Abstract
Bacterial pathogens use effector proteins to manipulate their hosts to propagate infection. These effectors divert host cell signaling pathways to the benefit of the pathogen and frequently target kinase signaling cascades. Notable pathways that are usurped include the nuclear factor κB (NF-κB), mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K)/Akt, and p21-activated kinase (PAK) pathways. Analyzing the functions of pathogenic effectors and their intersection with host kinase pathways has provided interesting insights into both the mechanisms of virulence and eukaryotic signaling.
Introduction
Bacterial pathogens are masters of survival. They usually distinguish themselves from other strains within their species by a drastic change in lifestyle. Although closely related strains dwell in the soil or aqueous environments, pathogenic strains have evolved to thrive within a host. This brings the advantage of a “niche” existence, but it also comes with many challenges. A pathogen must successfully infiltrate the host’s tissues by attaching to cells (sometimes entering them) and subverting the cell’s natural functions for its own ends, all while avoiding the host’s immune system and other defenses. In the ongoing evolutionary arms race between pathogens and their hosts, pathogens tend to “think big,” generally targeting pathways that control important cellular functions like cytoskeletal dynamics (Haglund and Welch, 2011), initiation of cell death (Ashida et al., 2011), or autophagy and cellular membrane homeostasis (Weinrauch and Zychlinsky, 1999; Bhavsar et al., 2007; Alix et al., 2011; Ham et al., 2011). It is no wonder that pathogens have evolved a large variety of tools to target one of the most ubiquitous of all eukaryotic signaling mechanisms: phosphorylation by protein kinases.
The human genome encodes ∼500 putative protein kinase genes, which correspond to 2% of all eukaryotic genes. This relatively small percentage of protein kinase–encoding genes is opposed by an enormous number of targets that are regulated by protein kinases. It has been estimated that roughly 30% of all cellular proteins may be modified by protein kinase activity (Manning et al., 2002). Consequently, kinases give rise to a vast network of interwoven signaling pathways, and the sheer complexity of eukaryotic kinase signaling networks still baffles scientists, biochemists, and system biologists alike. The field has made enormous progress toward understanding kinases on a biochemical and structural level by following specific “threads” of kinase regulatory cascades, starting from receptor activation to cellular consequences on the transcriptional level. However, in many cases, the field still lacks a deeper understanding about how seemingly “discrete” signaling pathways are communicating, thereby giving rise to a cellular fate that cannot be predicted based on additive outcomes of individual pathways.
An increasingly popular strategy to attempt to understand complex eukaryotic pathways is to study the mechanisms that pathogens use to subvert them. Invading pathogens specifically target signaling networks at a certain point, irreversibly provoking the downstream event they require to meet their needs. To this end, the study of how bacterial pathogens manipulate complex networks like the MAPK and nuclear factor κB (NF-κB) kinase pathways could illuminate otherwise inscrutable but important mechanisms of regulation and crosstalk in these pathways. Even a “simple” cascade can be regulated by diverse mechanisms, including dephosphorylation, binding to regulators, subcellular localization, and degradation. We will describe examples of all of these regulatory principles, show how pathogens have taken advantage of them to achieve a dominating up- or down-regulation of kinase activity (Table I), and present insights that we have gained into the pathways themselves.
Table I.
Effectors manipulating host kinase signaling
| Pathogen | Effector | Activity | Host targets | Pathway | Phenotype | Reference |
| P. gingivalis | Fimbriae | Binding | CXCR4/ TLR2 | NF-κB inhibition | Decreased ROS production | Hajishengallis et al., 2008 |
| M. tuberculosis | ? | Binding | CR-3 | p38 MAPK activation | Decreased CD1 expression | Gagliardi et al., 2009 |
| Shigella subspecies (ssp). | IpaH9.8 | E3 ligase | NEMO/ ABIN-1 | NF-κB inhibition | Inhibition of pro-inflammatory responses | Ashida et al., 2010 |
| Shigella ssp. | OspG | Kinase | UbcH5 | NF-κB inhibition | Inhibition of pro-inflammatory responses | Kim et al., 2005 |
| Shigella ssp. | OspF | Phosphothreonine lyase | ERK1/2 | ERK inhibition | Inhibition of pro-inflammatory responses | Arbibe et al., 2007; Kramer et al., 2007; Li et al., 2007 |
| Shigella ssp. | OspE | Binding | Focal adhesions | ILK activation | Stabilization of intestinal lining | Kim et al., 2009 |
| A. salmonicida | AopP | Acetyltransferase | ? | NF-κB inhibition | Inhibition of pro-inflammatory responses | Fehr et al., 2006 |
| V. parahaemolyticus | VopA/P | Acetyltransferase | MKKs | MAPK inhibition | Growth arrest | Trosky et al., 2007 |
| EHEC | EspG | Binding | ARFs/PAKs | Arf inhibition/PAK activation | Reprogramming of intracellular trafficking | Selyunin et al., 2011 |
| EPEC/EHEC | NleH | Binding | RPS3 | partial NF-κB inhibition | Increased bacterial colonization, decreased mortality of host | Wan et al., 2011 |
| EPEC | NleE | ? | IKKβ | NF-κB inhibition | Inhibition of pro-inflammatory responses | Nadler et al., 2010; Vossenkämper et al., 2010 |
| EPEC | NleC/NleD | Proteases | RelA | NF-κB inhibition | Inhibition of IL-8 secretion | Baruch et al., 2011; Pearson et al., 2011 |
| L. pneumophila | LegK1 | Kinase | IκB | NF-κB activation | Induction of pro-inflammatory responses | Ge at al., 2009 |
| Yersinia ssp. | YopJ | Acetyltransferase | MKKs | MAPK/ NF-κB inhibition | Induction of apoptosis/inhibition of pro-inflammatory responses | Mukherjee et al., 2006 |
| Yersinia ssp. | Invasin/YadA | Binding | β1-integrins | FAK activation | Actin rearrangements/bacterial uptake | Uliczka et al., 2009 |
| Yersinia ssp. | YopH | Phosphatase | Fyb | Fyn inhibition | Inhibition of phagocytosis | Yuan et al., 2005 |
| Yersinia ssp. | YopH | Phosphatase | FAK/p130Cas | FAK inhibition | Disruption of focal adhesions/inhibition of phagocytosis | Black and Bliska, 1997 |
| B. anthracis | Anthrax toxin | Protease | MKKs | MAPK inhibition | Induction of apoptosis | Ali et al., 2011 |
| B. anthracis | LF | Protease | MEK1 | MAPK inhibition | Induction of apoptosis | Park et al., 2002 |
| P. aeruginosa | ExoT | ADP-ribosyltransferase | Crk | Crk inhibition | Inhibition of invasion | Pielage et al., 2008 |
Targeting of the NF-κB and MAPK pathways
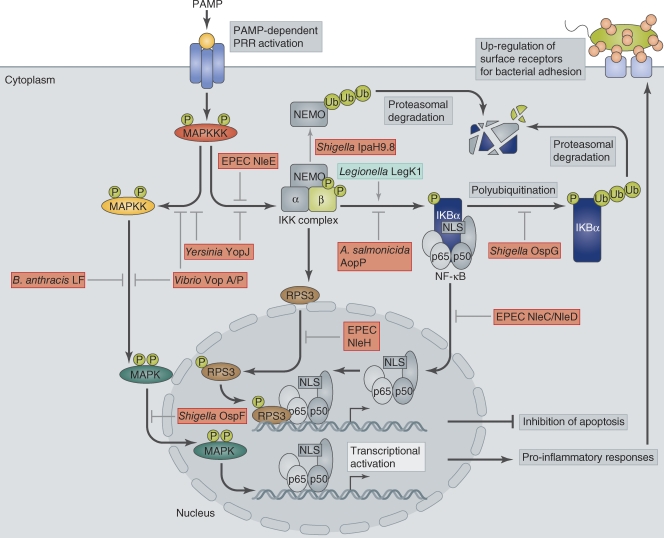
The evolution of eukaryotic immune recognition in the face of ever-changing bacterial phenotypes is a constant battle. The host’s innate immune system is poised to be triggered by signs of bacterial challenge, so-called pathogen-associated molecular patterns (PAMPs), which include characteristic molecules associated with infecting pathogens such as peptidoglycan, lipopolysaccharide, lipoteichoic acid, or flagellin (Lemaitre et al., 1996; Medzhitov and Janeway, 2002). These are detected by eukaryotic pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), and mannose receptors, or the family of cytoplasmic nucleotide-binding oligomerization domain containing protein (NOD)-like receptors (NLRs; Medzhitov et al., 1997; Inohara et al., 1999). When bound to their PAMP ligand, PRRs induce signaling cascades, most commonly NF-κB and MAPK pathways, which in turn trigger pro-inflammatory responses such as the up-regulation of cytokines and antimicrobial peptides or activation of the complement cascade (Ruco et al., 1978). For a recent review of innate immunity, please see Beutler (2009). However, pathogens have developed diverse mechanisms to counter the immune system, including disruption of immune recognition, inhibition of pro-inflammatory responses, or direct induction of immune cells (Fig. 1).
