad complex. (A) A schematic showing the missense mutations found in human pcdh19. Cyto, cytodomain; S, signal peptide; T, transmembrane domain. (B) Pull-downs showing the association of Fc fusions of selected EFMR mutants with NcadEC-6xHis. (C) Bead aggregation assays showing that selected EFMR mutations (L23P, L77R, and D117N) support calcium-dependent adhesion when coupled with NcadEC-6xHis, whereas the S135-N137 duplication does not. Bar, 50 µm.
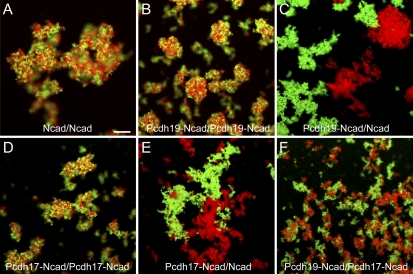
The aforementioned data suggest that Ncad may not play a direct role in adhesion by the Pcdh19–Ncad complex. Thus, the mechanism of adhesion and the adhesive interface may be distinct from that used by Ncad. To test this idea, we performed sorting assays using fluorescent protein A beads coated with Ncad or Pcdh19–Ncad. If Ncad contributes to Pcdh19–Ncad adhesion or is available for homophilic interactions, beads coated with Ncad should intermix with beads coated with Pcdh19–Ncad. Alternatively, if adhesion of Pcdh19–Ncad is incompatible with Ncad adhesion, the beads should segregate into distinct aggregates. When red and green beads were coated with Ncad alone and allowed to aggregate, aggregates exhibited extensive intermixing (Fig. 4 A). Similarly, Pcdh19–Ncad beads also formed mixed aggregates (Fig. 4 B). In contrast, Pcdh19–Ncad beads and Ncad beads both aggregated robustly when mixed but segregated into distinct populations (Fig. 4 C), indicating that they have incompatible homophilic binding specificities. These results support the conclusions that the adhesive interface of Pcdh19–Ncad is distinct from that of Ncad. Moreover, the homophilic adhesive capacity of Ncad must be masked when incorporated into the Pcdh19–Ncad complex. Thus, the formation of the Pcdh19–Ncad complex defines a novel adhesive unit.
Figure 4.
Pcdh19–Ncad adhesion is not compatible with Ncad homophilic binding. (A) Red or green fluorescent protein A beads were coated with NcadEC-Fc and mixed before the addition of calcium. After 30 min, mixed aggregates were observed. (B) Red or green fluorescent protein A beads were coated with the complex of Pcdh19EC-Fc–NcadEC-6xHis. As in A, mixed aggregates were observed after 30 min. (C) Green fluorescent protein A beads were coated with Pcdh19EC-Fc–NcadEC-6xHis and mixed with red fluorescent protein A beads coated with NcadEC-Fc. After mixing, calcium was added, and the beads were allowed to aggregate for 30 min. The beads did not form mixed aggregates, indicating that Pcdh19–Ncad does not cross-adhere with Ncad. (D) Pcdh17EC forms an adhesive complex with NcadEC. Mixed aggregates of red and green fluorescent protein A beads coated with the Pcdh17EC-Fc–NcadEC-6xHis complex are shown. (E) Aggregates of beads coated with Pcdh17EC-Fc–NcadEC-6xHis (green) segregate from beads coated with NcadEC-Fc (red). (F) Bead aggregation of Pcdh19EC–NcadEC (red) and Pcdh17EC–NcadEC (green) complexes. The distinct protocadherin complexes exhibit homophilic specificity, as they form distinct aggregates. However, there may be weak heterophilic association, as red and green clusters appear to have some ability to associate even though the beads do not exhibit extensive intermixing. Bar, 25 µm.
