Desmogleins and desmocollins are transported to the plasma membrane by different kinesin motors, providing a potential mechanism to tailor desmosome structure and function during development and epithelial remodeling.
Abstract
The desmosomal cadherins, desmogleins (Dsgs) and desmocollins (Dscs), comprise the adhesive core of intercellular junctions known as desmosomes. Although these adhesion molecules are known to be critical for tissue integrity, mechanisms that coordinate their trafficking into intercellular junctions to regulate their proper ratio and distribution are unknown. We demonstrate that Dsg2 and Dsc2 both exhibit microtubule-dependent transport in epithelial cells but use distinct motors to traffic to the plasma membrane. Functional interference with kinesin-1 blocked Dsg2 transport, resulting in the assembly of Dsg2-deficient junctions with minimal impact on distribution of Dsc2 or desmosomal plaque components. In contrast, inhibiting kinesin-2 prevented Dsc2 movement and decreased its plasma membrane accumulation without affecting Dsg2 trafficking. Either kinesin-1 or -2 deficiency weakened intercellular adhesion, despite the maintenance of adherens junctions and other desmosome components at the plasma membrane. Differential regulation of desmosomal cadherin transport could provide a mechanism to tailor adhesion strength during tissue morphogenesis and remodeling.
Introduction
Multicellular organisms depend on intercellular junctions—gap junctions, tight junctions, desmosomes, and adherens junctions—to physically and chemically link cells within a tissue. The coordinated assembly of these multiprotein complexes at the plasma membrane is essential for establishment and maintenance of epithelial polarity and tissue integrity during embryogenesis and in the adult. Defects in junction assembly and structure lead to human inherited and acquired disorders (Takeichi, 1995; Gumbiner, 1996; Nollet et al., 1999; Lai-Cheong et al., 2007). Despite their central importance in development and disease, surprisingly little is known about specific mechanisms driving plasma membrane targeting of the transmembrane building blocks of intercellular junctions.
One key question is how different transmembrane proteins destined for the same junction are synthesized, trafficked, and assembled into a single, complex, highly ordered structure. A good example of this problem is seen with desmosomes, whose correct assembly and function are critical for cell–cell integration in tissues that experience mechanical stress, such as skin and heart (Lai-Cheong et al., 2007). As with adherens junctions, desmosomal adhesion is mediated by members of the cadherin family (Garrod et al., 2002; Dusek et al., 2007). Although adherens junctions typically contain a single classical cadherin that anchors actin microfilaments to the membrane through a series of adapter proteins, desmosomes contain two cadherin types, desmogleins (Dsgs) and desmocollins (Dscs), which link intermediate filaments to the cell surface (Koch and Franke, 1994; Garrod et al., 2002; Dusek et al., 2007; Green et al., 2010). Both Dsgs and Dscs are required to confer adhesive properties on normally nonadherent cells, and both are required for normal desmosome function (Kowalczyk et al., 1996; Marcozzi et al., 1998; Tselepis et al., 1998; Getsios et al., 2004). However, the molecular machinery responsible for driving Dscs and Dsgs from a vesicular compartment to the plasma membrane and the extent to which these mechanisms are shared by the two types of desmosomal cadherin are unknown.
Microtubule (MT)-based motor proteins in the kinesin superfamily support vesicular transport toward the cell membrane (Hirokawa et al., 1991; Vale, 2003; Verhey and Hammond, 2009). Previous studies suggest that kinesins interact with classical cadherins and their associated binding partners. For instance, conditional knockout of KAP3, the nonmotor accessory subunit of kinesin-2, results in a decrease in levels of N-cadherin and β-catenin at cell–cell contacts in embryonic mouse neural precursors (Teng et al., 2005). An increase in cytoplasmic staining of N-cadherin was reported, without changes in overall expression, suggesting a defect in transport of N-cadherin to the cell surface. In another example, kinesin-1 was reported to interact with the N-terminal head domain of p120 catenin (Chen et al., 2003; Yanagisawa et al., 2004). In cells expressing wild-type p120, but not a kinesin binding-deficient mutant, endogenous kinesin-1 is recruited to vesicles containing classical cadherin to transport them to the plasma membrane. The p120-related molecule p0071 (plakophilin-4) has also been shown to interact with the kinesin-2 subunit KIF3B (Keil et al., 2009).
In the case of desmosomes, Dsgs and Dscs are synthesized as soluble proteins that subsequently become insoluble, followed by their transport to cell–cell contacts (Pasdar and Nelson, 1989; Gloushankova et al., 2003) and the development of cell–cell adhesion (Mattey et al., 1990) through homophilic or heterophilic interactions (Chitaev and Troyanovsky, 1997; Garrod and Chidgey, 2008; Nie et al., 2011). Early studies of calcium-mediated desmosome formation showed that desmosomal cadherins have different distributions during junction formation (Watt et al., 1984), and Dscs may initiate assembly of desmosomes, whereas Dsgs arrive later to stabilize the complex (Burdett and Sullivan, 2002). Data also support the idea that desmosomal cadherin transport to the plasma membrane is MT dependent (Pasdar et al., 1991), but the molecular machinery required for transporting vesicles containing desmosomal cadherins to intercellular junctions is unknown. Here, we show that the temporal and spatial coordination of Dsg2 and Dsc2 assembly into intercellular junctions is controlled by distinct mechanisms involving different kinesin motors. Although the Dsg2 cytoplasmic tail associates specifically with the kinesin-1 (KIF5B), the Dsc2 tail associates with kinesin-2. Furthermore, RNAi-mediated depletion of kinesin-1 or kinesin-2 blocked transport of Dsg2 and Dsc2, respectively, with little or no effect on the desmosomal cadherin counterpart. Genetic interference with either motor protein results in epithelial cell sheets with considerably weakened intercellular adhesion caused by the loss of the respective cadherin from intercellular contact sites. Separation of the trafficking pathways governing Dsc2 and Dsg2 exocytosis provides a potential mechanism to tailor desmosome structure and function during development and epithelial remodeling.
Results
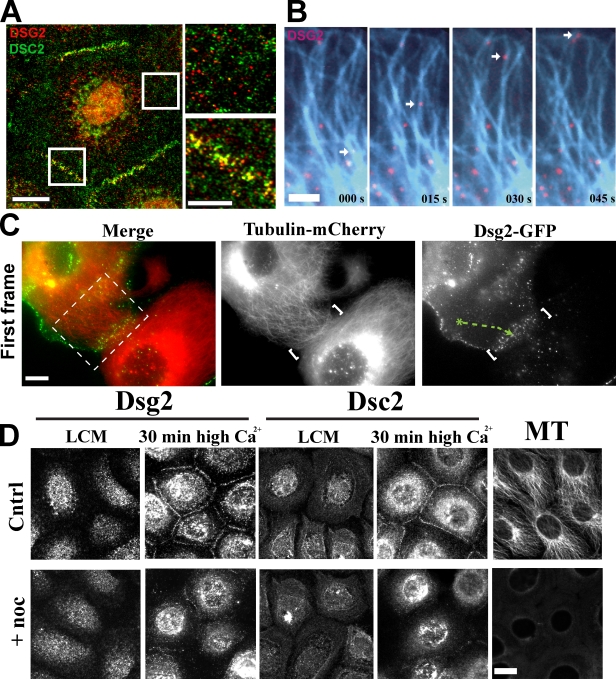
Dsg2 and Dsc2 exhibit distinct cellular distributions and require MTs for rapid accumulation at intercellular junctions
A previous study suggested that Dsc2- and Dsg2-containing vesicles exhibit different transport kinetics during calcium-induced desmosome formation, but the underlying basis for this difference was not addressed (Burdett and Sullivan, 2002). To determine the spatial and temporal behavior of Dsg/Dsc vesicular carriers, we analyzed the distribution of endogenous cadherins 30 min after a Ca2+ switch to trigger intercellular junction formation. At this time point, nascent desmosomes begin to appear at cell–cell interfaces, but intracellular desmosomal cadherins have not yet cleared the cytoplasm. Immunofluorescence analysis revealed that a proportion of Dsg2 and Dsc2 colocalized at cell–cell junctions as expected; however, Dsg2 and Dsc2 were frequently observed to be distributed in distinct particles within the cytoplasm (Fig. 1 A). Time-lapse imaging of Scc9 cells coexpressing Dsg2-mCherry and Dsc2-GFP revealed directly that Dsg2-mCherry and Dsc2-GFP localize to distinct vesicle populations that move independently in living cells (Video 1).
Figure 1.
Desmosomal cadherins require MTs for rapid accumulation at intercellular junctions. (A) Dual-label immunofluorescence revealed that Dsg2 and Dsc2 colocalize at cell–cell junctions but are present in separate cytoplasmic vesicles. Boxes indicate areas of magnification on the right. (B) Cells coexpressing Dsg2-GFP and tubulin-mCherry (blue) were imaged at 5-s intervals (Video 2). Dsg2-containing vesicles move along MTs toward the plasma membrane. (C) Contacting cells expressing Dsg2-GFP and tubulin-mCherry were imaged at 5-s intervals. Video 3 shows the cropped area in the left image. White brackets indicate cell–cell contact. Green track shows the path taken by a Dsg2-containing vesicle to the contact site. (D) Scc9s were switched to low-calcium medium (LCM) for 2 h before incubation with nocodazole for 1 h in low-calcium medium and then switched to normal Ca2+ (with or without nocodazole) for 30 min to trigger junction assembly. Cytoplasmic Dsg2 and Dsc2 vesicles are present in control (Cntrl) and nocodazole (noc)-treated cells in low-calcium medium. Disruption of MT delays Dsg2 and Dsc2 assembly at newly forming cell–cell interfaces. Bars: (A [left] and D) 20 µm; (A [right] and C) 10 µm; (B) 5 µm.
As an earlier study suggested that Dsg may be transported in an MT-dependent manner (Pasdar et al., 1991), we coexpressed Dsg2-GFP and tubulin-mCherry and performed time-lapse imaging. Analysis of these images revealed Dsg2-GFP particles moving along MT tracks toward to the plasma membrane (Fig. 1 B and Video 2) and to sites of nascent desmosome assembly in contacting cells (Fig. 1 C and Video 3). Accumulation of Dsg2 and Dsc2 proteins at cell–cell junctions was significantly inhibited in Scc9 cells treated with the MT antagonist nocodazole (10 µM) for 1 h before a 30 min Ca2+ switch compared with DMSO-treated controls (Fig. 1 D). Only after 1.5 h post-Ca2+ switch did both desmosomal cadherins begin to appear at the sites of cell–cell contact compared with 30 min for the control population (unpublished data). Together, these data show that Dsg2 and Dsc2 exhibit long-range MT-dependent trafficking in separate vesicles before reaching cell–cell interfaces where they incorporate into desmosomes.