Figure 1.
Regulation of MAPK and NF-κB pathways and their manipulation by bacterial effectors. Bacterial infection triggers PAMP-dependent activation of PRRs at the host cell membrane. Receptor activation triggers MKKK phosphorylation, which in turn activates MAPK pathways and the NF-κB pathway via activation of the IKK complex. IKK phosphorylates IκB, which then becomes polyubiquitinated and removed by proteasomal degradation. Degradation of IκB exposes the NLS on NF-κB, which subsequently undergoes nuclear translocation and binds to κB sites in target promoters to stimulate transcription of genes required for pro-inflammatory responses, such as cytokines and chemokines, and genes involved in cell proliferation. Activation of NF-κB also leads to surface expression of receptors such as CEACAMs, which are hijacked by pathogens to promote their attachment to the host cell surface. Some κB site-containing promoters require RPS3 as a transcriptional co-factor in addition to NF-κB. RPS3 nuclear translocation is dependent on its phosphorylation by IKKβ. Bacterial pathogens manipulate both MAPK and NF-κB pathways either by acting as inhibitors (red boxes) or activators (green box) of individual components.
Activation of the NF-κB and MAPK pathways results in a cascade of activated kinases that ultimately leads to a transcriptional response. For the MAPK signaling pathway, three types of kinases, herein referred to as MAP kinase kinase kinase (MKKK), MAP kinase kinase (MKK), and MAPK, are sequentially phosphorylated, ending with the phosphorylation of MAPK. The activated MAPK translocates to the nucleus to activate by phosphorylation proteins required for transcription of genes, including pro-inflammatory signaling molecules (Zheng and Guan, 1994). The NF-κB pathway is also activated by phosphorylation. In this case, it is activated by the IκB kinase kinase (IKK) complex, which in turn phosphorylates the α subunit of inhibitor of NF-κB (IκB), the cytoplasmic binding partner of NF-κB (Fig. 1). Phosphorylation of IκB leads to its poly-ubiquitination and proteolytic degradation, resulting in exposure of the NLS on the now unbound NF-κB (Alkalay et al., 1995). This promotes nuclear translocation of NF-κB and transcriptional activation of genes containing a κB site. Some NF-κB–dependent genes require an additional cofactor, the ribosomal protein S3 (RPS3), for their transcription (Wan et al., 2007). RPS3 nuclear translocation is promoted by IKKβ phosphorylation (Wan et al., 2011). The NF-κB pathway also activates genes involved in the pro-inflammatory response, and, possibly more importantly, genes involved in the antiapoptotic response (Arsura et al., 1996; Wu et al., 1996). Bacterial pathogens have devised a multitude of strategies to interfere with NF-κB signaling, often on more than one level, highlighting the importance of inhibiting this particular pathway for pathogen survival.
Manipulation of the NF-κB pathway
YopJ: an equal opportunity acetyltransferase.
Yersinia spp. has devised one of the most efficient strategies to date to disrupt the innate immune response and promote apoptosis in infected cells using one molecule, YopJ (Palmer et al., 1998, 1999). This 32-kD effector is injected from the pathogen directly into the host’s cytoplasm through a needle-like complex called type III secretion system (T3SS). YopJ (also termed YopP) blocks all the MAPK pathways and the NF-κB pathway by preventing the activation of all MKKs and IKKβ (but not IKKα; Fig. 1; Orth et al., 1999). Originally, bioinformatic tools identified YopJ as an effector that contained a catalytic domain similar to a cysteine protease (Orth et al., 2000). However, biochemical analysis of YopJ revealed that the protein requires an intact catalytic triad for its inhibitory acetyltransferase activity; a novel posttranslational modification that directly competes with phosphorylation. This effector inhibits kinase activation by modifying serine and/or threonine residues in the activation loop with an acetyl group from acetyl-CoA, thereby preventing their modification by phosphorylation (Orth et al., 1999; Trosky et al., 2004; Mittal et al., 2006; Mukherjee et al., 2006; Hao et al., 2008). During an infection with Yersinia, host cell signals for both survival and apoptosis are normally induced. However, with Yersinia strains containing YopJ, the default path will always be death because YopJ blocks the survival pathway. Using a yeast genetic screen, a conserved site on all MKKs and IKKβ, but not IKKα, was revealed to be the docking site for YopJ (Hao et al., 2008). As many signaling molecules are conserved in yeast, this genetically tractable system has become a very useful tool to characterize the molecular activity of bacterial effectors. The analysis of YopJ revealed Ser/Thr acetylation as a novel posttranslational modification and was one of the first examples to highlight the use of competitive posttranslational modifications for directing signaling (Mukherjee et al., 2006). Additionally, YopJ is an example of a protein that possesses an enzymatic activity different from what was predicted through bioinformatics, thereby demonstrating the limits of using any one tool for studying unknown signaling molecules.
Other homologues of YopJ have been analyzed but their targets are less clear. The Salmonella Typhimurium homologue AvrA seems to target different host proteins and play multiple roles that vary during the course of infection (Liu et al., 2010). For example, early on during infection, it inhibits NF-κB signaling (Collier-Hyams et al., 2002; Jones et al., 2008). During later stages of infection, it acetylates MKKs such as MKK7 to shut off the JNK pathway, which is activated early during S. Typhimurium infection (Du and Galán, 2009). Another YopJ homologue, the effector AopP from the fish-pathogen Aeromonas salmonicida, inhibits NF-κB signaling using the same catalytic mechanism on another phosphorylated target, possibly downstream of IκB phosphorylation (Fehr et al., 2006).
Binding NEMO: Ubiquitination by Shigella flexneri IpaH9.8 blocks NF-κB.
S. flexneri, an intracellular Gram-negative gastrointestinal pathogen capable of causing dysentery, blocks the NF-κB pathway on several levels to ensure silencing of immune responses. Upon invasion of epithelial cells, the NF-κB cascade would typically be activated when S. flexneri lipopolysaccharide binds to the cytoplasmic NLR NOD1, which causes enhanced oligomerization of NOD1 and recruitment of the serine/threonine kinase receptor-interacting protein 2/RIP-like interacting CLARP kinase (RIP2/RICK), and NF-κB essential modulator (NEMO), a regulatory subunit of the IKK complex (Girardin et al., 2001). However, S. flexneri possesses the effector IpaH9.8, a protein injected from the pathogen directly into the host’s cytoplasm through the T3SS and that possesses E3 ligase activity for NEMO. IpaH9.8 simultaneously binds NEMO and ABIN-1, an ubiquitin-binding adaptor protein, and promotes polyubiquitination of NEMO (Fig. 1). Polyubiquitinated NEMO undergoes proteolytic degradation, thus inhibiting NEMO-dependent activation of the IKK complex (Okuda et al., 2005; Rohde et al., 2007; Ashida et al., 2010). Interestingly, inhibition by IpaH9.8 is stronger in response to NOD1 than TNF, and further study may give insight to how upstream signals bias these pathways (Ashida et al., 2010).