In addition to our previous results showing both a functional synergism between Pcdh19 and Ncad and a physical interaction, other studies have shown interactions between δ-2 protocadherins and classical cadherins. Pcdh8/arcadlin associates with Ncad in cultured rat hippocampal neurons and promotes endocytosis of Ncad (Yasuda et al., 2007), and Xenopus paraxial protocadherin (Pcdh8 like) antagonizes C-cadherin adhesion in vivo and has been shown to interact with E-cadherin in vitro (Chen and Gumbiner, 2006; Chen et al., 2009). Similarly, the effects of overexpressing Pcdh10 are similar to depletion of Ncad in U251 cells (Nakao et al., 2008), suggesting an antagonistic relationship. Thus, our results may not be unique to Pcdh19 and could be generally applicable to δ-2 protocadherins. To test this idea, we repeated the sorting assays using the EC of zebrafish Protocadherin-17 (Pcdh17). Pcdh17 is also a member of the δ-2 subfamily and is closely related to Pcdh19. Pcdh17 associates with Ncad to form a Pcdh17–Ncad complex that mediates bead aggregation (Fig. 4 D). Although red and green fluorescent beads coated with the Pcdh17–Ncad complex exhibit extensive intermixing (Fig. 4 D), they segregate from Ncad beads (Fig. 4 E) or Pcdh19–Ncad beads (Fig. 4 F). These data support the idea that members of the δ-2 subfamily of protocadherins associate with Ncad to form adhesive cis-complexes. The adhesion of these complexes is not simply mediated by the Ncad component, as homophilic specificity is provided by the partner protocadherin.
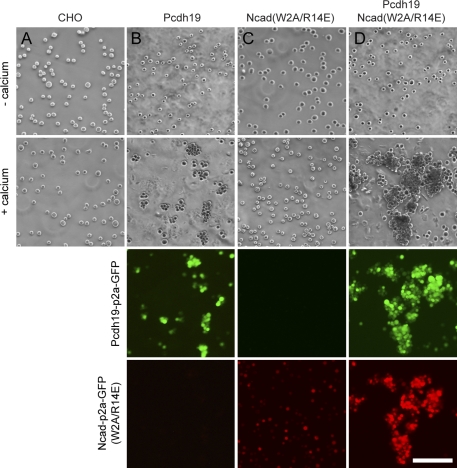
Although our experiments using secreted ECs uniformly support a role for Pcdh19 in adhesion as part of a Pcdh19–Ncad complex, it is important to demonstrate that the complex acts by a comparable mechanism in cells. To address this question, we performed cell aggregation assays using CHO cells that had been transfected with Pcdh19, Ncad(W2A/R14E), or both. To facilitate the identification and isolation of transfected cells, Pcdh19 and Ncad were expressed as Pcdh19-p2a-GFP and Ncad(W2A/R14E)-p2a-RFP fusions (Fig. S2). Self-cleavage by the p2a peptide releases soluble GFP or RFP to effectively mark transfected cells (Fig. S2, A and B). Naive CHO cells do not exhibit calcium-dependent cell aggregation (Fig. 5 A). We find that CHO cells expressing Pcdh19 exhibit low levels of calcium-dependent cell aggregation, consistent with assays performed with chicken Pcdh19 (Fig. 5 B; Tai et al., 2010). As expected, Ncad(W2A/R14E) does not induce any calcium-dependent cell aggregation (Fig. 5 C). However, coexpression of Pcdh19 and Ncad(W2A/R14E) results in vigorous cell aggregation (Fig. 5 D), supporting the conclusion that association with Ncad facilitates or enhances adhesion by Pcdh19.
Figure 5.
Ncad enhances Pcdh19 adhesion in cell aggregation assay. (A) CHO cells do not exhibit calcium-dependent cell aggregation. (B) When expressing Pcdh19-p2a-GFP, CHO cells form small aggregates, indicating low levels of adhesion. Expression of Pcdh19 is verified by GFP fluorescence. (C) CHO cells transfected with the Ncad(W2A/R14E) double mutant do not aggregate in the presence of calcium. Expression of the Ncad mutant is verified by RFP fluorescence. (D) CHO cells that express both Pcdh19 and Ncad(W2A/R14E) aggregate more robustly than cells that express only Pcdh19. Coexpression of Pcdh19 and Ncad is demonstrated by the presence of both GFP and RFP fluorescence. Bar, 100 µm.