Kinesin-1 is required for Dsg2, but not Dsc2, transport
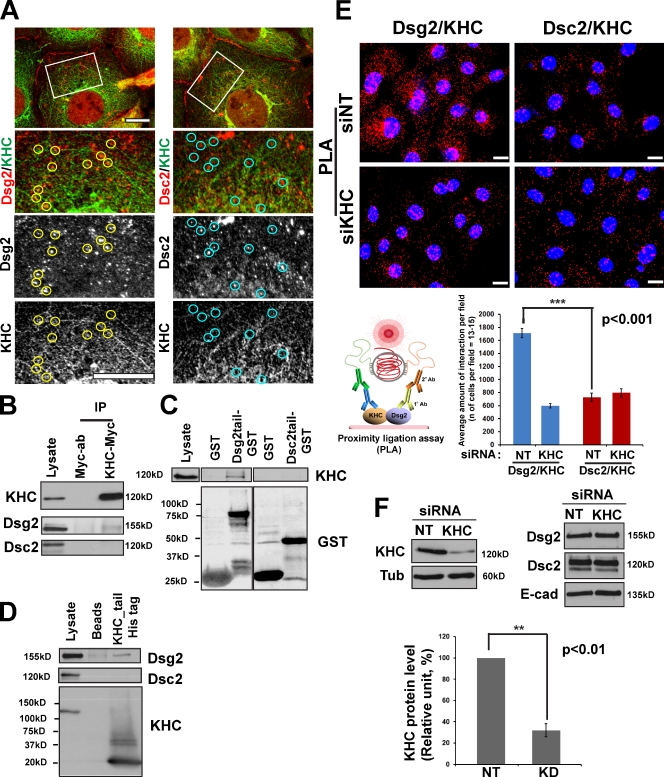
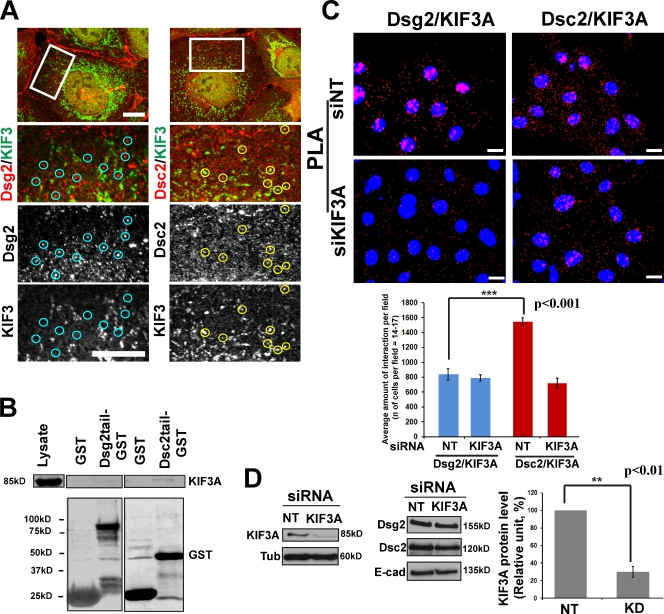
Kinesins comprise a large superfamily of motor proteins that drive vesicle transport along MTs, mostly toward the cell periphery (Vale, 2003; Hirokawa and Takemura, 2005). Kinesin-1, or conventional kinesin, is abundantly expressed in many cell types (Vale et al., 1985; Hirokawa et al., 1989) and was shown to be involved in vesicle transport from the trans-Golgi to the plasma membrane (Hirokawa et al., 1991; Lippincott-Schwartz et al., 1995; Woźniak and Allan, 2006; Jaulin et al., 2007). Kinesin-1 was also shown to play an important role in accumulation of N-cadherin at cell–cell junctions in fibroblasts (Chen et al., 2003). Immunofluorescence analysis of endogenous kinesin-1 heavy chain (KHC) and Dsg2 in Scc9 cells revealed partial colocalization of these proteins (Fig. 2 A). In addition, endogenous Dsg2, but not Dsc2, coimmunoprecipitated with ectopically expressed Myc-tagged KHC (Fig. 2 B) in Scc9 keratinocytes, demonstrating a selective association between Dsg2 and KHC. Furthermore, a recombinant Dsg2 cytoplasmic tail fused to GST, but not Dsc2-GST, pulled down endogenous KHC from Scc9 and A431 cell lysates (Fig. 2 C and Fig. S2 A). Finally, recombinant His-tagged KHC tail (KHC tail–His) associated with Dsg2, but not Dsc2, in keratinocyte cell lysates (Fig. 2 D). Together, these data support the idea that the cytoplasmic domain of Dsg2 specifically associates with the KHC tail, a region known to mediate binding of motors to vesicular cargo. To further examine whether endogenous Dsg2 and KHC interact in cells, we performed a proximity ligation assay (PLA), which serves as a reporter of proteins in a complex (see Materials and methods). In brief, primary antibodies to proteins of interest are derivatized with short nucleotide stretches that, when two molecules are within 20–100 nm of each other, form a closed DNA circle. The circle acts as a template for rolling circle amplification, incorporating fluorescently labeled nucleotides. Red fluorescent spots in cells indicate sites where Dsg2 and KHC interact in situ (Fig. 2 E). The interaction was abrogated by silencing KHC using siRNA oligonucleotides (oligos). Knockdown of KHC resulted in ∼70% reduction in the level of KHC without having a measurable effect on the total level of desmosomal or classical cadherin proteins (Fig. 2 F and Fig. S2 B).
Figure 2.
Dsg2 associates with kinesin-1. (A) Dual-label confocal microscopy showing Dsg2 (left) and Dsc2 (right) with KHC. Under these experimental conditions, the KHC antibody results in a combination of particulate and fibrous staining, the latter likely representing MTs. A subset of Dsg2-positive particles colocalize with KHC (yellow circles), whereas Dsc2-positive particles do not colocalize with KHC (blue circles). Boxes showed the zoomed areas. Bar, 10 µm. (B) Myc-tagged KIF5B was transfected into Scc9 cells, subjected to immunoprecipitation, and immunoblotted for Dsg2 and Dsc2. Dsg2, but not Dsc2, associated with KHC-Myc. (C) Recombinant Dsg2 cytoplasmic tail–GST or Dsc2 cytoplasmic tail–GST were incubated with Scc9 cell lysates. Retained proteins were immunoblotted for the presence of KHC. KHC associates with Dsg2 tail–GST but not Dsc2 tail–GST. (D) Recombinant His-tagged KHC tail was incubated with Scc9 cell lysates, and retained proteins were tested for the presence of desmosomal cadherins. Dsg2, but not Dsc2, associates with the KHC tail. (E) Proximity ligation assay (PLA) was performed as described in Materials and methods (bottom left) using primary antibodies directed against Dsg2 or Dsc2 coupled with KHC antibodies on nontargeting (NT) siRNA (siNT; top) or siKHC-treated Scc9 cells (middle). Red-labeled particles indicating protein complex formation were quantified (bottom right) to show significant Dsg2–KHC interactions compared with Dsc2–KHC (number of cells per condition = ∼195; ***, P < 0.001), which were abrogated upon KHC knockdown. Bars, 20 µm. (F) Level of KHC knockdown (KD) in PLA experiment shown above, representative of five experiments quantified below (**, P < 0.01). Protein expression levels of Dsg2, Dsc2, and E-cadherin (E-cad) shown in the top right do not change. Error bars represent SEM. Ab, antibody; Tub, tubulin.
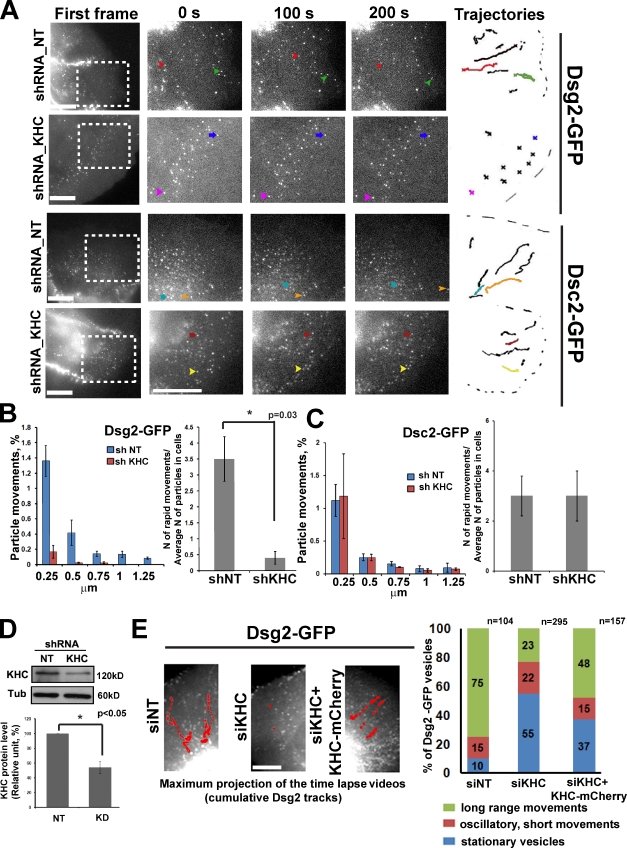
To test whether kinesin-1 drives Dsg2 within the cell, we doubly transfected Scc9 cells with Dsg2-GFP or Dsc2-GFP and short hairpin RNA (shRNA) targeting the same KHC sequences present in the siRNA oligos, in tandem with a fluorescent mCherry marker. Cadherin behavior was assessed 3 d after KHC knockdown at the cell periphery by time-lapse microscopy. This experiment and those that follow were performed under steady-state conditions (i.e., normal growth media and 1.2 mM Ca). shRNA-mediated silencing resulted in ∼50% knockdown of KHC in the cell population (Fig. 3 D); only those cells receiving the silencing vector were analyzed. Long-range Dsg2-GFP transport was completely blocked in KHC-deficient cells (Fig. 3, A and B and Video 4) compared with control cells. Dsc2-GFP dynamics were not inhibited, consistent with the idea that Dsc2 does not require kinesin-1 for its transport (Fig. 3, A and C; and Video 4). A dominant-negative (DN) mutant of KHC (KIF5B tail only) in Scc9 cells similarly impaired Dsg2 vesicle movement (Fig. S1, A–C; and Video 5).
Figure 3.
KHC knockdown blocked movement of Dsg2 but not Dsc2. (A) First frames show areas (boxes) analyzed by live-cell imaging in control or KHC-deficient cells 3 d after transfection (Video 4). The three magnified columns highlight selected vesicles (arrows) at three time points. Vesicle trajectories taken from the analyzed area are shown on the right, where colored tracks represent the movement of selected vesicles. Bars, 20 µm. (B) Bar graphs show quantification of distribution of particle movements (equal to instantaneous velocity; for each set of bar P < 0.05) and the ratio of rapid movements to the average number of analyzed particles (15–20 particles per cell) in each cell (three to four cells were analyzed per experiment) for Dsg2 in control and knockdown (KD) conditions (*, P = 0.03 for the ratio of rapid movements). In kinesin-1–deficient cells, the number of particle movements and the ratio of rapid movements for Dsg2 were decreased compared with controls. shNT, shRNA nontargeting. (C) Distribution of particle movements and the ratio of rapid movements to the average amount of analyzed particles for Dsc2 in KHC knockdown cells are similar to the control (P = 0.9). (D) Immunoblot showing level of KHC knockdown (∼50%, n = 3; *, P < 0.05). α-Tubulin (Tub) is the loading control. (E) Maximum projection of time-lapse analysis of cells into which cDNAs encoding Dsg2-GFP or Dsc2-GFP (not depicted) were coinjected with or without KHC cDNA into the nuclei of nontargeting (NT) siRNA (siNT) or siKHC-silenced cells (Video 6). Representative trajectories of Dsg2-GFP movements are shown in red. Bar, 5 µm. Bar graph on right is a population analysis showing particles engaging in long-range, short-range, or stationary movements as a percentage of Dsg2-GFP vesicles. The percentage of long-range movements is partially restored by the KHC-mCherry rescue construct (*, P < 0.05). n above the bars represents the number of vesicle trajectories that have been analyzed. Number of cells that been analyzed: siRNA nontargeting treated = 7, siKHC treated = 9, and siKHC with KHC-mCherry coinjection = 7. Error bars represent SEM.
To ensure that the observed defects in Dsg2 trafficking were specifically caused by loss of kinesin-1, we performed a rescue experiment in which cDNAs encoding Dsg2-GFP or Dsc2-GFP were coinjected with or without a cDNA encoding KHC fused with fluorescent mCherry (mCherry-KHC) into the nuclei of silenced cells. In control cells, newly synthesized Dsg2-GFP exited from the Golgi, and the majority of particles engaged in rapid long-range transport (Fig. 3 E and Video 6). In siRNA KHC (siKHC)–treated cells injected with desmosomal cadherin-GFP constructs only, the percentage of particles that exited from the Golgi and engaged in long-range movements was reduced from 75 to 23%, whereas the percentage of stationary particles increased from 10 to 55% (Fig. 3 E). However, in Dsg2-GFP/mCherry-KHC coinjected cells, long-range movements were restored up to 48%, and the stationary population dropped to 37%, a significant recovery. A smaller proportion of vesicles engaged in short oscillatory movements, and the percentage of these did not change significantly. Dsc2-GFP behavior was not altered by these treatments (unpublished data).