In contrast, the S. flexneri T3SS-injected effector, OspG, blocks the cascade further downstream by preventing polyubiquitination and thus degradation of IκB, the inhibitor of NF-κB (Kim et al., 2005). OspG is a protein kinase with an unknown phosphorylation target that binds ubiquitinylated E2 ubiquitin-conjugating enzymes, including the E2 specific for IκB—UbcH5—and prevents the transfer of ubiquitin onto phosphorylated IκBα by an E3 ubiquitin ligase (Fig. 1). Inappropriate association of protein domains is a subversive mechanism used to redirect signaling systems, and has led to a greater understanding of many signaling pathways. Examples of this are seen with oncogenic viruses (v-abl), lethal somatic chromosomal translocations (Bcr-Abl; Hunter, 2007), bacterial extracellular molecules (such as fimbriae), and other pathogenic effectors (e.g., EspG).
Modulation of host defenses by extracellular scaffolding.
Porphyromonas gingivalis, a periopathogen, uses proteinaceous extracellular structures—fimbriae (Pierce et al., 2009)—to scaffold both the chemokine receptor 4 (CXCR 4) and the Toll-like receptor TLR2 on human monocytes together in lipid rafts. The crosstalk between CXCR4 and TLR2 results in elevated levels of cAMP, which acts to stimulate the immunomodulatory cAMP-dependent protein kinase A (PKA) and induce a negative feedback loop to suppress CXCR4 function. The authors hypothesize that PKA acts downstream of CXCR4 to inhibit TLR2-dependent, NF-κB–induced TNF activation (inflammatory cytokine), and trigger increased expression of the immunosuppressive cytokine IL-10. The ultimate outcome of this subtle but efficient manipulation of NF-κB signaling is diminished production of nitric oxide (NO), which is usually generated by macrophages as an inflammatory response and secreted as free radicals to fight off pathogens. Suppression of the inflammatory response and NO secretion results in prolonged survival of P. gingivalis (Hajishengallis et al., 2008). Thus, the pathogen uses an extracellular scaffolding protein to redirect signaling pathways.
Inhibition of the NF-κB pathway by EPEC/EHEC Nle proteins.
The binding affinity of the NF-κB p50/p65 heterodimer to DNA is greatly increased by the NF-κB binding partner RPS3 (Wan et al., 2007). This regulator of NF-κB allows specificity in expression of target genes, such as those involved in cytokine production and host defense against enterohemorrhagic Escherichia coli (EHEC). NleH1 and NleH2 are a pair of highly conserved T3SS effectors from both enteropathogenic E. coli (EPEC) and EHEC. These effectors contain a conserved Ser/Thr kinase domain and a PDZ-binding motif, and are implicated in blocking apoptosis during infection (Fig. 1; Hemrajani et al., 2010; Martinez et al., 2010). Both NleH1 and NleH2 are also capable of binding RPS3 independent of their kinase activity (Gao et al., 2009). Binding of EHEC NleH1 to RPS3 inhibits RPS3 phosphorylation by IKKβ, which sequesters RPS3 in the cytoplasm and prevents its nuclear translocation, thereby disrupting transcriptional up-regulation of RPS3-dependent genes (Gao et al., 2009). The presence of NleH1 leads to increased bacterial colonization and diarrhea but, interestingly, decreases mortality in a piglet infection model, a paradoxical effect that perhaps promotes the spread of the bacteria to other hosts (Wan et al., 2011). Studies of NleH1 have highlighted the essentiality of RPS3 phosphorylation by IKKβ and have shown how altering only the specificity of NF-kB through RPS3 inhibition compromises defenses and affects cytokine expression (Wan et al., 2011)
Another EPEC effector, NleE, blocks NF-κB activation by inhibition of IKKβ phosphorylation. The activity of NleE is enhanced by a second effector, NleB (Nadler et al., 2010; Vossenkämper et al., 2010). Recently, NleC and NleD, members of a family of Zn-dependent metalloproteases, were identified in EPEC. These proteases specifically cleave and inactivate RelA (p65), a subunit of NF-κB, and JNK, respectively, thus blocking NF-κB and activator protein 1 (AP-1) activation. The net result of the inhibition of NF-κB by these bacterial effectors is strong inhibition of the pro-inflammatory cytokine IL-8 (Baruch et al., 2011; Pearson et al., 2011; Yen et al., 2010).
Activation of the NF-kB pathway by pathogenic effectors.
Listeria monocytogenes has been reported to cause local inflammation at the primary site of infection to attract monocytes, which they invade and use as a vehicle to spread throughout the host and cause systemic infection (Gray and Killinger, 1966). Other pathogens induce NF-κB activation to trigger enhanced surface expression of pathogen receptors on host cells. Examples of this include Chlamydophila pneumoniae, which causes an NF-κB–dependent increase in monocyte chemotactic protein 1 (MCP-1) by endothelial cells (Molestina et al., 2000) or Neisseria gonorrhoeae, which uses the NF-κB pathway to up-regulate carcinoembryonic antigen-related cellular adhesion molecules (CEACAMs; Muenzner et al., 2002). Further reasons for NF-κB activation include permeabilization of the vasculature and superficial cell layers to enable pathogens to invade deeper tissue layers or the circulatory system, or, as in the case of S. Typhimurium, to trigger the reactive oxygen species (ROS)-promoted formation of a new respiratory chain, which provides the pathogen with a growth advantage over the commensal microbiota (Winter et al., 2010).
Some pathogens secrete effector proteins into the host specifically to evoke NF-κB activation. An example is the Legionella pneumophila type IV secreted effector LegK1, which is a Ser/Thr kinase that specifically phosphorylates IκB to induce its ubiquitination and degradation, allowing NF-κB to activate downstream pro-inflammatory responses (Fig. 1; Ge et al., 2009). Interestingly, LegK1 also appears to have an antiapoptotic effect. It can be speculated that a wide range of pathogens cause a well-timed burst of pro-inflammatory responses upon initial infection to exploit the benefits described earlier, only to switch it off at a later stage of infection using specific effector proteins that inhibit the NF-κB pathway. LegK1 is an IKK mimic that does not require phosphorylation of its activation loop and thereby is regulated independently of the host signaling machinery (Ge et al., 2009).
Manipulation of the MAPK pathway
Inhibition of antigen presentation by MAPK activation.
Interference by mycobacteria of MAPK signaling pathways by usurping p38 to interfere with CD1 surface expression plays a pivotal role in many aspects of immune modulation (Gagliardi et al., 2009). CD1 belongs to a family of surface glycoproteins, which are expressed on antigen-presenting cells (APCs) and are responsible for presenting lipid antigens such as mycobacterial cell wall components to specific T cells. Mycobacteria such as Mycobacterium tuberculosis and Mycobacterium bovis interact with complement receptor 3 (CR-3) on the surface of APCs, leading to CR-3–mediated phagocytosis (Schlesinger et al., 1990). CR-3 binding results in phosphorylation of the MAPK p38, which, in turn, leads to phosphorylation of the downstream effector activating transcription factor (ATF-2). Activated ATF-2 is able to bind to the CD1A promoter region and decrease CD1 transcription. As a result, CD1 expression and surface presentation is reduced. Thus, the phagocytosed pathogen’s induction of p38 prevents surface display of mycobacterial antigens on APCs by inhibition of CD1 expression, which might allow mycobacteria to evade T cell surveillance during infection in vivo (Gagliardi et al., 2009). The manipulation of these signaling pathways also results in arrest of phagosome maturation in macrophages (Jo et al., 2007) and inhibition of class II MHC expression by macrophages (Pennini et al., 2006). These studies highlight a mechanism that uses MAPK signaling to reduce T cell recognition (Gagliardi et al., 2009).
Targeting of MKKs by anthrax toxin.