The diversity of protocadherins has suggested that they could contribute to differential cell–cell recognition through homophilic interactions, yet the evidence for protocadherin adhesion is modest. Our data indicate that Pcdh19 functions as a cell adhesion molecule by using Ncad as a cofactor. Within the Pcdh19–Ncad complex, Pcdh19 appears to play the dominant role, as functional Ncad is not required for adhesion. Association with Ncad may cause a conformational change in Pcdh19, possibly a bending or twisting of the EC, that unmasks an adhesion site. These data suggest a model in which Ncad switches between adhesive states, either directly mediating homophilic adhesion or acting as a cofactor for a partner protocadherin. Our data with Pcdh17 suggests that other δ-2 protocadherins can exhibit similar behavior, and it has recently been shown that Pcdh-γ isoforms can associate to form heteromeric cis-complexes (Schreiner and Weiner, 2010). Thus, protocadherins may function as part of larger macromolecular assemblies rather than as autonomous adhesive units. Our data also have implications for the regulation of cadherin function. Most studies of dynamic cadherin regulation focus on control of cadherin trafficking, association with catenins, or feedback through Rho GTPases and the actin cytoskeleton (Gumbiner, 2005; Niessen et al., 2011). Here, we show that extracellular cis-interactions can have profound effects on cadherin adhesion. It is possible that the adhesive activity of cadherins in vivo depends on the complement of protocadherins and other cofactors expressed by a given cell or tissue. It will be important for future studies to determine the functional significance of this and similar complexes in vivo.
Materials and methods
Bead aggregation assays
Bead aggregations were performed essentially as previously described (Sivasankar et al., 2009; Biswas et al., 2010). Pcdh19EC and NcadEC-Fc and -6xHis fusions were transfected independently or cotransfected into HEK293 cells using Fugene HD (Roche) according to the manufacturer’s instructions. After 24 h, cells were rinsed three times and allowed to grow in serum-free medium for an additional 48 h. The culture medium containing the secreted Fc and/or 6xHis fusion proteins was then collected. The media was filtered using 0.45-µm syringe filters, concentrated using Ultracel (Millipore), and incubated with 1.5 µl of protein A Dynabeads (Invitrogen) for 1 h with gentle agitation at 4°C. The beads were washed extensively in binding buffer (50 mM Tris, 100 mM NaCl, 10 mM KCl, and 0.2% BSA, pH 7.4) and split into two tubes, and either 2 mM EDTA or 2 mM CaCl2 was added to each tube. Beads were allowed to aggregate for 1 h in glass depression slides in a humidified chamber, and images were collected every 15 min on a microscope (AxioStar; Carl Zeiss) using a 10× objective and AxioCam MRc 5 (Carl Zeiss). Assays were performed at least three times using three independent transfections/protein preps.
Quantification of bead aggregation was performed using ImageJ software (National Institutes of Health), as previously described (Biswas et al., 2010). In brief, images were thresholded, and individual particles were detected using the ImageJ command Analyze/Analyze Particles. The size of aggregates was approximated as the area of detected particles in units of pixels. Mean aggregate size (±SEM) was calculated for each time point and was plotted using IGOR Pro software (WaveMetrics).
Bead sorting assays
Sorting assays were performed essentially as described for bead aggregation assays except that fluorescent beads were used. Protein was bound to either 1 µl of green beads (screenMAG/G–-protein A; Chemicell) or to red beads (screenMAG/O–Protein A; Chemicell). For the assays, beads were mixed, and either 2 mM CaCl2 or 2 mM EDTA was then added. Aggregates were allowed to form for 30 min, and fluorescent images were collected on a microscope (AxioStar) with a 10× objective and AxioCam MRc5.
Cell aggregation assays
The coding sequences of full-length Pcdh19 and Ncad were fused to those of p2a-GFP and p2a-RFP, respectively (provided by M. Meyer, King’s College London, England, UK). The p2a self-cleaving peptide liberates soluble GFP or RFP in transfected cells, providing a convenient marker of Pcdh19 or Ncad expression. CHO cells were transfected with plasmid-encoding Pcdh19-p2a-GFP, Ncad-p2a-RFP, or both using X-tremeGENE HP (Roche). Transfected cells were isolated by FACS and used in shaking cell aggregation assays. FACS-sorted cells were maintained and propagated while under G418 selection (800 µg/ml). For aggregation assays, cells were trypsinized in the presence of 2 mM CaCl2, pelleted, and resuspended in HCMF buffer (137 mM NaCl, 5.4 mM KCl, 0.63 mM Na2HPO4, 5.5 mM glucose, and 10 mM Hepes, pH 7.4) at a concentration of 2.5 × 105 cells/ml. For each condition, 0.5 ml of the cell suspension was added to 1% agar-coated wells in a 24-well plate, and either 2 mM EDTA or 2 mM CaCl2 was added. Cells were shaken for 6 h to overnight at 80 rpm at 37°C, and images of random fields were collected on a microscope (AxioStar) using a 10× objective. Cell aggregation assays were performed in triplicate.
