Abstract
Liposomes have been widely used as a drug delivery vehicle and currently, more than 10 liposomal formulations are approved by the Food and Drug Administration for clinical use. However, upon targeting, the release of the liposome-encapsulated contents is usually slow. We have recently demonstrated that contents from appropriately-formulated liposomes can be rapidly released by the cancer-associated enzyme matrix metalloproteinase-9 (MMP-9). Herein, we report our detailed studies to optimize the liposomal formulations. By properly selecting the lipopeptide, the major lipid component and their relative amounts, we demonstrate that the contents are rapidly released in the presence of cancer-associated levels of recombinant human MMP-9. We observed that the degree of lipid mismatch between the lipopepides and the major lipid component profoundly affects the release profiles from the liposomes. By utilizing the optimized liposomal formulations, we also demonstrate that cancer cells (HT-29) which secrete low levels of MMP-9 failed to release significant amount of the liposomal contents. Metastatic cancer cells (MCF7) secreting high levels of the enzyme rapidly release the encapsulated contents from the liposomes.
INTRODUCTION
Liposomes are lipid bilayers encapsulating an aqueous core (1, 2). Due to the ease of preparation in a size-controlled manner and biocompatibility, they are widely used as lipid-based nanoparticles for drug delivery (3, 4) Hydrophilic molecules are encapsulated in the aqueous interior of the liposomes and hydrophobic molecules partition into the lipid bilayers (5, 6). Incorporation of polyethylene glycol (PEG) containing lipids reduces opsonization and renders the liposomes long-circulating in the blood stream (7, 8). After intravenous administration, long-circulating liposomes passively accumulate into the tumor tissues due to the enhanced permeation and retention effect (9). For active targeting, antibodies (10), peptides (11) and small molecules (12) have been conjugated to the surface of the liposomes.
The side effects of anticancer drugs are considerably reduced by encapsulating them in the liposomes (13). There are currently about 10 liposomal drug formulations approved by the US Food and Drug Administration (FDA) for human use (14). Numerous clinical trials are currently in progress using liposome-encapsulated anti-cancer drugs (15). However, upon targeting, the release of the encapsulated drugs is usually very slow (16). An attractive solution to this problem is to formulate liposomes with appropriate lipids such that the contents are released in response to an external trigger. Temperature (17), pH (18), light (including near infrared, 19), ultrasound (20), metal ions (21), and enzymes (22, 23) have been used as triggers. We have recently demonstrated the release of liposomal contents by the cancer-associated enzyme matrix metalloproteinase-9 (MMP-9, 24, 25).
Matrix metalloproteinases (MMPs) are a family of 23 Zn2+ and Ca2+ dependents extracellular enzymes responsible for the degradation and remodeling of the extracellular matrix (26, 27). The expression levels and activation of these enzymes are regulated at multiple levels (28). Increased expression of MMPs is associated with many diseases, including various types of cancers (29, 30). MMP-9 is involved in the progression and metastasis of many cancers (31, 32).
Herein, we report our detailed mechanistic and optimization studies on the triggered release of liposomal contents by MMP-9. We have prepared triple helical peptide substrates for MMP-9 and conjugated them with fatty acids to synthesize the corresponding lipopeptides. Liposomes incorporating these lipopeptides released the encapsulated contents in response to cancer associated levels of recombinant MMP-9. We optimized the system by systematically varying the triple helicity of the lipopeptides, its amount and the structure of the major lipid component of the liposomes. We observed that the contents release is dependent on the degree of mismatch between the fatty acids of the lipopeptides and the major lipid of the liposomes. Using the optimized formulation, we demonstrate that the liposomal contents are released by MMP-9 secreted from the breast adenocarcinoma cells, MCF7. The colorectal adenocarcinoma cells HT29 do not secrete high levels of MMP-9 and failed to release significant amounts of the encapsulated contents from the liposomes.