Anthrax toxin, secreted by Bacillus anthracis, is another virulence factor that can inhibit MAPK signaling. It is a protease that reaches the host’s cytoplasm through lipid raft-mediated uptake and hydrolyses MKKs (Martchenko et al., 2010; Ali et al., 2011). B. anthracis also encodes a toxin called lethal factor (LF) that is a metalloproteinase. LF specifically cleaves the N-terminal extension of MKKs, resulting in a kinase that can no longer interact with its substrate (Duesbery et al., 1998; Vitale et al., 1998; Park et al., 2002). Importantly, this type of deregulation provided insight into the importance of scaffolding a kinase with a substrate in the MAPK signaling cascade and a potential target for a small molecule LF inhibitor (Joshi et al., 2011; Li et al., 2011).
VopA/P: YopJ’s more specific sibling.
VopA/P, a Vibrio parahaemolyticus effector, is different from its close homologue YopJ in that it can inhibit MAPK signaling pathways but not the NF-κB pathway (Fig. 1). Despite only inhibiting MAPK, it has a dual function in which it acetylates not only the activation loop of MKKs, thereby inhibiting activation, but also acetylates a conserved lysine in the catalytic loop of MKKs that is required for coordination of the γ-phosphate of ATP. This modification by VopA/P inhibits the binding of ATP (Trosky et al., 2004; Trosky et al., 2007). By analyzing homologues of different effectors, subtle mechanistic differences are observed that reveal dramatic differences in host cell responses. By inhibiting both NF-κB and MAPK pathways, Yersinia YopJ will promote apoptosis. In contrast to YopJ, by inhibiting only MAPK pathways, V. parahaemolyticus VopA/P permits survival by allowing activation of the NF-κB pathway.
Crosstalk between p38 and JNK revealed by OspF.
The phosphothreonine lyase OspF, a S. flexneri protein effector, is translocated into the host cell and localizes to the nucleus, where it eliminates a phosphate group from a phosphothreonine residue in the activation loop of the MAPKs ERK1/2 and p38 (Fig. 1; Li et al., 2007). This modification is irreversible and the kinase can no longer be activated by phosphorylation. OspF-catalyzed eliminylation inhibits extracellular signal–regulated kinase (ERK)-mediated activation of the mitogen- and stress-activated kinase 1 (MSK1) and MSK2, and thus prevents phosphorylation of histone H3, which is a prerequisite for chromatin reorganization and presentation of κB binding sites at NF-κB–regulated promoters (Brennan and Barford, 2009). The phosphothreonine lyase activity may be preferable to phosphatase activity because of the irreversible nature of the modification. As a consequence, transcriptional up-regulation of genes usually activated by NF-κB is blocked (Soloaga et al., 2003; Arbibe et al., 2007; Kramer et al., 2007). An unexpected side effect of irreversible p38 dephosphorylation is that JNK and NF-κB activity are actually increased, which suggests that inactivation of p38 may reduce negative feedback by the upstream activator Tak1, highlighting a layer of crosstalk between the pathways (Reiterer et al., 2011). Interestingly, at least some of the potential downstream effects of JNK and NF-κB activation by OspF are silenced, as OspF has the concomitant effect of reducing transcription of the JNK substrate c-jun, resulting in less phosphorylated c-jun and secreted IL-8 (Reiterer et al., 2011). It also reduces the accessibility of chromatin to transcription factors, perhaps silencing NF-κB activation. Silencing these potential drawbacks by irreversible deactivation of p38 by inhibiting transcription of downstream substrates shows an interesting mechanism that prevents a “backfire” result by targeting a central signaling pathway like MAPK.
Manipulation of host cytoskeleton and membranes by PI3K/Akt, p21-activated kinase (PAK), and other kinases
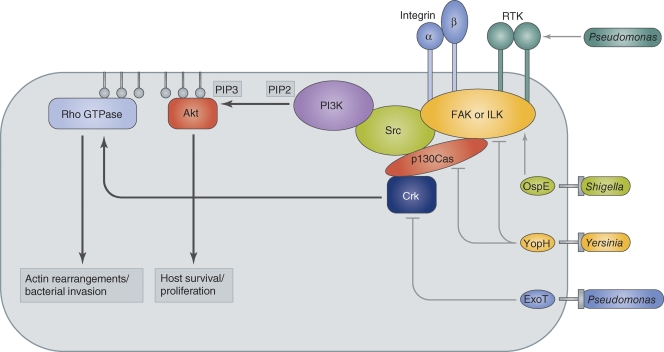
The phosphatidylinositol 3-kinase (PI3K)/Akt kinase signaling axis plays an important role in a variety of cellular processes, including cytoskeletal dynamics and migration as well as survival and proliferation (Hannigan et al., 1996; Penuel and Martin, 1999; Fayard et al., 2010). For this reason, the pathway is targeted by many pathogens to reinforce or destroy focal adhesions, which play an integral role in phagocytosis. The PI3K pathway can be triggered by activation of receptor tyrosine kinases or ligand binding to integrins, both leading to phosphorylation of downstream kinase targets (Fig. 2). Receptor activation leads to recruitment and activation of focal adhesion kinase (FAK) or integrin-linked kinase (ILK). These, in turn, recruit cytoplasmic components of the receptor-linked signaling complex, including the tyrosine kinase Src, the adapter protein Crk, and p130Cas (Crk-associated substrate). Crk phosphorylation leads to strong activation of the small GTPase Rac, which is required for cell motility. Src phosphorylation triggers the association of PI3K with the signaling complex. PI3K phosphorylates phosphoinositols at the D3 position and, depending on the substrate, generates either PI(3)P, PI(3,4)P2, or PI(3,4,5)P3. Generation of PI(3,4)P2 or PI(3,4,5)P3 recruits the Ser/Thr kinase Akt (also known as protein kinase B or PKB) to the membrane via its pleckstrin homology (PH) domain, which binds to these phosphoinositols with high affinity (Harlan et al., 1994). Akt regulates many cellular processes, including cell migration, proliferation, survival, and metabolism. Activation of PI3K can also activate small GTPases, such as Ras and Rac, which are linked to proliferation and cytoskeletal dynamics, respectively (Reedijk et al., 1990; Pleiman et al., 1993).
Figure 2.
Host signaling pathways exploited for bacterial invasion and colonization. Ligand-activated transmembrane receptors (integrins or receptor tyrosine kinases) recruit the cytoplasmic signaling proteins FAK or ILK. This in turn leads to binding of p130Cas and Crk, as well as Src and PI3K. PI3K converts PIP2 to PIP3, which binds the pleckstrin homology (PH) domain of Akt. Akt mediates host survival and proliferation signaling. PIP3 also recruits Rho GTPases to the membrane, where they activate actin rearrangements that lead to cellular motility or bacterial uptake through formation of membrane protrusions. Rho GTPases are also activated by Crk. P. aeruginosa activates PI3K signaling by integrin or RTK binding to promote cellular uptake. The S. flexneri T3SS-injected effector OspE activates PI3K signaling by recruiting ILK. Yersinia spp. and P. aeruginosa inject T3SS effectors YopH and ExoT to inhibit GTPase activation and phagocytosis (Yersinia spp.) or stop invasion at a later stage of infection (P. aeruginosa). Direct manipulation of small GTPases is a major target for a variety of bacterial effectors (Aktories et al., 2000; Alto, 2008).
YopH: Terminator of focal adhesion complexes.
Upon binding to macrophages, Yersinia spp. disables the phagocytosis machinery by introducing YopH, a tyrosine phosphatase injected through the T3SS (Rosqvist et al., 1988; Guan and Dixon, 1990; Bliska et al., 1991). Using a catalytically inactive mutant form as a substrate trap, YopH was shown to bind p130Cas and FAK, which are both members of the focal adhesion complex (Black and Bliska, 1997). YopH activity thereby disrupts focal adhesion complexes and inhibits phagocytosis, thus limiting the number of internalized bacteria (Mogemark et al., 2005). YopH also dephosphorylates Fyb (Fyn-binding protein), an immune cell–specific adaptor protein. This disrupts signaling via Fyn and PI3K to small GTPases to induce cytoskeletal rearrangements, which are required to initiate the phagocytic process (Yao et al., 1999; Yuan et al., 2005). Deletion of YopH severely decreases virulence of the mutant strains (Bliska et al., 1991). Thus, YopH targets and destroys focal adhesions by dephosphorylating key signaling molecules, and highlights the importance of focal adhesions in cell movement and phagocytosis.