Kinesin-1 is essential for normal Dsg2 accumulation in desmosomes
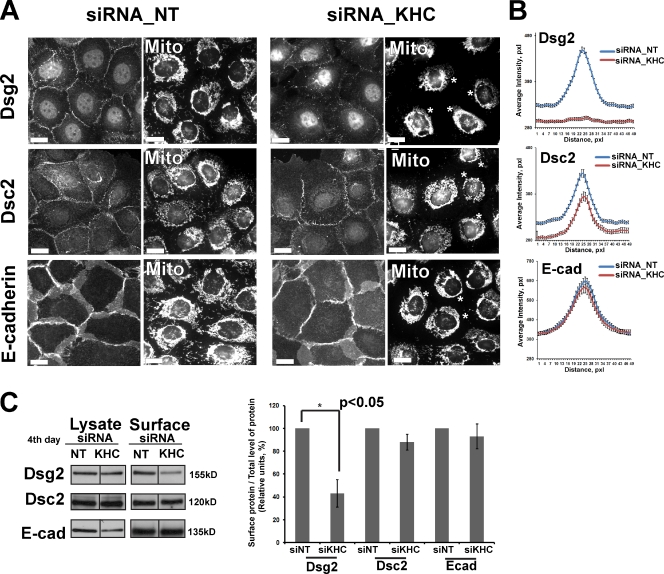
To determine whether Dsg2 incorporation into desmosomes is affected by kinesin-1 depletion in Scc9 cells, we performed immunofluorescence analysis. We used MitoTracker staining to find the cells with collapsed mitochondria as an indicator of kinesin-1 knockdown (Tanaka et al., 1998). Immunostaining of endogenous Dsg showed that KHC depletion resulted in a significant redistribution and reduction in Dsg2 fluorescence intensity in both Scc9 (Fig. 4, A and B) and A431 cells (Fig. S2, C and D). Although total pixel intensity of Dsc2 staining was modestly attenuated (by ∼10%), its organization and punctate distribution at cell–cell interfaces was not altered significantly (Fig. 4, A and B; and Fig. S2, C and D). Additionally, E-cadherin accumulation and organization were unaltered in KHC-deficient cells, suggesting that adherens junctions were maintained (Fig. 4, A and B; and Fig. S2, C and D). Both plakoglobin (Pg) and the desmosomal plaque protein desmoplakin (DP) retained their normal organization and intensity at cell junctions (Fig. S3, A and B).
Figure 4.
Knockdown of KHC impairs Dsg2 accumulation at the plasma membrane in Scc9 cells. (A) Scc9 cells in media with 1.2 mM Ca2+ were transfected with siRNA oligos against KHC or control nontargeted (NT) siRNA (siRNA_NT) for 2 d, replated and, after 24 h, stained with MitoTracker (Mito), fixed, and colabeled either for Dsg2, Dsc2, or E-cadherin (E-cad). Knockdown of kinesin-1 (indicated by mitochondria collapse, marked by asterisks) inhibits Dsg2 recruitment into the newly forming cell–cell interfaces but does not inhibit Dsc2 and E-cadherin accumulation. Level of knockdown in this experiment was comparable with that in Fig. 2 F. Bars, 20 µm. (B) Line scan analyses of border intensities for Dsg2, Dsc2, and E-cadherin were performed for ∼50 borders per each condition with three scans per border (n = ∼150 for each condition). Borders were chosen randomly from three independent experiments per condition. KHC knockdown results in loss of peak intensity for Dsg2 at cell–cell borders (P < 0.001) and decreased total pixel intensity of Dsc2 staining of ∼10% (P < 0.01). (C) Scc9 cells were transfected with siRNA oligos against KHC or nontargeting siRNA and were cell surface biotinylated 3 d after transfection. Biotinylated proteins were isolated by streptavidin pull-down, and levels of cell surface biotinylated Dsg2, Dsc2, and E-cadherin were identified by immunoblotting (compiled data from the same blot). Average of four independent experiments demonstrates a significant decrease in Dsg2 (*, P < 0.05) but not Dsc2 or E-cadherin cell surface expression. Black lines indicate that intervening lanes have been spliced out. Error bars represent SEM. pxl, pixel.
To confirm that plasma membrane–associated Dsg2 was reduced in KHC-deficient cells, we performed cell surface biotinylation. Streptavidin pull-down of biotinylated proteins followed by immunoblotting for Dsg2 revealed a significant decrease in the surface-associated pool of this cadherin compared with control-treated cells, whereas Dsc2 and E-cadherin surface labeling was not significantly affected (Fig. 4 C). Together, these data suggest that kinesin-1 is important specifically for Dsg2 intracellular transport and accumulation at desmosomes.
Kinesin-2 is required for normal Dsc2 accumulation in desmosomes
The fact that kinesin-1 was not involved in Dsc2 intracellular transport prompted us to consider other candidates for cadherin trafficking among the kinesin superfamily. In contrast to the broad distribution pattern of kinesin-1, kinesin-2 (KIF3) is abundantly expressed in the brain and to a lesser extent in other tissues (Aizawa et al., 1992; Cole et al., 1993; Kondo et al., 1994; Teng et al., 2005). Kinesin-2 has also been reported to govern intracellular vesicular transport (Yamazaki et al., 1995; Hirokawa, 2000; Brown et al., 2005). Importantly, kinesin-2 was implicated in the transport of N-cadherin to the cell periphery in neuroepithelium (Teng et al., 2005) and was shown to be essential for adherens junction positioning in Drosophila melanogaster photoreceptors (Mukhopadhyay et al., 2010).
Confocal fluorescence analysis revealed partial colocalization of KIF3A and Dsc2, but not Dsg2, in Scc9 cells, consistent with an interaction between Dsc2 and kinesin-2 (Fig. 5 A). In addition, KIF3A associated with the recombinant Dsc2 tail–GST fusion protein but not with Dsg2 tail–GST (Fig. 5 B and Fig. S2 A for A431 cells). Finally, Dsc2, but not Dsg2, showed a significant interaction with KIF3A in PLA experiments, which was inhibited in siKIF3A-treated cells exhibiting ∼70% knockdown (using two independent targeting sequences; Fig. 5 C).
Figure 5.
Dsc2 associates with kinesin-2. (A) Dual-label immunofluorescence confocal microscopy showing Dsc2 (right) and Dsg2 (left) with KIF3A. A subset of Dsc2-positive particles colocalize with KIF3A (yellow circles), whereas Dsg2-positive particles do not colocalize with KIF3A (blue circles). Boxes showed the zoomed areas. Bars, 10 µm. (B) Recombinant Dsg2 cytoplasmic tail–GST or Dsc2 cytoplasmic tail–GST were incubated with Scc9 cell lysates, and retained proteins were immunoblotted for the presence of KIF3A. KIF3A associates with Dsc2 tail–GST but not Dsg2 tail–GST. Note that the blot is the same as on Fig. 2 C. (C) PLA was performed using primary antibodies directed against Dsg2 or Dsc2 coupled with KIF3A antibodies on nontargeting (NT) siRNA (siNT; top) or siKIF3A-treated Scc9 cells (middle). Red particles were quantified, showing significant Dsc2–KIF3A interactions (***, P < 0.001) compared with Dsg2–KIF3A, which were abrogated upon KIF3A knockdown (bottom). Number of cells per condition = ∼240. Bars, 20 µm. (D) Level of KIF3A knockdown (KD) in the PLA experiment shown above, representative of three experiments on the right (**, P < 0.01). Protein expression level of Dsg2, Dsc2, and E-cadherin (E-cad) shown in the bottom middle were unchanged. Error bars represent SEM. Tub, tubulin.
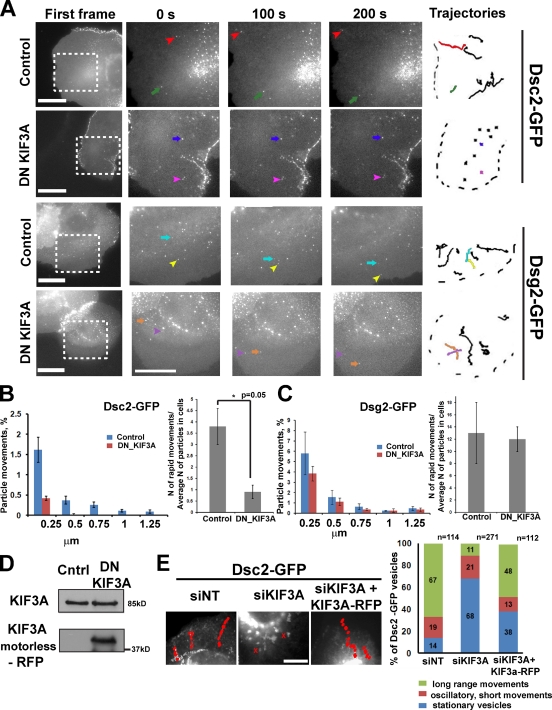
To determine whether kinesin-2 (KIF3) serves as the Dsc2 motor in epithelial cells, we analyzed Dsc2 dynamic behavior in control cells or cells expressing a motorless DN KIF3A mutant (Jaulin et al., 2007) to inhibit kinesin-2 function (Fig. 6 D). Time-lapse analysis of Scc9 cells after ∼12 h of double transfection with Dsc2-GFP and RFP–DN KIF3A showed that long-range translocation of Dsc2 particles was abrogated. Dsc2 particles remained stationary or were reduced to short-range oscillatory movements (Fig. 6, A and B; and Video 7), whereas Dsg2 trafficking was unaffected (Fig. 6, A and C; and Video 7). KIF3A silencing also interfered with Dsc2 transport, and this movement was rescued by nuclear coinjection of full-length RFP-KIF3A cDNA (Fig. 6 E and Video 8).
Figure 6.
Intracellular trafficking of Dsc2, but not Dsg2, was affected by blocking kinesin-2 function with DN KIF3A. (A) Representative videos show behavior of Dsg2- and Dsc2-GFP in cells imaged 12 h after double transfection with motorless DN KIF3A–RFP constructs. First frames show the areas (boxes) analyzed (Video 7), and the three magnified columns show selected vesicles (colored arrows) at three time points. Vesicle trajectories taken from the analyzed area are shown on the left, where colored tracks represent the movement of selected vesicles. Bars, 20 µm. (B) Distribution of particle movements (equal to instantaneous velocity; *, P < 0.05 for each set of bars) and the ratio of rapid movements to the average amount of analyzed particles (15–20 particles per cell) in each cell (three cells were analyzed per experiment) for Dsc2 in control cells and cells expressing the DN mutant (*, P = 0.05 for the ratio of rapid movements). Particle movements and the ratio of rapid movements for Dsc2 were decreased in cells with inhibited kinesin-2 function compared with control cells. (C) Distribution of long vectors and the ratio of long vectors to the average amount of analyzed particles (15–20 particles in each cell) for Dsg2 in cells expressing DN KIF3A are similar to that observed in control cells (P = 0.8). (D). Immunoblot showing level of DN KIF3A–RFP expression and total level of endogenous KIF3A for experiments in A and B. Cntrl, control. (E) Maximum projection of time-lapse analysis of cells into which cDNAs encoding Dsc2-GFP or Dsg2-GFP (not depicted) were coinjected with or without KIF3A cDNA into the nuclei of nontargeting (NT) siRNA (siNT) or siKIF3A-silenced cells (Video 8). Bar, 5 µm. Representative trajectories of Dsc2-GFP movements are shown in red. Bar graph on the right is a population analysis showing particles engaging in long-range, short-range, or stationary movements as a percentage of Dsc2-GFP vesicles. The percentage of long-range movements is partially restored by the KHC-mCherry rescue construct (P = 0.001). n above the bars represents the number of vesicles trajectories that have been analyzed. Number of cells: nontargeting siRNA = 8, siKIF3A = 10, and siKIF3A + KIF3A-RFP = 8.