Pull-down assays and Western blotting
For pull-down assays, HEK293 cells were transiently transfected, and the culture medium was collected as described for bead aggregation assays. Media was concentrated using Ultracel and incubated with protein A Sepharose (GE Healthcare) or TALON metal affinity resin (Takara Bio Inc.) for 1 h with gentle agitation at 4°C. Alternatively, beads were collected after aggregation assays and processed. The resin was washed with binding buffer, resuspended in loading buffer, and boiled for 5 min. Samples were loaded onto 10% Bis-Tris NuPAGE gels (Invitrogen) and subjected to electrophoresis. Proteins were then transferred (Bio-Rad Laboratories) to PVDF (GE Healthcare), blocked with 5% nonfat milk in TBS with Tween 20 (0.1%), and incubated overnight with primary antibodies anti–human IgG (1:400; Jackson ImmunoResearch Laboratories, Inc.) and anti-6xHis (1:1,000; NeuroMab Clone no. N144/14; University of California Davis/National Institutes of Health NeuroMab Facility). HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used at 1:5,000, and the chemiluminescent signal was amplified using Western Lightning Ultra (PerkinElmer). Blots were imaged on a molecular imaging system (Omega 12iC; UltraLum, Inc.).
Immunocytochemistry
CHO cells expressing Pcdh19-p2a-GFP or Ncad-p2a-RFP were seeded on glass coverslips, fixed in 4% PFA in PBS, permeabilized in PBS + 0.25% Triton X-100, and blocked in PBS + 2% normal goat serum + 3% BSA. Cells were then incubated in a primary antibody against Pcdh19 (1:100; Emond et al., 2009) or pan-Cadherin (1:100; Cell Signaling Technology) for 2 h. Anti–rabbit secondary antibodies conjugated either to DyLight 488 or DyLight 549 were used at 1:500 (Jackson ImmunoResearch Laboratories, Inc.). Coverslips were mounted in Fluoromount G (Electron Microscopy Sciences) and imaged on a microscope (AxioStar) as previously described.
Protein purification
For the isolation of purified protein, stable HEK293 cell lines expressing either Pcdh19EC-Fc or NcadEC-6xHis were used. Culture medium was collected as described for bead aggregation assays. The filtered media was concentrated and loaded onto a protein A HP SpinTrap (GE Healthcare) or TALONspin (Takara Bio Inc.) columns for Pcdh19EC-Fc and NcadEC-6xHis, respectively. Purifications were performed following the manufacturer’s instructions.
DNA constructs
Pcdh19EC-Fc and NcadEC-Fc were previously reported (Biswas et al., 2010). The 6xHis tag was generated using PCR and was subcloned in place of the Fc coding sequence. Pcdh19 and Ncad mutants were generated by PCR. Mutated segments of Pcdh19 or Ncad were then subcloned into Pcdh19EC and NcadEC plasmids, respectively. All clones were sequenced.
Online supplemental material
Fig. S1 shows an adhesion assay performed with individually purified Pcdh19 and Ncad that were mixed and added to protein A beads. Fig. S2 shows coexpression of Pcdh19 and GFP or Ncad and RFP using a self-cleaving p2a linker. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201108115/DC1.
Acknowledgments
We would like to thank Candice Askwith for assistance with CHO cells, Martin Meyer for the p2a plasmids, Greg Phillips for comments on the manuscript, and James Nelson for suggestions and helpful discussions. The 6xHis antibody (N144/14) was obtained form the University of California Davis/National Institutes of Health NeuroMab Facility.
This work was supported by the National Science Foundation (ARRA/IOS 0920357 to J.D. Jontes) and a Neurosciences Core grant (P30 NS045758).