Stabilization of the intestinal lining by S. flexneri.
Enteropathogenic S. flexneri bind to the intestinal mucosal layer and invade epithelial cells, where they spread from cell to cell to avoid any extracellular steps. This mode of infection provokes extensive tissue damage and would normally lead to shedding of epithelial cells. However, S. flexneri actively stabilizes the epithelial lining by promoting cell adherence. The T3SS effector OspE localizes to focal adhesion sites, where it recruits ILK to promote cell adhesion by inhibiting focal adhesion disassembly. Although ILK appears to act as a scaffold for OspE localization, mutation of its putative kinase residues leads to a reduction in focal adhesion formation. Despite the controversies on whether ILK is actually a kinase, OspE “activation” of ILK leads to an increase in the relative number of focal adhesions, thereby preventing exfoliation of infected mucosal epithelium (Miura et al., 2006; Kim et al., 2009). Several other enteropathogens, such as E. coli or S. Typhimurium, have been found to possess OspE homologues, and it is hypothesized that they use a similar mechanism for enhanced ILK recruitment to stabilize the intestinal lining during infection to promote bacterial dissemination (Van Nhieu and Guignot, 2009).
EspG, a catalytic … scaffold?
The EHEC T3SS effector EspG is another example of how bacteria can manipulate the host by “catalytic scaffolding.” Both ADP-ribosylation factor (ARF) GTPases and PAKs are substrates of EspG. Arf GTPases are mediators of intracellular membrane trafficking and organelle remodeling, whereas PAKs are Ser/Thr kinases that are activated by Cdc42 and Rac GTPases, thereby regulating actin remodeling and cellular polarity. EspG inhibits Arf GTPase activity by blocking GTPase-activating protein (GAP) binding and thus GTP hydrolysis, while at the same time acting as an allosteric activator of PAK. Both Arf and PAK interact with EspG simultaneously by binding to different surfaces of the effector protein. By recruiting and inactivating Arf, EspG localizes PAK activity to specific membrane compartments, thereby reprogramming processes such as cellular polarity, receptor trafficking, and protein secretion to the pathogen’s advantage (Germane and Spiller, 2011; Selyunin et al., 2011). The study of this pathogenic effector has provided insight into host mechanisms involved in intracellular trafficking.
The complex nature of Pseudomonas aeruginosa internalization.
P. aeruginosa is another example of a pathogen taking advantage of the interplay between kinase and GTPase signaling pathways to manipulate the cytoskeletal machinery and promote its own internalization. Recent studies with P. aeruginosa have revealed the potential interplay of several cytoskeleton-associated GTPases and kinases. Pielage et al. (2008) used an RNAi screen of Drosophila S2 cells to identify host factors required for P. aeruginosa invasion. The screen identified Abl tyrosine kinase, the adaptor protein Crk, the GTPases Rac1 and Cdc42, as well as PAK as components of a host signaling pathway leading to internalization of P. aeruginosa, and the pathways were subsequently validated by different complementary approaches. Interestingly, the P. aeruginosa T3SS effector ExoT takes control of the pathway after pathogen internalization by inhibiting pathogen-induced Abl-dependent Crk phosphorylation, presumably to block further invasion (Pielage et al., 2008). Internalization studies with P. aeruginosa demonstrated that the phagocytic nature of S2 cells make them ideal for use in identification of host molecules important for bacterial internalization using RNAi-mediated gene inactivation (Pielage et al., 2008). Their studies uncovered the diversity of host molecules required for successful invasion by intracellular bacteria.
P. aeruginosa also provides an interesting example of how the PI3K pathway can be exploited to promote bacterial tissue invasion. P. aeruginosa preferably attaches to and invades epithelial cell layers from the basolateral side. To gain entry from the exposed apical surface, it subverts the PI3K pathway by binding to an unknown receptor present in tight junctions, which recruits and activates PI3K. This triggers the generation of PIP3 at the apical membrane and recruitment of Akt, which in turn redirects trafficking of basolateral membrane components to the apical side by transcytosis. Consequently, PIP3-rich membrane protrusions form on the apical side and facilitate P. aeruginosa attachment and entry into the epithelial cell layer (Kierbel et al., 2007). Studies on this pathogen have thus revealed several points of crosstalk between kinase pathways and signaling cascades mediating either cytoskeletal dynamics or membrane trafficking, all exploited to achieve entry of P. aeruginosa into host cells. These studies also highlight the importance of cell polarity in dictating invasion, as it creates a suitable PIP3-rich microenvironment at the apical surface to allow bacterial entry into the host cell (Kierbel et al., 2007).
Conclusion
Studying the mechanisms of virulence that pathogens use to propagate in a host not only provides potential targets for treatment, but also often provides insight into the host molecules affected. Although some of the future benefits of knowledge gained about eukaryotic signaling may be elusive at this point, the potential for discovery is obvious. The complex and central kinase pathways outlined in this review highlight the importance of using novel angles of investigation. Seeing signaling pathways through the lens of a pathogen reveals unknown intricacies of kinase-mediated signaling pathways.
Acknowledgments
We thank the Orth laboratory for critical reading and comments on the manuscript. Illustrations were provided by Neil Smith, neil@neilsmithillustration.co.uk.
K. Orth, A.M. Krachler, and A.R. Woolery are supported by grants from National Institutes of Health, National Institute for Allergy and Infectious Disease (R01-AI056404 and R01-AI087808) and the Welch Foundation (I-1561). K. Orth is a Burroughs Wellcome Investigator in Pathogenesis of Infectious Disease and a W.W. Caruth Jr. Biomedical Scholar.