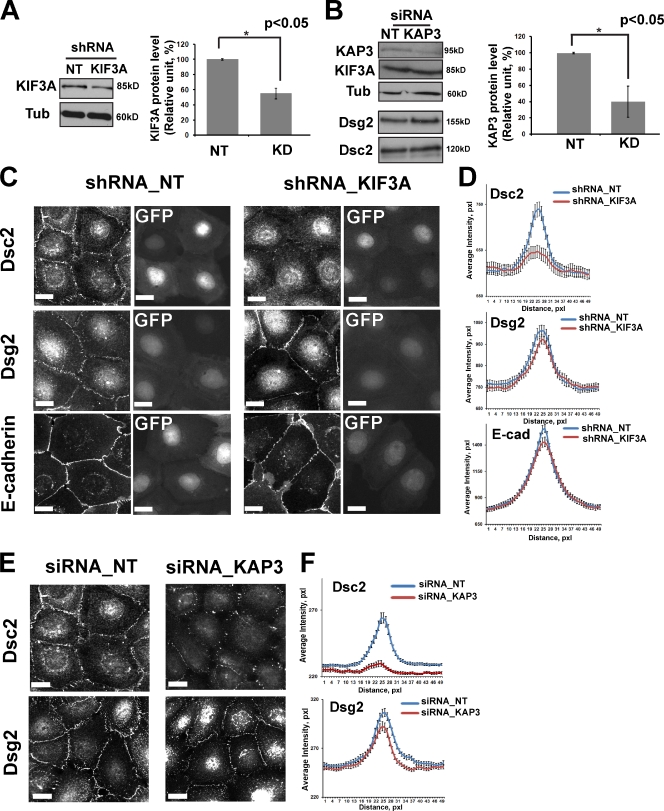
To determine the impact kinesin-2 interference has on Dsc2 desmosome accumulation, we silenced either KIF3A (Fig. 7 A) or the kinesin-associated protein KAP3 (Fig. 7 B). KAP3 is responsible for KIF3 binding to most cargoes via its armadillo repeat domain. The overall total protein levels of the desmosomal and classical cadherins were not affected by KIF3A or KAP3 siRNA treatment (Fig. 5 D and Fig. 7 B). After KIF3A depletion, the intensity of Dsc2 signals distributed within desmosomes was significantly reduced in both Scc9 and A431 cells (Fig. 7, C and D; and Fig. S4, B and C). The same was observed in KAP3-silenced cells (Fig. 7, E and F). The accumulation of Dsg2 and E-cadherin at the cell junctions was not affected by KIF3A or KAP3 depletion (Fig. 7, C–F; and Fig. S4, B and C), whereas Pg and DP localization was only modesty altered (by ∼15% for Pg and ∼10% for DP) in KIF3A-deficient cells (Fig. S5, A and B).
Figure 7.
Knockdown of KIF3A or KAP3 affects Dsc2, but not Dsg2, accumulation at junctions in Scc9 cells. KIF3A and KAP3 (kinesin-2 intermediate chain) functions were targeted with shRNA or siRNA, respectively. (A) Immunoblot demonstrating ∼50% reduction of KIF3A in shRNA-transfected cells (n = 3; *, P < 0.05). α-Tubulin (Tub) is the loading control. (B) Immunoblot demonstrating ∼60% decrease in KAP3 in siRNA-transfected cells (n = 5; *, P < 0.05). No change in expression of Dsg2 and Dsc2 was observed. (C) Scc9 cells were transiently transfected with a GFP-shRNA construct against KIF3A or nontargeting (NT) shRNA for 2 d, replated and, after 24 h, fixed and labeled either for Dsg2, Dsc2, or E-cadherin (E-cad). Knockdown (KD) of KIF3A (cells with GFP) delays Dsc2 recruitment into newly forming junctions but does not have a measurable effect on Dsg2 or E-cadherin localization at cell–cell borders. (D) Line scan analysis of border intensities for Dsc2, Dsg2, and E-cadherin were performed for ∼50 borders per each condition with three times scans per border (n = ∼150 for each condition). Borders were chosen randomly from three independent experiments per condition. KIF3A knockdown results in a decrease of peak intensity for Dsc2 at cell–cell borders compared with the control cells (P = 0.01). (E) Scc9 cells were transfected with siRNA oligos against KAP3 or control siRNA for 2 d, replated, and after 24 h, labeled either for Dsg2 or Dsc2. (F) KAP3 knockdown results in loss of the peak intensity for Dsc2 at the cell–cell border (P < 0.001). pxl, pixel. Bars, 20 µm.
Altogether, these findings demonstrate that desmosomal cadherins Dsg2 and Dsc2 use distinct mechanisms for long-range transport from a vesicular compartment to sites of junction assembly at the plasma membrane. Furthermore, specific impairment of kinesin-1 or -2 leads to the formation of junctions depleted of the respective desmosomal cadherin cargo.
PKP2 (plakophilin 2) is required for long-range transport of Dsc2 but not Dsg2
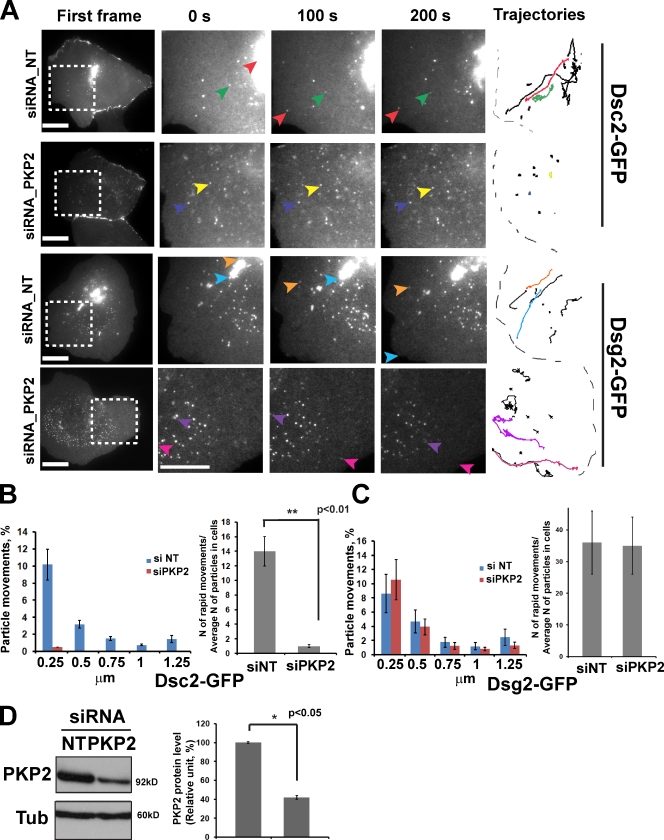
The cadherin-associated armadillo protein p120 catenin provides a link between kinesin and N-cadherin during its trafficking to the plasma membrane (Chen et al., 2003). To test whether p120-related desmosomal proteins called plakophilins participate in Dsg2 or Dsc2 transport, we performed live-cell imaging experiments in Scc9 cells in which PKP2 or PKP3 were silenced (Fig. 8 D and not depicted). PKP1 is not present in junctions in this cell type; therefore, we did not carry out silencing experiments for this family member. PKP2 knockdown inhibited Dsc2 (Fig. 8, A and B), but not Dsg2 (Fig. 8, A and C), movements, whereas PKP3 knockdown did not block desmosomal cadherin trafficking (not depicted). Desmosome assembly and other PKP2 functions are rescued by a silencing-resistant construct in Scc9 cells treated with this siRNA (Bass-Zubek et al., 2008; Godsel et al., 2010). We also silenced a related protein p0071 (PKP4), which had previously been shown to interact with the kinesin-2 subunit KIF3B (Keil et al., 2009). p0071 knockdown had no effect on the transport of either desmosomal cadherin (unpublished data).
Figure 8.
PKP2 is required for long-range transport of Dsc2 but not Dsg2. (A) Scc9 cells were treated with control or siPKP2 oligos mixed with siGLO (positive transfection indicator) and imaged 3 d after transfection. First frames show areas (boxes) analyzed in control or PKP2-deficient cells, and the three magnified columns show selected vesicles (colored arrows) at three time points. Vesicle trajectories taken from the analyzed area are shown on the right, where colored tracks represent the movement of selected vesicles. Bars, 20 µm. (B) Bar graphs show quantification of distribution of particle movements (equal to instantaneous velocity; for each set of bars **, P < 0.01) and the ratio of rapid movements to the average amount of analyzed particles (10–15 particles per cell) in each cell (three to four cells were analyzed per experiment) for Dsc2 in control and knockdown conditions. In PKP2-deficient cells, the amount of the particle movements as well as the ratio of rapid movements for Dsc2 was dramatically decreased compared with the control cells (**, P < 0.01 for the ratio of rapid movements). (C) Distribution of long vectors and the ratio of long vectors to the average amount of analyzed particles (10–15 particles in each cell) for Dsg2 in cells with PKP2 knockdown are similar to that observed in control cells (P = 0.9). (D) Immunoblot showing level of PKP2 knockdown (∼60%, n = 3; *, P < 0.05) for experiments in A and B. NT, nontargeting; Tub, tubulin.
Loss of kinesin-1 or kinesin-2 weakens intercellular adhesion
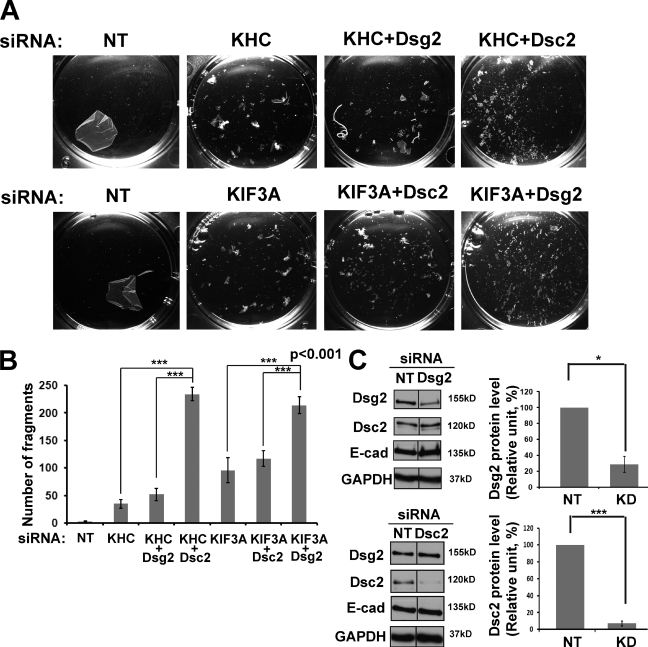
Previous studies demonstrated that to form robust cell–cell adhesion, coexpression of both desmosomal cadherins proteins, Dsgs and Dscs, is required (Amagai et al., 1994; Chidgey et al., 1996; Kowalczyk et al., 1996; Marcozzi et al., 1998). To test whether interference with kinesin-1 or kinesin-2 trafficking impairs intercellular adhesion, we subjected silenced cultures to a mechanical dissociation assay. In each experiment, a confluent monolayer of cells was enzymatically released from the cell culture dish and subjected to mechanical stress to generate fragments of the epithelial sheet (Fig. 9 A). Silencing either kinesin-1 or kinesin-2 function weakened intercellular adhesive strength, as indicated by the increased number of fragments observed in mechanically challenged kinesin-1– and kinesin-2–deficient epithelial sheets (Fig. 9 B). Importantly, double knockdown of KHC and Dsg2 did not significantly increase tissue fragility over kinesin-1 silencing only, whereas double knockdown of kinesin-1 and Dsc2 weakened adhesion by approximately fivefold. Similarly, double knockdown of KIF3A and Dsc2 did not increase tissue fragility over KIF3A silencing only, whereas double knockdown of KIF3A and Dsg2 weakened adhesion to about the same extent as KHC plus Dsc2 knockdown. Interestingly, targeting the Dsc2 system had a more dramatic impact on adhesion strength, whether this was through Dsc2 knockdown or kinesin-2 knockdown. These data provide compelling support for the functional specificity of these motor–cadherin pairings in cell–cell adhesion.
Figure 9.
Loss of kinesin-1 or -2 results in weakened cell–cell adhesion. (A) Confluent monolayers of Scc9 keratinocytes after 72-h siRNA transfection with either kinesin-1 (KHC) or kinesin-2 (KIF3A) or doubly silenced for kinesin-1/Dsg2, kinesin-1/Dsc2, kinesin-2/Dsg2, or kinesin-2/Dsc2 were subjected to a dispase mechanical dissociation assay in triplicate. Cells were incubated with dispase enzyme to lift the cells from the dish, and the intact monolayer was subjected to mechanical stress. Increasing amounts of fragments indicate the loss of cell–cell adhesion. (B) Bar graphs show quantification of fragmentation as an average from at least three experiments. ***, P < 0.001 for pairs siKHC/siKHC+siDsc2, siKHC+siDsg2/siKHC+siDsc2, siKIF3A/siKIF3A+siDsg2, and siKIF3A+siDsc2/siKIF3A+siDsg2. P < 0.01 for pairs nontargeting (NT) siRNA/siKHC and nontargeting siRNA/siKIF3A. P < 0.05 for the pair siKHC/siKIF3A. (C) Level of Dsg2 and Dsc2 knockdowns (KD) in dispase experiments shown by Western blots (compiled data from the same blots). Level of knockdown for Dsg2 was ∼70% (*, P < 0.05) and ∼90% for Dsc2 (***, P < 0.001; n = 3 for both). The protein expression levels of the partner desmosomal cadherins and E-cadherin (E-cad) were unchanged. Levels of KHC and KIF3A knockdowns were comparable with that shown in Fig. 2 F and Fig. 5 D. Black lines indicate that intervening lanes have been spliced out. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Discussion
Desmosomes are highly dynamic structures that assemble and disassemble during the morphogenesis and remodeling of tissues. Although protein–protein interactions that give rise to the final form of this organelle have been well studied, the machinery responsible for transporting desmosomal cadherins to the plasma membrane had not been addressed. Here, we demonstrate that molecular motors in the kinesin superfamily govern long-range MT-dependent trafficking of both Dsg2 and Dsc2, the most widely expressed desmosomal cadherins. Transport of these junctional proteins depends on the activities of two distinct motors, kinesin-1 and kinesin-2, and both motors are required to form a functional adhesive interface.