Footnotes
Abbreviations used in this paper:
- EC
- ectodomain
- Ncad
- N-cadherin
References
- Biswas S., Emond M.R., Jontes J.D. 2010. Protocadherin-19 and N-cadherin interact to control cell movements during anterior neurulation. J. Cell Biol. 191:1029–1041 10.1083/jcb.201007008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggon T.J., Murray J., Chappuis-Flament S., Wong E., Gumbiner B.M., Shapiro L. 2002. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 296:1308–1313 10.1126/science.1071559 [DOI] [PubMed] [Google Scholar]
- Chen X., Gumbiner B.M. 2006. Paraxial protocadherin mediates cell sorting and tissue morphogenesis by regulating C-cadherin adhesion activity. J. Cell Biol. 174:301–313 10.1083/jcb.200602062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Koh E., Yoder M., Gumbiner B.M. 2009. A protocadherin-cadherin-FLRT3 complex controls cell adhesion and morphogenesis. PLoS ONE. 4:e8411 10.1371/journal.pone.0008411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C., Bouteiller D., Keren B., Cheuret E., Poirier K., Trouillard O., Benyahia B., Quelin C., Carpentier W., Julia S., et al. 2009. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 5:e1000381 10.1371/journal.pgen.1000381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C., Trouillard O., Bouteiller D., Gourfinkel-An I., Poirier K., Rivier F., Berquin P., Nabbout R., Chaigne D., Steschenko D., et al. 2011. Mutations and deletions in PCDH19 account for various familial or isolated epilepsies in females. Hum. Mutat. 32:E1959–E1975 10.1002/humu.21373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens L.M., Tarpey P.S., Hynes K., Bayly M.A., Scheffer I.E., Smith R., Bomar J., Sutton E., Vandeleur L., Shoubridge C., et al. 2008. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat. Genet. 40:776–781 10.1038/ng.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emond M.R., Biswas S., Jontes J.D. 2009. Protocadherin-19 is essential for early steps in brain morphogenesis. Dev. Biol. 334:72–83 10.1016/j.ydbio.2009.07.008 [DOI] [PubMed] [Google Scholar]
- Frank M., Ebert M., Shan W., Phillips G.R., Arndt K., Colman D.R., Kemler R. 2005. Differential expression of individual gamma-protocadherins during mouse brain development. Mol. Cell. Neurosci. 29:603–616 10.1016/j.mcn.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Gumbiner B.M. 2005. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 6:622–634 10.1038/nrm1699 [DOI] [PubMed] [Google Scholar]
- Harrison O.J., Bahna F., Katsamba P.S., Jin X., Brasch J., Vendome J., Ahlsen G., Carroll K.J., Price S.R., Honig B., Shapiro L. 2010. Two-step adhesive binding by classical cadherins. Nat. Struct. Mol. Biol. 17:348–357 10.1038/nsmb.1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison O.J., Jin X., Hong S., Bahna F., Ahlsen G., Brasch J., Wu Y., Vendome J., Felsovalyi K., Hampton C.M., et al. 2011. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 19:244–256 10.1016/j.str.2010.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulpiau P., van Roy F. 2009. Molecular evolution of the cadherin superfamily. Int. J. Biochem. Cell Biol. 41:349–369 10.1016/j.biocel.2008.09.027 [DOI] [PubMed] [Google Scholar]
- Hulpiau P., van Roy F. 2011. New insights into the evolution of metazoan cadherins. Mol. Biol. Evol. 28:647–657 10.1093/molbev/msq233 [DOI] [PubMed] [Google Scholar]
- Hynes K., Tarpey P., Dibbens L.M., Bayly M.A., Berkovic S.F., Smith R., Raisi Z.A., Turner S.J., Brown N.J., Desai T.D., et al. 2010. Epilepsy and mental retardation limited to females with PCDH19 mutations can present de novo or in single generation families. J. Med. Genet. 47:211–216 10.1136/jmg.2009.068817 [DOI] [PubMed] [Google Scholar]
- Jamal S.M., Basran R.K., Newton S., Wang Z., Milunsky J.M. 2010. Novel de novo PCDH19 mutations in three unrelated females with epilepsy female restricted mental retardation syndrome. Am. J. Med. Genet. A. 152A:2475–2481 10.1002/ajmg.a.33611 [DOI] [PubMed] [Google Scholar]
- Morishita H., Umitsu M., Murata Y., Shibata N., Udaka K., Higuchi Y., Akutsu H., Yamaguchi T., Yagi T., Ikegami T. 2006. Structure of the cadherin-related neuronal receptor/protocadherin-alpha first extracellular cadherin domain reveals diversity across cadherin families. J. Biol. Chem. 281:33650–33663 10.1074/jbc.M603298200 [DOI] [PubMed] [Google Scholar]