Footnotes
Abbreviations used in this paper:
- APC
- antigen-presenting cell
- EHEC
- enterohemorrhagic E. coli
- EPEC
- enteropathogenic E. coli
- ERK
- extracellular signal–regulated kinase
- FAK
- focal adhesion kinase
- IκB
- inhibitor of NF-κB
- IKK
- IκB kinase kinase
- ILK
- integrin-linked kinase
- LF
- lethal factor
- MKK
- MAP kinase kinase
- NF-κB
- nuclear factor κB
- PAK
- p21-activated kinase
- PAMP
- pathogen-associated molecular pattern
- PI3K
- phosphatidylinositol 3-kinase
- PRR
- pattern recognition receptor
- ssp
- subspecies
- T3SS
- type III secretion system
- TLR
- Toll-like receptor
References
- Aktories K., Schmidt G., Just I. 2000. Rho GTPases as targets of bacterial protein toxins. Biol. Chem. 381:421–426 10.1515/BC.2000.054 [DOI] [PubMed] [Google Scholar]
- Ali S.R., Timmer A.M., Bilgrami S., Park E.J., Eckmann L., Nizet V., Karin M. 2011. Anthrax toxin induces macrophage death by p38 MAPK inhibition but leads to inflammasome activation via ATP leakage. Immunity. 35:34–44 10.1016/j.immuni.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix E., Mukherjee S., Roy C.R. 2011. Subversion of membrane transport pathways by vacuolar pathogens. J. Cell Biol. 195:943–952 10.1083/jcb.201105019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkalay I., Yaron A., Hatzubai A., Orian A., Ciechanover A., Ben-Neriah Y. 1995. Stimulation-dependent I kappa B alpha phosphorylation marks the NF-kappa B inhibitor for degradation via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA. 92:10599–10603 10.1073/pnas.92.23.10599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto N.M. 2008. Mimicking small G-proteins: an emerging theme from the bacterial virulence arsenal. Cell. Microbiol. 10:566–575 10.1111/j.1462-5822.2007.01110.x [DOI] [PubMed] [Google Scholar]
- Arbibe L., Kim D.W., Batsche E., Pedron T., Mateescu B., Muchardt C., Parsot C., Sansonetti P.J. 2007. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 8:47–56 10.1038/ni1423 [DOI] [PubMed] [Google Scholar]
- Arsura M., Wu M., Sonenshein G.E. 1996. TGF beta 1 inhibits NF-kappa B/Rel activity inducing apoptosis of B cells: transcriptional activation of I kappa B alpha. Immunity. 5:31–40 10.1016/S1074-7613(00)80307-6 [DOI] [PubMed] [Google Scholar]
- Ashida H., Kim M., Schmidt-Supprian M., Ma A., Ogawa M., Sasakawa C. 2010. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat. Cell Biol. 12:66–73: 1–9 10.1038/ncb2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H., Mimuro H., Ogawa M., Kobayashi T., Sanada T., Kim M., Sasakawa C. 2011. Cell death and infection: A double-edged sword for host and pathogen survival. J. Cell Biol. 195:931–942 10.1083/jcb.201108081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch K., Gur-Arie L., Nadler C., Koby S., Yerushalmi G., Ben-Neriah Y., Yogev O., Shaulian E., Guttman C., Zarivach R., Rosenshine I. 2011. Metalloprotease type III effectors that specifically cleave JNK and NF-κB. EMBO J. 30:221–231 10.1038/emboj.2010.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B. 2009. Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases. Immunol. Rev. 227:248–263 10.1111/j.1600-065X.2008.00733.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar A.P., Guttman J.A., Finlay B.B. 2007. Manipulation of host-cell pathways by bacterial pathogens. Nature. 449:827–834 10.1038/nature06247 [DOI] [PubMed] [Google Scholar]
- Black D.S., Bliska J.B. 1997. Identification of p130Cas as a substrate of Yersinia YopH (Yop51), a bacterial protein tyrosine phosphatase that translocates into mammalian cells and targets focal adhesions. EMBO J. 16:2730–2744 10.1093/emboj/16.10.2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska J.B., Guan K.L., Dixon J.E., Falkow S. 1991. Tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc. Natl. Acad. Sci. USA. 88:1187–1191 10.1073/pnas.88.4.1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan D.F., Barford D. 2009. Eliminylation: a post-translational modification catalyzed by phosphothreonine lyases. Trends Biochem. Sci. 34:108–114 10.1016/j.tibs.2008.11.005 [DOI] [PubMed] [Google Scholar]
- Collier-Hyams L.S., Zeng H., Sun J., Tomlinson A.D., Bao Z.Q., Chen H., Madara J.L., Orth K., Neish A.S. 2002. Cutting edge: Salmonella AvrA effector inhibits the key proinflammatory, anti-apoptotic NF-kappa B pathway. J. Immunol. 169:2846–2850 [DOI] [PubMed] [Google Scholar]
- Du F., Galán J.E. 2009. Selective inhibition of type III secretion activated signaling by the Salmonella effector AvrA. PLoS Pathog. 5:e1000595 10.1371/journal.ppat.1000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesbery N.S., Webb C.P., Leppla S.H., Gordon V.M., Klimpel K.R., Copeland T.D., Ahn N.G., Oskarsson M.K., Fukasawa K., Paull K.D., Vande Woude G.F. 1998. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 280:734–737 10.1126/science.280.5364.734 [DOI] [PubMed] [Google Scholar]
- Fayard E., Xue G., Parcellier A., Bozulic L., Hemmings B.A. 2010. Protein kinase B (PKB/Akt), a key mediator of the PI3K signaling pathway. Curr. Top. Microbiol. Immunol. 346:31–56 10.1007/82_2010_58 [DOI] [PubMed] [Google Scholar]
- Fehr D., Casanova C., Liverman A., Blazkova H., Orth K., Dobbelaere D., Frey J., Burr S.E. 2006. AopP, a type III effector protein of Aeromonas salmonicida, inhibits the NF-kappaB signalling pathway. Microbiology. 152:2809–2818 10.1099/mic.0.28889-0 [DOI] [PubMed] [Google Scholar]
- Gagliardi M.C., Teloni R., Giannoni F., Mariotti S., Remoli M.E., Sargentini V., Videtta M., Pardini M., De Libero G., Coccia E.M., Nisini R. 2009. Mycobacteria exploit p38 signaling to affect CD1 expression and lipid antigen presentation by human dendritic cells. Infect. Immun. 77:4947–4952 10.1128/IAI.00607-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Wan F., Mateo K., Callegari E., Wang D., Deng W., Puente J., Li F., Chaussee M.S., Finlay B.B., et al. 2009. Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLoS Pathog. 5:e1000708 10.1371/journal.ppat.1000708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J., Xu H., Li T., Zhou Y., Zhang Z., Li S., Liu L., Shao F. 2009. A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc. Natl. Acad. Sci. USA. 106:13725–13730 10.1073/pnas.0907200106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germane K.L., Spiller B.W. 2011. Structural and functional studies indicate that the EPEC effector, EspG, directly binds p21-activated kinase. Biochemistry. 50:917–919 10.1021/bi1020138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin S.E., Tournebize R., Mavris M., Page A.L., Li X., Stark G.R., Bertin J., DiStefano P.S., Yaniv M., Sansonetti P.J., Philpott D.J. 2001. CARD4/Nod1 mediates NF-kappaB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2:736–742 10.1093/embo-reports/kve155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M.L., Killinger A.H. 1966. Listeria monocytogenes and listeric infections. Bacteriol. Rev. 30:309–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K.L., Dixon J.E. 1990. Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science. 249:553–556 10.1126/science.2166336 [DOI] [PubMed] [Google Scholar]
- Haglund C.M., Welch M.D. 2011. Pathogens and polymers: Microbe-host interactions illuminate the cytoskeleton. J. Cell Biol. 195:7–17 10.1083/jcb.201103148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G., Wang M., Liang S., Triantafilou M., Triantafilou K. 2008. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc. Natl. Acad. Sci. USA. 105:13532–13537 10.1073/pnas.0803852105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham H., Sreelatha A., Orth K. 2011. Manipulation of host membranes by bacterial effectors. Nat. Rev. Microbiol. 9:635–646 10.1038/nrmicro2602 [DOI] [PubMed] [Google Scholar]
- Hannigan G.E., Leung-Hagesteijn C., Fitz-Gibbon L., Coppolino M.G., Radeva G., Filmus J., Bell J.C., Dedhar S. 1996. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature. 379:91–96 10.1038/379091a0 [DOI] [PubMed] [Google Scholar]