Our data are consistent with previous studies suggesting that desmosome components exhibit distinct biochemical and kinetic behaviors. Metabolic labeling and pulse–chase experiments previously demonstrated that desmosomal cadherins turn over at different rates (Penn et al., 1987), and newly synthesized components of the cytoplasmic plaque and membrane core are assembled with different kinetics (Pasdar and Nelson, 1989). Ultrastructural observations suggest that the initial phases of desmosome assembly occur in two steps in MDCK cells: in the first 30 min after a calcium switch, Dsc2-enriched vesicles are transported via small ∼60-nm vesicles to the plasma membrane, where they are gradually stabilized by a second wave of ∼200-nm vesicles enriched with Dsg2 and associated armadillo proteins (Burdett and Sullivan, 2002). Live-cell imaging allowed us to directly observe the dynamic behaviors of the two types of desmosomal cadherin and their response to specific genetic perturbations. The identification of distinct molecular machinery responsible for governing Dsg2 and Dsc2 trafficking provides a potential explanation for the previously described differences in desmosomal cadherin behavior.
Although MT organization in keratinocytes has recently been demonstrated to be regulated by desmosomes (Lechler and Fuchs, 2007), the extent to which MTs contribute to the formation of intercellular junctions is not well understood. Several lines of evidence support the idea that biogenesis of cell–cell contacts requires not only MTs but also MT-dependent membrane transport. Depolymerization of MTs with nocodazole in mammary tumor cell lines perturbs the ability of cells to accumulate E-cadherin at cell–cell contacts (Stehbens et al., 2006). Similarly, nocodazole treatment of fibroblasts prevents N-cadherin transport from the Golgi to the plasma membrane and abolishes cell–cell contact formation (Mary et al., 2002). In addition, cell–cell adhesion was disrupted after MT depolymerization in newt lung epithelial cells (Waterman-Storer et al., 2000). Finally, an intact MT cytoskeleton is required for proper trafficking of connexins, the proteins that form gap junctions, and in vivo, nocodazole treatment markedly reduced connexin concentration at the cell–cell borders (Shaw et al., 2007).
Here, we demonstrate the importance of the MT network for rapid desmosomal cadherin translocation toward the plasma membrane. After nocodazole treatment, the appearance of both desmosomal cadherins at the newly formed junctions was dramatically delayed compared with DMSO-treated cells. Dsg2 and Dsc2 began to recover and appear at contact sites within ∼1.5 h after the initiation of junction assembly, a time period that was three times slower than in control cells. One possible explanation for this recovery may be the delivery of proteins derived from Golgi ministacks near the plasma membrane, as a result of Golgi fragmentation and redistribution reported to occur under these experimental conditions (Cole et al., 1996; Storrie et al., 1998). Because some previous work suggested that MTs might not be required for desmosome assembly (Pasdar et al., 1992), we also cannot completely exclude the possible contribution of the cortical actin cytoskeleton in desmosomal cadherin trafficking. It is expected that this type of transport would be slower and be used mostly for translocation of recycled copies of desmosomal cadherins from the cortical actin network to the plasma membrane during desmosome reorganization.
Our data support a model whereby the cytoplasmic tail domains of Dsg2 and Dsc2 provide a scaffold for the association of kinesin-1 and -2, respectively, which are responsible for the transport of their specific cadherin cargoes (Fig. 10). However, coimmunoprecipitation to identify interactions between endogenous desmosomal cadherins and kinesins was hampered by the limited availability of a soluble pool of Dsg2 and Dsc2. To overcome this challenge, we used the PLA technique, which identifies endogenous protein–protein interactions in situ (Söderberg et al., 2006; Leuchowius et al., 2009). These experiments demonstrated specific interactions between Dsg2-KHC and Dsc2-KIF3A, both of which require the presence of the respective motor proteins.
Figure 10.
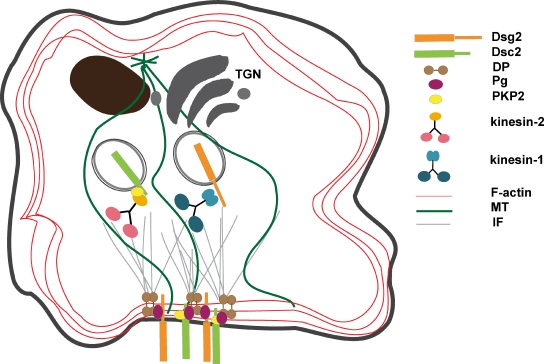
Model of Dsg2 and Dsc2 intracellular trafficking to the plasma membrane. Desmosomal cadherins exist in separate intracellular vesicular compartments that are transported using distinct motors. Dsg2-containing vesicles move inside the cell using the kinesin-1 motor protein, whereas kinesin-2, in cooperation with PKP2, is responsible for Dsc2 dynamic behavior. IF, intermediate filament.
It was previously determined that p120 catenin forms a complex with KHC and provides the link between kinesin and N-cadherin during its trafficking to the plasma membrane. Disruption of the interaction between the cadherin and p120, or the interaction between p120 and kinesin-1, leads to decreased N-cadherins at cell–cell contacts during junction reassembly (Chen et al., 2003). Here, we demonstrate that silencing the p120-related proteins PKP3 or p0071; the latter, which was previously shown to associate with KIF3B, does not affect either Dsg2 or Dsc2 transport. However, PKP2 is required for the rapid intracellular trafficking of Dsc2 but not Dsg2. Although our previous data support an association between PKP2 and both desmosomal cadherin tails in epithelial cells (Chen et al., 2002), the current work suggests that the interaction between PKP2 and the Dsc2 cytoplasmic tail plays a specific role in MT-dependent transport of this cadherin. The mechanism by which Dsg2 couples to KHC is not known but could potentially be through kinesin light chains, as it was previously reported that kinesin light chain can modulate the cargo-binding affinity to the motor (Hirokawa et al., 1989; Stenoien and Brady, 1997).
Kinesin deficiency dramatically weakened the cell–cell adhesive strength of confluent epithelial sheets. This was in spite of the fact that loss of Dsg2 or Dsc2 at the cell junction area in kinesin-1– or -2–depleted cells had either no measurable, or only modest, effects on the accumulation of E-cadherin, the partner desmosomal cadherin, or plaque components at the plasma membrane. The data are consistent with the idea that the remaining desmosomal cadherin interactions are sufficient to stabilize other junctional molecules at the site of cell–cell contacts but are insufficient to build normal functional desmosomes that provide strong intercellular adhesion. These findings differ somewhat from a recent observation that interference with a Sec3-containing exocyst complex has a broad effect on the morphology and function of desmosomes, accompanied by defects in the distribution of multiple components (Andersen and Yeaman, 2010).
Although we cannot rule out a functional loss of an unrelated membrane adhesion protein, overall, loss of kinesin trafficking appears to be surprisingly specific in its effects on membrane organization. Indeed, kinesin-1/Dsg2 double knockdown did not significantly alter adhesion strength, supporting the idea that Dsg2 likely depends almost exclusively on this motor protein for its cell surface presentation. In dramatic contrast, kinesin-1/Dsc2 double knockdown decreased adhesion strength approximately fivefold, which would be predicted if these manipulations target mechanistically distinct adhesion mechanisms. Similar results were seen for kinesin-2/Dsc2 double knockdowns. Interestingly, targeting the Dsc2 system had a more dramatic impact on adhesion strength, possibly offering one potential explanation for why separate mechanisms for regulating the cell surface levels of Dsgs and Dscs have evolved.
Independent modulation of distinct kinesin motors in vivo could provide a way to tailor desmosome adhesive properties. Indeed, by varying the ectopic expression level of Dsg1 relative to Dsc1 in mouse L cell fibroblasts, we previously demonstrated that the Dsg1/Dsc1 ratio is a critical determinant of desmosomal adhesion in fibroblasts (Getsios et al., 2004). Independent regulation of cell surface desmosomal cadherins could thus serve as an adhesion rheostat during dynamic cell movements that occur during epithelial morphogenesis and remodeling. It also seems possible that kinesin-dependent trafficking of desmosomal cadherins could be a target in human disease. Deficiency in kinesin transport is associated with the development and progression of neurological disorders (Tanaka et al., 1998), including Alzheimer’s (Stokin et al., 2005; Terada et al., 2010) and Huntington’s disease (Colin et al., 2008; Twelvetrees et al., 2010). Furthermore, alterations in the expression and function of kinesins have been associated with epithelial and neuronal cancers (Teng et al., 2005; Nindl et al., 2006). Both skin and heart disease (i.e., arrhythmogenic right ventricular cardiomyopathy) are caused by mutations in desmosome molecules, and it will be interesting to determine whether any of these mutations interfere with kinesin binding and desmosomal cadherin trafficking.
Materials and methods
cDNA and RNAi constructs
Dsg2-FLAG-EGFP (Klessner et al., 2009) has been described previously. In brief, cDNA of human Dsg2 fused with a C-terminal FLAG epitope was subcloned into pEGFP-N2 (Takara Bio Inc.) using the BamH1 restriction site. Dsg2-mCherry was created by PCR amplifying the Dsg2 into pmCherry N1 XhoI and SacII restriction sites. pmCherry was generated by excising EGFP cDNA from the pEGFP N1 vector (Takara Bio Inc.) using AgeI and BsrGI and incorporating PCR-amplified mCherry cDNA into the AgeI–BsrGI gap of the pEGFP N1 vector. Dsc2-GFP was a gift from S. Troyanovsky (Northwestern University, Chicago, IL; Windoffer et al., 2002). Dsg2 tail–GST has been described previously (Brennan et al., 2011). Dsc2 tail–GST was generated by incorporating PCR-amplified cDNA encoding intracellular domain of Dsc2 (nt 2,603–3,158 available from GenBank/EMBL/DDBJ under accession no. NM_024422.3) into BamH1 and NotI restriction sites of the PGEX4T-1 vector (GE Healthcare).
mCherry-KIF5B was generated by incorporating PCR-amplified cDNA encoding full-length KIF5B into mCherry vector by standard Gateway protocol: KIF5B forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCGGACCTGGCCGAG-3′, and reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCACTTGTTTGCCTCCTCCACCTCG-3′. Monomeric RFP (mRFP)-KIF3A was generated by incorporating full-length KIF3A inserted into His/mRFP SacI and KpnI restriction sites. KIF5B tail–His (Jaulin et al., 2007) has been described previously. In brief, the KIF5B tail was amplified by PCR from human A549 cells and cloned into the mammalian Gateway (Invitrogen) expression vector. pSuper-mCherry shRNA KHC (shKHC) and pSuper nontargeting negative control were gifts from J. Sznajder (Northwestern University, Chicago, IL). pMIRAGE-G shKIF3A was generated by cloning a microRNA annealed duplex (as was previously described in Brummelkamp et al., 2002) into the pMIRAGE-G BstXI site. pMIRAGE-G and the construct encoding mCherry-tagged β-tubulin were gifts from S. Kojima (Northwestern University, Evanston, IL). A construct encoding Myc-tagged rat KHC was provided by K.J. Verhey (University of Michigan Medical School, Ann Arbor, MI; Verhey et al., 1998). The DN mutants for the kinesins used were the RFP-tagged KIF5B tail construct (aa 780–963) and the RFP-tagged KIF3A motorless construct (aa 357–702; Jaulin et al., 2007).