- Nakao S., Platek A., Hirano S., Takeichi M. 2008. Contact-dependent promotion of cell migration by the OL-protocadherin–Nap1 interaction. J. Cell Biol. 182:395–410 10.1083/jcb.200802069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen C.M., Leckband D., Yap A.S. 2011. Tissue organization by cadherin adhesion molecules: Dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol. Rev. 91:691–731 10.1152/physrev.00004.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet F., Kools P., van Roy F. 2000. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J. Mol. Biol. 299:551–572 10.1006/jmbi.2000.3777 [DOI] [PubMed] [Google Scholar]
- Redies C. 2000. Cadherins in the central nervous system. Prog. Neurobiol. 61:611–648 10.1016/S0301-0082(99)00070-2 [DOI] [PubMed] [Google Scholar]
- Schreiner D., Weiner J.A. 2010. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc. Natl. Acad. Sci. USA. 107:14893–14898 10.1073/pnas.1004526107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L., Fannon A.M., Kwong P.D., Thompson A., Lehmann M.S., Grübel G., Legrand J.F., Als-Nielsen J., Colman D.R., Hendrickson W.A. 1995. Structural basis of cell-cell adhesion by cadherins. Nature. 374:327–337 10.1038/374327a0 [DOI] [PubMed] [Google Scholar]
- Sivasankar S., Zhang Y., Nelson W.J., Chu S. 2009. Characterizing the initial encounter complex in cadherin adhesion. Structure. 17:1075–1081 10.1016/j.str.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg M.S. 1963. Reconstruction of tissues by dissociated cells. Some morphogenetic tissue movements and the sorting out of embryonic cells may have a common explanation. Science. 141:401–408 10.1126/science.141.3579.401 [DOI] [PubMed] [Google Scholar]
- Steinberg M.S. 1996. Adhesion in development: An historical overview. Dev. Biol. 180:377–388 10.1006/dbio.1996.0312 [DOI] [PubMed] [Google Scholar]
- Steinberg M.S. 2007. Differential adhesion in morphogenesis: A modern view. Curr. Opin. Genet. Dev. 17:281–286 10.1016/j.gde.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Tai K., Kubota M., Shiono K., Tokutsu H., Suzuki S.T. 2010. Adhesion properties and retinofugal expression of chicken protocadherin-19. Brain Res. 1344:13–24 10.1016/j.brainres.2010.04.065 [DOI] [PubMed] [Google Scholar]
- Takeichi M. 1977. Functional correlation between cell adhesive properties and some cell surface proteins. J. Cell Biol. 75:464–474 10.1083/jcb.75.2.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. 1988. The cadherins: Cell-cell adhesion molecules controlling animal morphogenesis. Development. 102:639–655 [DOI] [PubMed] [Google Scholar]
- Takeichi M. 1990. Cadherins: A molecular family important in selective cell-cell adhesion. Annu. Rev. Biochem. 59:237–252 10.1146/annurev.bi.59.070190.001321 [DOI] [PubMed] [Google Scholar]
- Triana-Baltzer G.B., Blank M. 2006. Cytoplasmic domain of protocadherin-alpha enhances homophilic interactions and recognizes cytoskeletal elements. J. Neurobiol. 66:393–407 10.1002/neu.20228 [DOI] [PubMed] [Google Scholar]
- Urushihara H., Takeichi M. 1980. Cell-cell adhesion molecule: Identification of a glycoprotein relevant to the Ca2+-independent aggregation of Chinese hamster fibroblasts. Cell. 20:363–371 10.1016/0092-8674(80)90622-4 [DOI] [PubMed] [Google Scholar]
- Vendome J., Posy S., Jin X., Bahna F., Ahlsen G., Shapiro L., Honig B. 2011. Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins. Nat. Struct. Mol. Biol. 18:693–700 10.1038/nsmb.2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S., Tanaka H., Sugiura H., Okamura K., Sakaguchi T., Tran U., Takemiya T., Mizoguchi A., Yagita Y., Sakurai T., et al. 2007. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2beta and p38 MAP kinases. Neuron. 56:456–471 10.1016/j.neuron.2007.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C., Takeichi M. 1982. Teratocarcinoma cell adhesion: Identification of a cell-surface protein involved in calcium-dependent cell aggregation. Cell. 28:217–224 10.1016/0092-8674(82)90339-7 [DOI] [PubMed] [Google Scholar]
- Yoshida K. 2003. Fibroblast cell shape and adhesion in vitro is altered by overexpression of the 7a and 7b isoforms of protocadherin 7, but not the 7c isoform. Cell. Mol. Biol. Lett. 8:735–741 [PubMed] [Google Scholar]