- Hao Y.H., Wang Y., Burdette D., Mukherjee S., Keitany G., Goldsmith E., Orth K. 2008. Structural requirements for Yersinia YopJ inhibition of MAP kinase pathways. PLoS ONE. 3:e1375 10.1371/journal.pone.0001375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan J.E., Hajduk P.J., Yoon H.S., Fesik S.W. 1994. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature. 371:168–170 10.1038/371168a0 [DOI] [PubMed] [Google Scholar]
- Hemrajani C., Berger C.N., Robinson K.S., Marchès O., Mousnier A., Frankel G. 2010. NleH effectors interact with Bax inhibitor-1 to block apoptosis during enteropathogenic Escherichia coli infection. Proc. Natl. Acad. Sci. USA. 107:3129–3134 10.1073/pnas.0911609106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. 2007. Treatment for chronic myelogenous leukemia: the long road to imatinib. J. Clin. Invest. 117:2036–2043 10.1172/JCI31691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N., Koseki T., del Peso L., Hu Y., Yee C., Chen S., Carrio R., Merino J., Liu D., Ni J., Núñez G. 1999. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J. Biol. Chem. 274:14560–14567 10.1074/jbc.274.21.14560 [DOI] [PubMed] [Google Scholar]
- Jo E.K., Yang C.S., Choi C.H., Harding C.V. 2007. Intracellular signalling cascades regulating innate immune responses to Mycobacteria: branching out from Toll-like receptors. Cell. Microbiol. 9:1087–1098 10.1111/j.1462-5822.2007.00914.x [DOI] [PubMed] [Google Scholar]
- Jones R.M., Wu H., Wentworth C., Luo L., Collier-Hyams L., Neish A.S. 2008. Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe. 3:233–244 10.1016/j.chom.2008.02.016 [DOI] [PubMed] [Google Scholar]
- Joshi A., Kate S., Poon V., Mondal D., Boggara M.B., Saraph A., Martin J.T., McAlpine R., Day R., Garcia A.E., et al. 2011. Structure-based design of a heptavalent anthrax toxin inhibitor. Biomacromolecules. 12:791–796 10.1021/bm101396u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierbel A., Gassama-Diagne A., Rocha C., Radoshevich L., Olson J., Mostov K., Engel J. 2007. Pseudomonas aeruginosa exploits a PIP3-dependent pathway to transform apical into basolateral membrane. J. Cell Biol. 177:21–27 10.1083/jcb.200605142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.W., Lenzen G., Page A.L., Legrain P., Sansonetti P.J., Parsot C. 2005. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. USA. 102:14046–14051 10.1073/pnas.0504466102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Ogawa M., Fujita Y., Yoshikawa Y., Nagai T., Koyama T., Nagai S., Lange A., Fässler R., Sasakawa C. 2009. Bacteria hijack integrin-linked kinase to stabilize focal adhesions and block cell detachment. Nature. 459:578–582 10.1038/nature07952 [DOI] [PubMed] [Google Scholar]
- Kramer R.W., Slagowski N.L., Eze N.A., Giddings K.S., Morrison M.F., Siggers K.A., Starnbach M.N., Lesser C.F. 2007. Yeast functional genomic screens lead to identification of a role for a bacterial effector in innate immunity regulation. PLoS Pathog. 3:e21 10.1371/journal.ppat.0030021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B., Nicolas E., Michaut L., Reichhart J.M., Hoffmann J.A. 1996. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 86:973–983 10.1016/S0092-8674(00)80172-5 [DOI] [PubMed] [Google Scholar]
- Li H., Xu H., Zhou Y., Zhang J., Long C., Li S., Chen S., Zhou J.M., Shao F. 2007. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 315:1000–1003 10.1126/science.1138960 [DOI] [PubMed] [Google Scholar]
- Li F., Terzyan S., Tang J. 2011. Subsite specificity of anthrax lethal factor and its implications for inhibitor development. Biochem. Biophys. Res. Commun. 407:400–405 10.1016/j.bbrc.2011.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Lu R., Xia Y., Wu S., Sun J. 2010. Eukaryotic signaling pathways targeted by Salmonella effector protein AvrA in intestinal infection in vivo. BMC Microbiol. 10:326 10.1186/1471-2180-10-326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. 2002. The protein kinase complement of the human genome. Science. 298:1912–1934 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- Martchenko M., Jeong S.Y., Cohen S.N. 2010. Heterodimeric integrin complexes containing beta1-integrin promote internalization and lethality of anthrax toxin. Proc. Natl. Acad. Sci. USA. 107:15583–15588 10.1073/pnas.1010145107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E., Schroeder G.N., Berger C.N., Lee S.F., Robinson K.S., Badea L., Simpson N., Hall R.A., Hartland E.L., Crepin V.F., Frankel G. 2010. Binding to Na(+) /H(+) exchanger regulatory factor 2 (NHERF2) affects trafficking and function of the enteropathogenic Escherichia coli type III secretion system effectors Map, EspI and NleH. Cell. Microbiol. 12:1718–1731 10.1111/j.1462-5822.2010.01503.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C.A., Jr 2002. Decoding the patterns of self and nonself by the innate immune system. Science. 296:298–300 10.1126/science.1068883 [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Preston-Hurlburt P., Janeway C.A., Jr 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 388:394–397 10.1038/41131 [DOI] [PubMed] [Google Scholar]
- Mittal R., Peak-Chew S.Y., McMahon H.T. 2006. Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc. Natl. Acad. Sci. USA. 103:18574–18579 10.1073/pnas.0608995103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura M., Terajima J., Izumiya H., Mitobe J., Komano T., Watanabe H. 2006. OspE2 of Shigella sonnei is required for the maintenance of cell architecture of bacterium-infected cells. Infect. Immun. 74:2587–2595 10.1128/IAI.74.5.2587-2595.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogemark L., McGee K., Yuan M., Deleuil F., Fällman M. 2005. Disruption of target cell adhesion structures by the Yersinia effector YopH requires interaction with the substrate domain of p130Cas. Eur. J. Cell Biol. 84:477–489 10.1016/j.ejcb.2004.11.009 [DOI] [PubMed] [Google Scholar]
- Molestina R.E., Miller R.D., Lentsch A.B., Ramirez J.A., Summersgill J.T. 2000. Requirement for NF-kappaB in transcriptional activation of monocyte chemotactic protein 1 by Chlamydia pneumoniae in human endothelial cells. Infect. Immun. 68:4282–4288 10.1128/IAI.68.7.4282-4288.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenzner P., Billker O., Meyer T.F., Naumann M. 2002. Nuclear factor-kappa B directs carcinoembryonic antigen-related cellular adhesion molecule 1 receptor expression in Neisseria gonorrhoeae-infected epithelial cells. J. Biol. Chem. 277:7438–7446 10.1074/jbc.M108135200 [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Keitany G., Li Y., Wang Y., Ball H.L., Goldsmith E.J., Orth K. 2006. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 312:1211–1214 10.1126/science.1126867 [DOI] [PubMed] [Google Scholar]
- Nadler C., Baruch K., Kobi S., Mills E., Haviv G., Farago M., Alkalay I., Bartfeld S., Meyer T.F., Ben-Neriah Y., Rosenshine I. 2010. The type III secretion effector NleE inhibits NF-kappaB activation. PLoS Pathog. 6:e1000743 10.1371/journal.ppat.1000743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda J., Toyotome T., Kataoka N., Ohno M., Abe H., Shimura Y., Seyedarabi A., Pickersgill R., Sasakawa C. 2005. Shigella effector IpaH9.8 binds to a splicing factor U2AF(35) to modulate host immune responses. Biochem. Biophys. Res. Commun. 333:531–539 10.1016/j.bbrc.2005.05.145 [DOI] [PubMed] [Google Scholar]
- Orth K., Palmer L.E., Bao Z.Q., Stewart S., Rudolph A.E., Bliska J.B., Dixon J.E. 1999. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science. 285:1920–1923 10.1126/science.285.5435.1920 [DOI] [PubMed] [Google Scholar]
- Orth K., Xu Z., Mudgett M.B., Bao Z.Q., Palmer L.E., Bliska J.B., Mangel W.F., Staskawicz B., Dixon J.E. 2000. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science. 290:1594–1597 10.1126/science.290.5496.1594 [DOI] [PubMed] [Google Scholar]
- Palmer L.E., Hobbie S., Galán J.E., Bliska J.B. 1998. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-alpha production and downregulation of the MAP kinases p38 and JNK. Mol. Microbiol. 27:953–965 10.1046/j.1365-2958.1998.00740.x [DOI] [PubMed] [Google Scholar]