Nontargeting siRNA (negative control); siGLO (positive transfection indicator; Thermo Fisher Scientific); siRNA against KHC (Sigma-Aldrich) with targeting sequence (the same used for the pSuper-mCherry shKHC construct) 5′-AGATGTACTTGAAGGATAT-3′; siRNA against KIF3A (Integrated DNA Technologies) with targeting sequences (the same used for the pMIRAGE-G shKIF3A construct) 5′-GCAGATCAGATTGTCCTATGTTGCG-3′ and 5′-CCAAGATGCCGATCAATAAATCAGA-3′; siRNA against KAP3 (Sigma-Aldrich) with targeting sequence 5′-GTGTCGAGTTAGCTACAAA-3′; siRNA against hDsg2 (Integrated DNA Technologies) with targeting sequence 5′-CCTGGAAGCAGAGACAGTGTGGTCCTT-3′; and siRNA against hDsc2 with targeting sequence 5′-AGAGAGACACTATAAACAAGTACAC-3′ were used for siRNA experiments. hPKP2 siRNAs were previously described (SMART pool; Thermo Fisher Scientific) with targeting sequences 5′-GAGGAACCATTGCAGATTA-3′, 5′-CAACGTCACTGGATGCCTA-3′, 5′-GCACGCGACCTTCTAAACA-3′, and 5′-GAGTATGTCTACAACCTAC-3′, and silencing-resistant constructs were shown to rescue desmosome assembly and other PKP2-dependent functions for these targeting sequences (Bass-Zubek et al., 2008; Godsel et al., 2010).
Cell culture and transfections
Scc9 and A431 cell lines were grown in DME/F12 (50:50 mix; for Scc9) or in DME (for A431) media supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin (Mediatech). All experiments were performed in normal growth media with 1.2 mM Ca2+, unless if other conditions state specifically. For transient transfection of cDNAs, Lipofectamine 2000 (Invitrogen) was used according to the manufacturer’s protocol, and cells were assayed 24 h later. siRNAs (20–50 nM per experiment) were transiently transfected using DharmaFECT 1 (Thermo Fisher Scientific). In this case, cells were analyzed 72–96 h after transfection. For microinjection experiments, Scc9 cells (80–90% confluent) were trypsinized and electroporated with 20 nM control, KHC-, or KIF3A-targeting siRNAs using the Nucleofector System (Lonza). 2 d later, cells were split and seeded onto coverslips and into 6-well plates. On the next day, cells on coverslips were injected for time-lapse imaging. For retrovirus infections, cells at 20% confluency were incubated in growth media containing 4 µg/ml polybrene hexadimethrine bromide (Sigma-Aldrich) and retrovirus supernatants. Retrovirus supernatants were generated as previously described in Getsios et al. (2004). In brief, Phoenix cells transiently transfected with retroviral cDNAs constructs were harvested at 70% confluency for 12–24 h at 32°C, and then retrovirus supernatants were collected and concentrated using a column (Centricon Plus-20; Millipore).
Antibodies and chemical reagents
The following primary antibodies were used: mouse monoclonal 4B2 (Getsios et al., 2004) against Dsg2; rabbit polyclonal RB5 against Dsg2 (PROGEN Biotechnik); mouse monoclonal 6D8 against Dsg2 and 7G6 against Dsc2 (gifts from J. Wahl III, University of Nebraska Medical Center, Omaha, NE); mouse monoclonal HECD1 against E-cadherin (gift from M. Takeichi and O. Abe, RIKEN Center for Developmental Biology, Kobe, Japan); rabbit polyclonal NW6 against DP (Angst et al., 1990); chicken polyclonal 1407 against Pg (Aves Labs, Inc.); mouse monoclonal MAB6013S against PKP2 (BioDesign, Inc.); mouse monoclonal H2 (Millipore) and goat monoclonal anti-KIF5B (Sigma-Aldrich) against KHC; mouse monoclonal K 2.4 (Covance) and rabbit polyclonal Anti-KIF3A (Abcam) against KIF3A; goat polyclonal N-19 against the KAP3 subunit (Santa Cruz Biotechnology, Inc.); mouse monoclonal DM1-α (Sigma-Aldrich) against α-tubulin; rabbit polyclonal anti–glyceraldehyde 3-phosphate dehydrogenase (Abcam); rabbit polyclonal anti-Myc (Bethyl Laboratories, Inc.); and goat polyclonal anti-GST (GE Healthcare). Secondary antibodies used for Western blotting included goat anti–mouse, –rabbit, and –chicken peroxidase and rabbit anti–goat peroxidase (Rockland Immunochemicals, Inc.; Kirkegaard & Perry Laboratories, Inc.). Secondary antibodies used for immunofluorescence included goat anti–mouse, –rabbit, and –chicken conjugated with Alexa Fluor 488 or 568 nm (Invitrogen) and donkey anti–goat and –mouse conjugated with Alexa Fluor 488 or 568 nm (Invitrogen). For staining mitochondria, 200 nM MitoTracker red CMXRos was added to the culture medium, and the cells were incubated for 30 min at 37°C before being washed with PBS and fixed.
Calcium switch and nocodazole treatment
Cells were incubated in low-calcium medium (DME with 0.05 mM Ca2+) for 3 h and switched to normal growth media containing ∼1.2 mM Ca2+ to induce cell junction assembly for ≤30 min or 1.5 h and processed for immunofluorescence analysis. For experiments to depolymerize MTs, cells were treated with 10 µM nocodazole (Sigma-Aldrich) for 1 h at 37°C, added to low-calcium medium after 2 h incubation, switched to normal growth media with the same nocodazole concentration, and fixed for immunofluorescence after 30 min.
Immunofluorescence analysis and image acquisition
Cells were seeded onto glass coverslips for ≥12 h after splitting. For immunofluorescence analysis, cells were washed in PBS, fixed either in anhydrous ice-cold methanol for 2 min at −20°C or 4% formalin solution for 20 min at room temperature, and processed for indirect immunofluorescence using Rb5 (1:100), 4B2 (1:100), 7G6 (1:100), HECD1 (1:100), NW6 (1:50), 1407 (1:100), or DM1-α (1:100) for 1 h at room temperature. When using 4% formalin, fixed cells were extracted with 0.1% Triton X-100 and 1 mg/ml BSA for 30 min at room temperature immediately after fixation. After incubation with secondary antibodies (1:300 dilution for 30 min at 37°C) coverslips were mounted in polyvinyl alcohol (Sigma-Aldrich). Cells were visualized with a wide-field microscope (DMR; Leica) or a confocal microscope (LSM 510; Carl Zeiss). The DMR microscope images were generated using 40 and 63× objectives (PL Fluotar, NA 1.0), a digital camera (ORCA-100 model C4742-95; Hamamatsu Photonics), and MetaMorph 7.6 software (Molecular Devices). The LSM 510 confocal microscope was fitted with 63 and 100× objectives (Plan Apochromat, NA 1.32 and NA 1.4; Carl Zeiss) and ZEN software (version 5,5,0,375; Carl Zeiss). Images were further processed using Photoshop (CS3; Adobe) and compiled using Illustrator (CS3; Adobe).
PLA
Reagents for the PLA of kinesin–cadherin interaction in situ were purchased from Olink Biosciences through Axxora LLC. PLA was performed according to the manufacturer’s protocol (Duolink In Situ PLA; Olink Bioscience). In brief, cells were stained with primary antibodies (dilution 1:200) as described in the Immunofluorescence analysis and image acquisition section followed by incubation for 1 h at 37°C with oligo-linked secondary antibodies. Close proximity (∼20–100 nm) of target antigens allows their respective secondary antibody nucleotide sequences to hybridize. 30-min incubation at 37°C with ligase and additional oligos provide a closed DNA circle formation. A subsequent step involving polymerase-driven rolling circle amplification incorporates fluorescently labeled nucleotides, which fluoresce in the red channel when excited. Fluorescent spots appearing at the site of kinesin–cadherin interactions were detected by wide-field microscopy (DMR) and compiled by ImageJ software (National Institutes of Health).
Time-lapse fluorescence imaging
Cells growing on 2-well Laboratory-Tek chambered coverglass (Thermo Fisher Scientific) were transiently transfected or retrovirally infected. 24 h later, the normal growth medium was changed to imaging medium (Hank’s balanced salt solution, 20 mM Hepes, 10% FBS, 2 mM l-glutamine, 4.5 g/liter glucose, and amino acids) at 37°C for 15–30 min and then processed for time-lapse imaging. Time-lapse image recordings were acquired using two imaging systems: (1) an inverted microscope (DMI 6000B; Leica) with mercury illumination fitted with ORCA-ER (for dual-color imaging) and ImagEM electron multiplier charge-coupled device (for one-color time-lapse imaging) cameras (Hamamatsu Photonics) and a 100× objective (HCX Plan Apochromat, oil, NA 1.40) and (2) an Application Solution Multidimensional Workstation (Leica) equipped with an inverted microscope (DMIRE2; Leica), 100× objective (HCX Plan Apochromat, oil, NA 1.35), and a camera (CoolSNAP HQ; Roper Scientific). Both are housed inside a temperature-controlled 37°C climate chamber. Each frame was captured at 1-s intervals for 3 min using Simple PCI 6 software (Hamamatsu Photonics) or 5-s intervals for 2 and 5 min using Application Solution Multidimensional Workstation. All videos were compiled using MetaMorph 7.6 imaging software and ImageJ.
Microinjection and time-lapse imaging
Cells were seeded onto sterilized coverslips and grown 24 h before microinjection. Microinjection was performed essentially as previously described (Jaulin et al., 2007). In brief, cells were pressure microinjected intranuclearly with cDNAs in HKCl (10 mM Hepes and 140 mM KCl, pH 7.4) using a micromanipulator (Narishige). cDNA concentrations in the needle were 50 µg/ml−1 for GFP-Dsg2 and GFP-Dsc2 and 20 µg/ml−1 for full-length mCherry-KIF5B and mRFP-KIF3A. Cells were incubated at 37°C for 90–120 min to allow for expression of cDNAs, transferred to recording medium (Hank’s balanced salt solution with 1% FBS, 20 mM Hepes, and 4.5 g/liter glucose), and placed in a temperature-controlled recording chamber at 37°C on a microscope (TiE; Nikon) for time-lapse imaging. For rescue experiments, cDNAs encoding full-length kinesins and Dsg-2 or Dsc-2 were coinjected into cells transfected with siRNAs targeting KHC or KIF3A. Images were acquired at 2-s intervals for 3 min with a 60× water immersion objective and a charge-coupled device camera (ORCAII-ER; Hamamatsu Photonics). All devices were controlled by MetaMorph.
Protein expression, purification, and pull-down assay
pGEX-GST, pGEX-GST-Dsg2, pGEX-GST-Dsc2, and pDEST17-His-Kif5B were transformed into Rosetta competent cells (EMD). Cells were grown at 37°C until OD600 ≈ 0.9 and cooled down to 22°C. 100–200 µM IPTG was added, and culture grew for another 4 h at 22°C. Cells were harvested and resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, and 0.5% Triton X-100) plus protease inhibitor cocktail (Roche). Cells were further lysed by sonication on ice (eight times with 15-s burst with 30-s interval). Unlysed cells and debris were removed by centrifugation (10,000 g for 20 min at 4°C). Cleared cell lysate were incubated with glutathione–Sepharose 4B (GE Healthcare) for GST-tagged proteins or Ni–nitrilotriacetic acid agarose (QIAGEN) for His-tagged protein for 2 h at 4°C. Beads with pulled down complexes were washed with PBS (four times for 10 min).
Scc9 or A431 cells were lysed with 1 ml ice-cold lysis buffer (1% NP-40, 145 mM NaCl, 10 mM Tris-HCl, pH 7.4, 10% glycerol, 5 mM EDTA, 2 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail ± 1% phosphatase inhibitor cocktail IV [EMD]). Lysates were incubated with equal amount of GST/GST-fused proteins or Ni-agarose/His-tagged proteins conjugated with agarose overnight at 4°C. Beads were washed extensively (three times for 10 min each) with lysis buffer, and bound proteins were fractionated by 10 or 7.5/12% SDS-PAGE followed by immunoblotting as described previously (Chen et al., 2002).