MATERIALS AND METHODS
Synthesis of the Lipopeptides
The lipopeptides were synthesized by employing a microwave assisted peptide synthesizer (Liberty, CEM corporation, Matthews, NC) using standard Fmoc protected amino acids (0.1 mmol scale). The commercially available Fmoc-Gly-CLEAR acid resin was used as the solid support. The reactions were carried out in a 30 mL reaction vessel housed within the microwave chamber (CEM Discover) of the peptide synthesizer. A mixture of HOBT/HBTU in five fold excess was used as the coupling reagent. Each coupling step (with the exception of Arg) was performed at 50 °C under 20 W microwave power for 5 min. The coupling of amino acid Arg was done twice at 25 °C for 25 min without the use of microwave. The N-terminal fatty acid was also subjected to double coupling. The activator base used was 5 equivalents of diisopropylethylamine dissolved in N-methylpyrrolidone. The Fmoc deprotection was accomplished with 5% (by weight) of piperazine in DMF. Cleavage from the solid support was performed inside the microwave chamber for 35 min using a cocktail of TFA-triisopropylsilane and water (95%-2.5%-2.5%). The crude peptides were precipitated as white solids with ice-cold ether, centrifuged and purified by a semi-preparatory RP-HPLC. The monoisotopic masses of purified peptides were determined with an Applied Biosystems/MDS SCIEX 4800 MALDI ToF/ToF analyzer.
Conditions for analytical HPLC
The analytical HPLC was performed with Vydac analytical diphenyl column (RP219TP5415) with a linear gradient of 0–70% acetonitrile in water as eluant, with each solvent containing 0.1% TFA. The flow rate was maintained at 1.5 mL/min for 25 min. The UV detection wavelength was kept at 214 nm.
Conditions for Semi-preparatory HPLC
Vydac diphenyl (RP219TP510) RP HPLC column was used. A linear gradient of 0–70% acetonitrile in water was maintained for 60 min at flow rate 8mL/min. Each solvent contained 0.1 % TFA. The purification was monitored at a detector wavelength of 235 nm.
Liposome formation
Stock solutions of lipopeptides were prepared by weighing appropriate amounts of the lipopeptide and then dissolving in 5:1 chloroform/methanol to a final concentration of 1 mg/mL. Stock solutions of phospholipids were prepared in chloroform to obtain a final concentration of 2 mg/mL. The lipopeptide and the phospholipid were mixed in appropriate volumes in a 10 mL round bottom flask. A thin lipid film was obtained by evaporating this mixture in a rotary evaporator at 40 °C. The thin film was kept under high vacuum overnight to remove any residual organic solvents. The dry lipid film was hydrated with 1.5 mL buffer containing 6-carboxyfluorescein at 60 °C for 1 – 2 h. The hydrated solution was stored for 3–4 h to ensure complete hydration and to allow for proper folding of the lipopeptides into triple helices. In the next step, the large multilamellar vesicles were broken into unilamellar vesicles of appropriate sizes by probe sonication in a heating block for 1 h at 56 °C. After sonication, the liposome solution was centrifuged to remove any suspended metal particles from the probe tip and extruded through a 100 nm polycarbonate filter (Avanti Polar lipids, AL). The unencapsulated carboxyfluorescein was removed from the liposomes by gel filtration through a Sephadex G-100 column. The Sphadex G-100 was hydrated for 24 h at 60 °C and equilibrated with 5 times the volume of the assay buffer before loading of the liposomes. The osmolarity of this elunat buffer was made equal to the osmolarity of the carboxyfluorescein solution used for hydration by adding appropriate amount of NaCl. This ensured minimum background leakage of carboxyfluorescein due to osmotic shock during the actual leakage experiment.
For CD experiments, the thin film prepared in a similar manner as discussed above was hydrated with 1 mL of 5 mM phosphate buffer (pH 8.0) at 60 °C for 1 h. The liposomes after 2 h were subjected to probe sonication for 30 min. The CD spectra were recorded at 25 °C immediately after the probe sonication.
Buffer solutions
The buffer used in making carboxyfluorescein solution was 25 mM HEPES (pH = 8.0) containing 100 mM carboxyfluorescein, 10 mM CaCl2, 10 μM ZnCl2 and 100 mM NaCl. The buffer used for column equilibration, elution of liposomes from the column and release experiment consist of 25 mM HEPES (pH 8.0) with 10 mM CaCl2, 10 μM ZnCl2 and between 100–150 mM NaCl. The buffer prepared for the liposome CD experiments was 5 mM phosphate buffer at pH 8.0.