- Palmer L.E., Pancetti A.R., Greenberg S., Bliska J.B. 1999. YopJ of Yersinia spp. is sufficient to cause downregulation of multiple mitogen-activated protein kinases in eukaryotic cells. Infect. Immun. 67:708–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.M., Greten F.R., Li Z.W., Karin M. 2002. Macrophage apoptosis by anthrax lethal factor through p38 MAP kinase inhibition. Science. 297:2048–2051 10.1126/science.1073163 [DOI] [PubMed] [Google Scholar]
- Pearson J.S., Riedmaier P., Marchès O., Frankel G., Hartland E.L. 2011. A type III effector protease NleC from enteropathogenic Escherichia coli targets NF-κB for degradation. Mol. Microbiol. 80:219–230 10.1111/j.1365-2958.2011.07568.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennini M.E., Pai R.K., Schultz D.C., Boom W.H., Harding C.V. 2006. Mycobacterium tuberculosis 19-kDa lipoprotein inhibits IFN-gamma-induced chromatin remodeling of MHC2TA by TLR2 and MAPK signaling. J. Immunol. 176:4323–4330 [DOI] [PubMed] [Google Scholar]
- Penuel E., Martin G.S. 1999. Transformation by v-Src: Ras-MAPK and PI3K-mTOR mediate parallel pathways. Mol. Biol. Cell. 10:1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielage J.F., Powell K.R., Kalman D., Engel J.N. 2008. RNAi screen reveals an Abl kinase-dependent host cell pathway involved in Pseudomonas aeruginosa internalization. PLoS Pathog. 4:e1000031 10.1371/journal.ppat.1000031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce D.L., Nishiyama S., Liang S., Wang M., Triantafilou M., Triantafilou K., Yoshimura F., Demuth D.R., Hajishengallis G. 2009. Host adhesive activities and virulence of novel fimbrial proteins of Porphyromonas gingivalis. Infect. Immun. 77:3294–3301 10.1128/IAI.00262-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiman C.M., Clark M.R., Gauen L.K., Winitz S., Coggeshall K.M., Johnson G.L., Shaw A.S., Cambier J.C. 1993. Mapping of sites on the Src family protein tyrosine kinases p55blk, p59fyn, and p56lyn which interact with the effector molecules phospholipase C-gamma 2, microtubule-associated protein kinase, GTPase-activating protein, and phosphatidylinositol 3-kinase. Mol. Cell. Biol. 13:5877–5887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedijk M., Liu X.Q., Pawson T. 1990. Interactions of phosphatidylinositol kinase, GTPase-activating protein (GAP), and GAP-associated proteins with the colony-stimulating factor 1 receptor. Mol. Cell. Biol. 10:5601–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiterer V., Grossniklaus L., Tschon T., Kasper C.A., Sorg I., Arrieumerlou C. 2011. Shigella flexneri type III secreted effector OspF reveals new crosstalks of proinflammatory signaling pathways during bacterial infection. Cell. Signal. 23:1188–1196 10.1016/j.cellsig.2011.03.006 [DOI] [PubMed] [Google Scholar]
- Rohde J.R., Breitkreutz A., Chenal A., Sansonetti P.J., Parsot C. 2007. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 1:77–83 10.1016/j.chom.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Rosqvist R., Bölin I., Wolf-Watz H. 1988. Inhibition of phagocytosis in Yersinia pseudotuberculosis: a virulence plasmid-encoded ability involving the Yop2b protein. Infect. Immun. 56:2139–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruco L.P., Meltzer M.S., Rosenstreich D.L. 1978. Macrophage activation for tumor cytotoxicity: control of macrophage tumoricidal capacity by the LPS gene. J. Immunol. 121:543–548 [PubMed] [Google Scholar]
- Schlesinger L.S., Bellinger-Kawahara C.G., Payne N.R., Horwitz M.A. 1990. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J. Immunol. 144:2771–2780 [PubMed] [Google Scholar]
- Selyunin A.S., Sutton S.E., Weigele B.A., Reddick L.E., Orchard R.C., Bresson S.M., Tomchick D.R., Alto N.M. 2011. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature. 469:107–111 10.1038/nature09593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloaga A., Thomson S., Wiggin G.R., Rampersaud N., Dyson M.H., Hazzalin C.A., Mahadevan L.C., Arthur J.S. 2003. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 22:2788–2797 10.1093/emboj/cdg273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trosky J.E., Mukherjee S., Burdette D.L., Roberts M., McCarter L., Siegel R.M., Orth K. 2004. Inhibition of MAPK signaling pathways by VopA from Vibrio parahaemolyticus. J. Biol. Chem. 279:51953–51957 10.1074/jbc.M407001200 [DOI] [PubMed] [Google Scholar]
- Trosky J.E., Li Y., Mukherjee S., Keitany G., Ball H., Orth K. 2007. VopA inhibits ATP binding by acetylating the catalytic loop of MAPK kinases. J. Biol. Chem. 282:34299–34305 10.1074/jbc.M706970200 [DOI] [PubMed] [Google Scholar]
- Uliczka F., Kornprobst T., Eitel J., Schneider D., Dersch P. 2009. Cell invasion of Yersinia pseudotuberculosis by invasin and YadA requires protein kinase C, phospholipase C-gamma1 and Akt kinase. Cell. Microbiol. 11:1782–1801 10.1111/j.1462-5822.2009.01371.x [DOI] [PubMed] [Google Scholar]
- Van Nhieu G.T., Guignot J. 2009. When Shigella tells the cell to hang on. J Mol Cell Biol. 1:64–65 10.1093/jmcb/mjp013 [DOI] [PubMed] [Google Scholar]
- Vitale G., Pellizzari R., Recchi C., Napolitani G., Mock M., Montecucco C. 1998. Anthrax lethal factor cleaves the N-terminus of MAPKKs and induces tyrosine/threonine phosphorylation of MAPKs in cultured macrophages. Biochem. Biophys. Res. Commun. 248:706–711 10.1006/bbrc.1998.9040 [DOI] [PubMed] [Google Scholar]
- Vossenkämper A., Marchès O., Fairclough P.D., Warnes G., Stagg A.J., Lindsay J.O., Evans P.C., Luong A., Croft N.M., Naik S., et al. 2010. Inhibition of NF-κB signaling in human dendritic cells by the enteropathogenic Escherichia coli effector protein NleE. J. Immunol. 185:4118–4127 10.4049/jimmunol.1000500 [DOI] [PubMed] [Google Scholar]
- Wan F., Anderson D.E., Barnitz R.A., Snow A., Bidere N., Zheng L., Hegde V., Lam L.T., Staudt L.M., Levens D., et al. 2007. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 131:927–939 10.1016/j.cell.2007.10.009 [DOI] [PubMed] [Google Scholar]
- Wan F., Weaver A., Gao X., Bern M., Hardwidge P.R., Lenardo M.J. 2011. IKKβ phosphorylation regulates RPS3 nuclear translocation and NF-κB function during infection with Escherichia coli strain O157:H7. Nat. Immunol. 12:335–343 10.1038/ni.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrauch Y., Zychlinsky A. 1999. The induction of apoptosis by bacterial pathogens. Annu. Rev. Microbiol. 53:155–187 10.1146/annurev.micro.53.1.155 [DOI] [PubMed] [Google Scholar]
- Winter S.E., Thiennimitr P., Winter M.G., Butler B.P., Huseby D.L., Crawford R.W., Russell J.M., Bevins C.L., Adams L.G., Tsolis R.M., et al. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 467:426–429 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Lee H., Bellas R.E., Schauer S.L., Arsura M., Katz D., FitzGerald M.J., Rothstein T.L., Sherr D.H., Sonenshein G.E. 1996. Inhibition of NF-kappaB/Rel induces apoptosis of murine B cells. EMBO J. 15:4682–4690 [PMC free article] [PubMed] [Google Scholar]
- Yao T., Mecsas J., Healy J.I., Falkow S., Chien Y. 1999. Suppression of T and B lymphocyte activation by a Yersinia pseudotuberculosis virulence factor, yopH. J. Exp. Med. 190:1343–1350 10.1084/jem.190.9.1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H., Ooka T., Iguchi A., Hayashi T., Sugimoto N., Tobe T. 2010. NleC, a type III secretion protease, compromises NF-κB activation by targeting p65/RelA. PLoS Pathog. 6:e1001231 10.1371/journal.ppat.1001231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Deleuil F., Fällman M. 2005. Interaction between the Yersinia tyrosine phosphatase YopH and its macrophage substrate, Fyn-binding protein, Fyb. J. Mol. Microbiol. Biotechnol. 9:214–223 10.1159/000089649 [DOI] [PubMed] [Google Scholar]
- Zheng C.F., Guan K.L. 1994. Cytoplasmic localization of the mitogen-activated protein kinase activator MEK. J. Biol. Chem. 269:19947–19952 [PubMed] [Google Scholar]