Immunoprecipitation and immunoblotting
Whole-cell lysates were generated using urea sample buffer (8 M deionized urea, 1% SDS, 10% glycerol, 60 mM Tris, pH 6.8; 0.1% pyronin-Y, and 5% β-mercaptoethanol). Immunoprecipitation and immunoblotting were described previously (Chen et al., 2002). In brief, cells were lysed with 1 ml ice-cold lysis buffer (1% NP-40, 145 mM NaCl, 10 mM Tris-HCl, pH 7.4, 10% glycerol, 5 mM EDTA, 2 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail ± 1% phosphatase inhibitor cocktail IV). Myc-tagged KHC was immunoprecipitated with anti-Myc–agarose (Sigma-Aldrich). Immunoreactive proteins were visualized using enhanced chemiluminescence. Antibody dilutions are as follows: 6D8 (1:500), 7G6 (1:1,000), HECD1 (1:1,000), DM1-α (1:1,000), anti-KIF5B (1:500), K2.4 (1:2,000), N-19 (1:500), and anti–glyceraldehyde 3-phosphate dehydrogenase (1:2,000).
Biotinylation assay
Cells were rinsed two times in ice-cold PBS with 1.2 mM Ca2+ and incubated with 2 mg/ml Sulfo-NHS-SS-biotin (EZ-Link; Thermo Fisher Scientific) for 30 min at 4°C in a dark room. Excess biotin was removed with 100 mM glycine in PBS by washing three times, 10 min each. Cells were washed one more time with PBS and lysed in radioimmunoprecipitation assay buffer (10 mM Tris, pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 5 mM EDTA, and 2 mM EGTA) containing protease inhibitor. Lysates were spun down at 14,000 rpm for 30 min, and supernatants were normalized for total protein. 30 µl of each lysate was saved for an input loading. Biotinylated proteins were pulled down with 40 µl streptavidin beads (Thermo Fisher Scientific) at 4°C overnight followed by several washes with radioimmunoprecipitation assay buffer. Complexes were released using Laemmli buffer at 95°C and analyzed by immunoblotting (see Immunoprecipitation and immunoblotting section).
Dispase mechanical dissociation assay
Dispase assays were performed as described previously (Hudson et al., 2004; Hobbs et al., 2011). Cells were transfected with the appropriate siRNA, and 48 h after, Ca2+ concentration of medium was reduced from 1.2 to 0.5 mM. Confluent cell cultures were rinsed with PBS and incubated with 2.4 U/ml dispase (Roche) for 30 min at 37°C. Released monolayers were rotated on an orbital shaker (150 rpm) for 20 min before imaging. Fragments were visualized with a dissecting microscope (MZ6; Leica), and the final images were obtained with a camera (C4742-95; Hamamatsu Photonics) attached to a stereomicroscope (MZ FL III; Leica). The number of fragments was counted by the ImageJ cell counter function (note that fragments <4 pixels in diameter were excluded from the quantification) and processed in Photoshop CS3.
Quantification of fluorescence intensity of cell–cell borders and particle kinetics
The fluorescence intensity of cell borders was determined by line scan analysis using ImageJ software. For each border, three independent areas with the same length and width were drawn at random positions, and the average pixel intensity was collected. At least three independent experiments were performed for each condition, and ∼50 borders from those experiments were chosen randomly for analysis. All images were captured using the exact same parameters. Particle movement was analyzed using MetaMorph 7.6 software. The distance that each particle traveled between two consecutive frames was defined as movement and equaled instantaneous velocity for the particles that were analyzed. A threshold of 0.25 µm was applied for determining rapid movement event, and movements >0.25 µm were used for calculations. The number of movements was normalized by the total number of particles that were analyzed in each video. At least three independent experiments were performed for each condition. Four to five cells from the experiments were chosen randomly for analysis, and ≥15 particles were analyzed in each video. The maximum projections of time-lapse videos were made using the stack arithmetic function in MetaMorph 7.6 software, and distances of vesicle projections were analyzed by MetaMorph 7.6.
Statistical analysis
For all experiments, error bars represent SEM, and statistical analysis was performed using an unpaired two-tailed t test.
Online supplemental material
Fig. S1 shows that the intracellular trafficking of Dsg2 is blocked by overexpression of a DN mutant of KHC in Scc9 cells. Fig. S2 shows that kinesin-1 associates with Dsg2, whereas kinesin-2 associates with Dsc2 in A431 cells, and knockdown of kinesin-1 impairs Dsg2 accumulation at the plasma membrane. Fig. S3 shows that silencing of KHC does not affect DP and Pg accumulation at sites of cell–cell contacts in Scc9 cells. Fig. S4 shows that knockdown of KIF3A (kinesin-2) prevents Dsc2, but not Dsg2, accumulation at intercellular junctions in A431 cells. Fig. S5 shows that blocking kinesin-2 function by silencing KIF3A does not impair DP and Pg accumulation in intercellular junctions in Scc9 cells. Video 1 shows that Dsg2- and Dsc2-enriched vesicles traffic independently within living Scc9 cells. Videos 2 and 3 correspond to Fig. 1 (B and C), respectively, in which Scc9 cells were cotransfected with Dsg2-GFP and tubulin-mCherry and show that Dsg2-enriched vesicles move along MTs. Videos 4 and 6 correspond to Fig. 3 (A and E), respectively. Video 4 shows intracellular trafficking of Dsg2 and Dsc2 in either control cells or cells with knockdown of KHC. Video 6 shows a maximum projection time-lapse video of Scc9 cells that were transfected with nontargeting RNA or siKHC and subsequently microinjected with Dsg2-GFP/+KHC-mCherry. Video 5 corresponds to Fig. S1 in which Scc9 cells were cotransfected with Dsg2-GFP and with or without a DN mutant of KHC and shows that trafficking of Dsg2-containing vesicles was blocked in cells expressing the DN mutant of KHC. Videos 7 and 8 correspond to Fig. 6 (A and E), respectively. Video 7 shows intracellular trafficking of Dsg2 and Dsc2 in either control cells or cells with knockdown of KIF3A. Video 8 shows a maximum projection time-lapse video of Scc9 cells transfected with nontargeting RNA or siKIF3A and subsequently microinjected with Dsc2-GFP/+KIF3A-RFP. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106057/DC1.
Acknowledgments
We would like to thank those who generously contributed reagents, including S. Troyanovsky, S. Kojima, K. Verhey, J. Sznajder, J. Wahl, M. Takeichi, and O. Abe. Thank you to Dr. V. Gelfand and C. Espenel for help and for useful discussions.
This work was supported by National Institutes of Health grants R01 AR41836 and CA122151 with partial support from R37AR44380 to K.J. Green, National Institutes of Health grant R01GM087575 to G.E. Kreitzer, and an American Heart Association Postdoctoral Fellowship 09POST2060718 to O. Nekrasova.
Footnotes
Abbreviations used in this paper:
- DN
- dominant negative
- DP
- desmoplakin
- Dsc
- desmocollin
- Dsg
- desmoglein
- KHC
- kinesin-1 heavy chain
- mRFP
- monomeric RFP
- MT
- microtubule
- Pg
- plakoglobin
- PLA
- proximity ligation assay
- shRNA
- short hairpin RNA
References
- Aizawa H., Sekine Y., Takemura R., Zhang Z., Nangaku M., Hirokawa N. 1992. Kinesin family in murine central nervous system. J. Cell Biol. 119:1287–1296 10.1083/jcb.119.5.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amagai M., Kàrpàti S., Klaus-Kovtun V., Udey M.C., Stanley J.R. 1994. Extracellular domain of pemphigus vulgaris antigen (desmoglein 3) mediates weak homophilic adhesion. J. Invest. Dermatol. 102:402–408 10.1111/1523-1747.ep12372164 [DOI] [PubMed] [Google Scholar]
- Andersen N.J., Yeaman C. 2010. Sec3-containing exocyst complex is required for desmosome assembly in mammalian epithelial cells. Mol. Biol. Cell. 21:152–164 10.1091/mbc.E09-06-0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angst B.D., Nilles L.A., Green K.J. 1990. Desmoplakin II expression is not restricted to stratified epithelia. J. Cell Sci. 97:247–257 [DOI] [PubMed] [Google Scholar]
- Bass-Zubek A.E., Hobbs R.P., Amargo E.V., Garcia N.J., Hsieh S.N., Chen X., Wahl J.K., III, Denning M.F., Green K.J. 2008. Plakophilin 2: a critical scaffold for PKCα that regulates intercellular junction assembly. J. Cell Biol. 181:605–613 10.1083/jcb.200712133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan D., Peltonen S., Dowling A., Medhat W., Green K.J., Wahl J.K., III, Del Galdo F., Mahoney M.G. 2011. A role for caveolin-1 in desmoglein binding and desmosome dynamics. Oncogene. 10.1038/onc.2011.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.L., Maier K.C., Stauber T., Ginkel L.M., Wordeman L., Vernos I., Schroer T.A. 2005. Kinesin-2 is a motor for late endosomes and lysosomes. Traffic. 6:1114–1124 10.1111/j.1600-0854.2005.00347.x [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- Burdett I.D., Sullivan K.H. 2002. Desmosome assembly in MDCK cells: transport of precursors to the cell surface occurs by two phases of vesicular traffic and involves major changes in centrosome and Golgi location during a Ca(2+) shift. Exp. Cell Res. 276:296–309 10.1006/excr.2002.5509 [DOI] [PubMed] [Google Scholar]
- Chen X., Bonne S., Hatzfeld M., van Roy F., Green K.J. 2002. Protein binding and functional characterization of plakophilin 2. Evidence for its diverse roles in desmosomes and beta -catenin signaling. J. Biol. Chem. 277:10512–10522 10.1074/jbc.M108765200 [DOI] [PubMed] [Google Scholar]
- Chen X., Kojima S., Borisy G.G., Green K.J. 2003. p120 catenin associates with kinesin and facilitates the transport of cadherin–catenin complexes to intercellular junctions. J. Cell Biol. 163:547–557 10.1083/jcb.200305137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgey M.A., Clarke J.P., Garrod D.R. 1996. Expression of full-length desmosomol glycoproteins (desmocollins) is not sufficient to confer strong adhesion on transfected L929 cells. J. Invest. Dermatol. 106:689–695 10.1111/1523-1747.ep12345525 [DOI] [PubMed] [Google Scholar]
- Chitaev N.A., Troyanovsky S.M. 1997. Direct Ca2+-dependent heterophilic interaction between desmosomal cadherins, desmoglein and desmocollin, contributes to cell–cell adhesion. J. Cell Biol. 138:193–201 10.1083/jcb.138.1.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D.G., Chinn S.W., Wedaman K.P., Hall K., Vuong T., Scholey J.M. 1993. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 366:268–270 10.1038/366268a0 [DOI] [PubMed] [Google Scholar]
- Cole N.B., Sciaky N., Marotta A., Song J., Lippincott-Schwartz J. 1996. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol. Biol. Cell. 7:631–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X.J., Saudou F., Humbert S. 2008. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 27:2124–2134 10.1038/emboj.2008.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusek R.L., Godsel L.M., Green K.J. 2007. Discriminating roles of desmosomal cadherins: beyond desmosomal adhesion. J. Dermatol. Sci. 45:7–21 10.1016/j.jdermsci.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Garrod D., Chidgey M. 2008. Desmosome structure, composition and function. Biochim. Biophys. Acta. 1778:572–587 10.1016/j.bbamem.2007.07.014 [DOI] [PubMed] [Google Scholar]
- Garrod D.R., Merritt A.J., Nie Z. 2002. Desmosomal cadherins. Curr. Opin. Cell Biol. 14:537–545 10.1016/S0955-0674(02)00366-6 [DOI] [PubMed] [Google Scholar]
- Getsios S., Amargo E.V., Dusek R.L., Ishii K., Sheu L., Godsel L.M., Green K.J. 2004. Coordinated expression of desmoglein 1 and desmocollin 1 regulates intercellular adhesion. Differentiation. 72:419–433 10.1111/j.1432-0436.2004.07208008.x [DOI] [PubMed] [Google Scholar]
- Gloushankova N.A., Wakatsuki T., Troyanovsky R.B., Elson E., Troyanovsky S.M. 2003. Continual assembly of desmosomes within stable intercellular contacts of epithelial A-431 cells. Cell Tissue Res. 314:399–410 10.1007/s00441-003-0812-3 [DOI] [PubMed] [Google Scholar]
- Godsel L.M., Dubash A.D., Bass-Zubek A.E., Amargo E.V., Klessner J.L., Hobbs R.P., Chen X., Green K.J. 2010. Plakophilin 2 couples actomyosin remodeling to desmosomal plaque assembly via RhoA. Mol. Biol. Cell. 21:2844–2859 10.1091/mbc.E10-02-0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.J., Getsios S., Troyanovsky S., Godsel L.M. 2010. Intercellular junction assembly, dynamics, and homeostasis. Cold Spring Harb. Perspect. Biol. 2:a000125 10.1101/cshperspect.a000125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B.M. 1996. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 84:345–357 10.1016/S0092-8674(00)81279-9 [DOI] [PubMed] [Google Scholar]
- Hirokawa N. 2000. Stirring up development with the heterotrimeric kinesin KIF3. Traffic. 1:29–34 10.1034/j.1600-0854.2000.010105.x [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Takemura R. 2005. Molecular motors and mechanisms of directional transport in neurons. Nat. Rev. Neurosci. 6:201–214 10.1038/nrn1624 [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Pfister K.K., Yorifuji H., Wagner M.C., Brady S.T., Bloom G.S. 1989. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 56:867–878 10.1016/0092-8674(89)90691-0 [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Sato-Yoshitake R., Kobayashi N., Pfister K.K., Bloom G.S., Brady S.T. 1991. Kinesin associates with anterogradely transported membranous organelles in vivo. J. Cell Biol. 114:295–302 10.1083/jcb.114.2.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs R.P., Amargo E.V., Somasundaram A., Simpson C.L., Prakriya M., Denning M.F., Green K.J. 2011. The calcium ATPase SERCA2 regulates desmoplakin dynamics and intercellular adhesive strength through modulation of PKCalpha signaling. FASEB J. 25:990–1001 10.1096/fj.10-163261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson T.Y., Fontao L., Godsel L.M., Choi H.J., Huen A.C., Borradori L., Weis W.I., Green K.J. 2004. In vitro methods for investigating desmoplakin-intermediate filament interactions and their role in adhesive strength. Methods Cell Biol. 78:757–786 10.1016/S0091-679X(04)78026-7 [DOI] [PubMed] [Google Scholar]
- Jaulin F., Xue X., Rodriguez-Boulan E., Kreitzer G. 2007. Polarization-dependent selective transport to the apical membrane by KIF5B in MDCK cells. Dev. Cell. 13:511–522 10.1016/j.devcel.2007.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil R., Kiessling C., Hatzfeld M. 2009. Targeting of p0071 to the midbody depends on KIF3. J. Cell Sci. 122:1174–1183 10.1242/jcs.045377 [DOI] [PubMed] [Google Scholar]
- Klessner J.L., Desai B.V., Amargo E.V., Getsios S., Green K.J. 2009. EGFR and ADAMs cooperate to regulate shedding and endocytic trafficking of the desmosomal cadherin desmoglein 2. Mol. Biol. Cell. 20:328–337 10.1091/mbc.E08-04-0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P.J., Franke W.W. 1994. Desmosomal cadherins: another growing multigene family of adhesion molecules. Curr. Opin. Cell Biol. 6:682–687 10.1016/0955-0674(94)90094-9 [DOI] [PubMed] [Google Scholar]
- Kondo S., Sato-Yoshitake R., Noda Y., Aizawa H., Nakata T., Matsuura Y., Hirokawa N. 1994. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J. Cell Biol. 125:1095–1107 10.1083/jcb.125.5.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A.P., Borgwardt J.E., Green K.J. 1996. Analysis of desmosomal cadherin-adhesive function and stoichiometry of desmosomal cadherin-plakoglobin complexes. J. Invest. Dermatol. 107:293–300 10.1111/1523-1747.ep12363000 [DOI] [PubMed] [Google Scholar]
- Lai-Cheong J.E., Arita K., McGrath J.A. 2007. Genetic diseases of junctions. J. Invest. Dermatol. 127:2713–2725 10.1038/sj.jid.5700727 [DOI] [PubMed] [Google Scholar]
- Lechler T., Fuchs E. 2007. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J. Cell Biol. 176:147–154 10.1083/jcb.200609109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuchowius K.J., Weibrecht I., Landegren U., Gedda L., Söderberg O. 2009. Flow cytometric in situ proximity ligation analyses of protein interactions and post-translational modification of the epidermal growth factor receptor family. Cytometry A. 75:833–839 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Cole N.B., Marotta A., Conrad P.A., Bloom G.S. 1995. Kinesin is the motor for microtubule-mediated Golgi-to-ER membrane traffic. J. Cell Biol. 128:293–306 10.1083/jcb.128.3.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcozzi C., Burdett I.D., Buxton R.S., Magee A.I. 1998. Coexpression of both types of desmosomal cadherin and plakoglobin confers strong intercellular adhesion. J. Cell Sci. 111:495–509 [DOI] [PubMed] [Google Scholar]
- Mary S., Charrasse S., Meriane M., Comunale F., Travo P., Blangy A., Gauthier-Rouvière C. 2002. Biogenesis of N-cadherin-dependent cell-cell contacts in living fibroblasts is a microtubule-dependent kinesin-driven mechanism. Mol. Biol. Cell. 13:285–301 10.1091/mbc.01-07-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattey D.L., Burdge G., Garrod D.R. 1990. Development of desmosomal adhesion between MDCK cells following calcium switching. J. Cell Sci. 97:689–704 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay B., Nam S.C., Choi K.W. 2010. Kinesin II is required for cell survival and adherens junction positioning in Drosophila photoreceptors. Genesis. 48:522–530 10.1002/dvg.20642 [DOI] [PubMed] [Google Scholar]
- Nie Z., Merritt A., Rouhi-Parkouhi M., Tabernero L., Garrod D.R. 2011. Membrane-impermeable cross-linking provides evidence for homophilic, isoform-specific binding of desmosomal cadherins in epithelial cells. J. Biol. Chem. 286:2143–2154 10.1074/jbc.M110.192245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nindl I., Dang C., Forschner T., Kuban R.J., Meyer T., Sterry W., Stockfleth E. 2006. Identification of differentially expressed genes in cutaneous squamous cell carcinoma by microarray expression profiling. Mol. Cancer. 5:30 10.1186/1476-4598-5-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet F., Berx G., van Roy F. 1999. The role of the E-cadherin/catenin adhesion complex in the development and progression of cancer. Mol. Cell Biol. Res. Commun. 2:77–85 10.1006/mcbr.1999.0155 [DOI] [PubMed] [Google Scholar]
- Pasdar M., Nelson W.J. 1989. Regulation of desmosome assembly in epithelial cells: kinetics of synthesis, transport, and stabilization of desmoglein I, a major protein of the membrane core domain. J. Cell Biol. 109:163–177 10.1083/jcb.109.1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasdar M., Krzeminski K.A., Nelson W.J. 1991. Regulation of desmosome assembly in MDCK epithelial cells: coordination of membrane core and cytoplasmic plaque domain assembly at the plasma membrane. J. Cell Biol. 113:645–655 10.1083/jcb.113.3.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasdar M., Li Z., Krzeminski K.A. 1992. Desmosome assembly in MDCK epithelial cells does not require the presence of functional microtubules. Cell Motil. Cytoskeleton. 23:201–212 10.1002/cm.970230304 [DOI] [PubMed] [Google Scholar]
- Penn E.J., Burdett I.D., Hobson C., Magee A.I., Rees D.A. 1987. Structure and assembly of desmosome junctions: biosynthesis and turnover of the major desmosome components of Madin-Darby canine kidney cells in low calcium medium. J. Cell Biol. 105:2327–2334 10.1083/jcb.105.5.2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw R.M., Fay A.J., Puthenveedu M.A., von Zastrow M., Jan Y.N., Jan L.Y. 2007. Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell. 128:547–560 10.1016/j.cell.2006.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Stehbens S.J., Paterson A.D., Crampton M.S., Shewan A.M., Ferguson C., Akhmanova A., Parton R.G., Yap A.S. 2006. Dynamic microtubules regulate the local concentration of E-cadherin at cell-cell contacts. J. Cell Sci. 119:1801–1811 10.1242/jcs.02903 [DOI] [PubMed] [Google Scholar]
- Stenoien D.L., Brady S.T. 1997. Immunochemical analysis of kinesin light chain function. Mol. Biol. Cell. 8:675–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokin G.B., Lillo C., Falzone T.L., Brusch R.G., Rockenstein E., Mount S.L., Raman R., Davies P., Masliah E., Williams D.S., Goldstein L.S. 2005. Axonopathy and transport deficits early in the pathogenesis of Alzheimer’s disease. Science. 307:1282–1288 10.1126/science.1105681 [DOI] [PubMed] [Google Scholar]
- Storrie B., White J., Röttger S., Stelzer E.H., Suganuma T., Nilsson T. 1998. Recycling of golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 143:1505–1521 10.1083/jcb.143.6.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. 1995. Morphogenetic roles of classic cadherins. Curr. Opin. Cell Biol. 7:619–627 10.1016/0955-0674(95)80102-2 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Kanai Y., Okada Y., Nonaka S., Takeda S., Harada A., Hirokawa N. 1998. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 93:1147–1158 10.1016/S0092-8674(00)81459-2 [DOI] [PubMed] [Google Scholar]
- Teng J., Rai T., Tanaka Y., Takei Y., Nakata T., Hirasawa M., Kulkarni A.B., Hirokawa N. 2005. The KIF3 motor transports N-cadherin and organizes the developing neuroepithelium. Nat. Cell Biol. 7:474–482 10.1038/ncb1249 [DOI] [PubMed] [Google Scholar]
- Terada S., Kinjo M., Aihara M., Takei Y., Hirokawa N. 2010. Kinesin-1/Hsc70-dependent mechanism of slow axonal transport and its relation to fast axonal transport. EMBO J. 29:843–854 10.1038/emboj.2009.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tselepis C., Chidgey M., North A., Garrod D. 1998. Desmosomal adhesion inhibits invasive behavior. Proc. Natl. Acad. Sci. USA. 95:8064–8069 10.1073/pnas.95.14.8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twelvetrees A.E., Yuen E.Y., Arancibia-Carcamo I.L., MacAskill A.F., Rostaing P., Lumb M.J., Humbert S., Triller A., Saudou F., Yan Z., Kittler J.T. 2010. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron. 65:53–65 10.1016/j.neuron.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D. 2003. The molecular motor toolbox for intracellular transport. Cell. 112:467–480 10.1016/S0092-8674(03)00111-9 [DOI] [PubMed] [Google Scholar]
- Vale R.D., Reese T.S., Sheetz M.P. 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 42:39–50 10.1016/S0092-8674(85)80099-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K.J., Hammond J.W. 2009. Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Lizotte D.L., Abramson T., Barenboim L., Schnapp B.J., Rapoport T.A. 1998. Light chain–dependent regulation of kinesin’s interaction with microtubules. J. Cell Biol. 143:1053–1066 10.1083/jcb.143.4.1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Salmon W.C., Salmon E.D. 2000. Feedback interactions between cell-cell adherens junctions and cytoskeletal dynamics in newt lung epithelial cells. Mol. Biol. Cell. 11:2471–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F.M., Mattey D.L., Garrod D.R. 1984. Calcium-induced reorganization of desmosomal components in cultured human keratinocytes. J. Cell Biol. 99:2211–2215 10.1083/jcb.99.6.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R., Borchert-Stuhlträger M., Leube R.E. 2002. Desmosomes: interconnected calcium-dependent structures of remarkable stability with significant integral membrane protein turnover. J. Cell Sci. 115:1717–1732 [DOI] [PubMed] [Google Scholar]
- Woźniak M.J., Allan V.J. 2006. Cargo selection by specific kinesin light chain 1 isoforms. EMBO J. 25:5457–5468 10.1038/sj.emboj.7601427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H., Nakata T., Okada Y., Hirokawa N. 1995. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J. Cell Biol. 130:1387–1399 10.1083/jcb.130.6.1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa M., Kaverina I.N., Wang A., Fujita Y., Reynolds A.B., Anastasiadis P.Z. 2004. A novel interaction between kinesin and p120 modulates p120 localization and function. J. Biol. Chem. 279:9512–9521 10.1074/jbc.M310895200 [DOI] [PubMed] [Google Scholar]