Circular Dichromism spectroscopy
CD spectra were recorded in a Jasco J815 CD spectrometer using 1 mm path length quartz cuvette. For temperature dependant CD spectra, the concentration of the lipopeptides used was 1 mg/mL in aqueous solution. The solutions were stored for 12 h at 4 °C before recording the spectra. For each sample, the equilibration time was set for 15 min at each temperature. For each temperature an average of 3 scans were recorded at a scan speed of 50 nm/min. The response time set was 0.25 s. The melting temperature was obtained by plotting the CD intensity maxima at 225 nm vs. temperature. For the liposome CD spectra, the average of 50 scans was recorded with a scan speed of 100 nm/min.
Release studies
The kinetics of carboxyfluorescence release from the liposomes was monitored by recording the intensity of fluorescence emission at 518 nm using a spectrophotometer (Spectrmax, Molecular Devices) with an excitation wavelength at 480 nm. Each experiment was conducted in triplicate wells on a 96 standard opaque fluorescent well plate. The final reading taken was the average of reading from 3 wells. For experiments involving recombinant human MMP-9, the activity of the enzyme was tested with a commercially available fluorogenic substrate prior to the experiment to ensure the enzyme is fully active. The fluorescence emission was recorded at 20 s interval for 1 h. After 1 h, the liposomes were ruptured using optimized concentration of Triton-X to allow complete leakage of the dye. The fluorescence emission corresponding to this leakage was taken as the maximum emission possible in calculating the percentage release. The percentage release was calculated using a formula reported previously (25).
Growth and maintenance of cells
All cells were obtained from American Type Culture Collection (Manassas, VA, USA). MCF7 (human breast adenocarcinoma) and HT-29 (human colorectal adenocarcinoma) cell lines were cultured in Eagle's Minimum Essential medium, and McCoy's 5a medium, respectively at 37 °C in humidified atmosphere containing 5% CO2. All media were supplemented with 5% penicillin and streptomycin solution and 10% fetal bovine serum. When the cells achieved 70% of confluency, the old culture media were replaced with fresh culture media without phenol red. The cells were further incubated for 24 h, collected, and centrifuged. The supernatants were collected for further experiments. For cell based liposomal release experiments, 50 μL of cell media was added to a total volume of 200 μL in each well and the intensity of emission spectra of carboxyfluorescein at 518 nm (λex = 480 nm) was monitored as discussed in the recombinant MMP-9 release studies.
RESULTS AND DISCUSSION
Synthesis of the lipopeptides
The natural substrate for the MMP-9 is gelatin (33). Gelatin and collagen-mimetic triple-helical peptides have been used as models to study substrate selectivity of the MMPs (34), as scaffolds and functional biomaterials for tissue engineering (35), and as targeting moieties for liposomes (36). Structurally, the collagen-mimetic peptides contain repeat units of the amino acid triad Gly-Pro-Hyp (GPO). For these studies, we prepared homo-trimeric collagen-mimetic peptides containing the cleavage site for MMP-9 employing a microwave-assisted, solid-phase peptide synthesizer (37). The peptides have the general structure of CH3(CH2)16CONH-GPQGIAGQR(GPO)nGG-COOH, where the MMP-9 cleavage site is the peptide bond connecting Gly and Ile. In order to vary the triple-helicity, the number of GPO repeat units in the peptides (i.e., n) were systematically varied (Table 1). We employed the microwave-assisted peptide synthesizer to conjugate the fatty acids to the peptides.
Table 1.
The synthesized lipopeptides, their calculated and observed molecular masses, the calculated Rpn values at 5 °C, and the melting temperatures (Tm).
| No. | Name | n | Calcd. M+ | Obs. M+ | Rpn | Tm (°C) |
|---|---|---|---|---|---|---|
| 1 | GPO3 | 3 | 2065.06 | 2066.07 | 0.08 | 43 |
| 2 | GPO4 | 4 | 2332.24 | 2332.27 | 0.11 | 56 |
| 3 | GPO5 | 5 | 2600.37 | 2599.40 | 0.13 | 64 |
In order to determine the structural resemblance of the synthesized lipopeptides with collagen (and gelatin), we recorded the CD spectra in aqueous solution (1mg/mL). In CD spectra, the presence of a positive peak at 225 nm and a large negative peak at 195 – 200 nm confirmed the triple helical structure of the lipopeptides (Figure 1A, 38). The calculated Rpn values (38) at 5 °C and the melting temperatures (Tm) of the lipopeptides are also shown in Table 1. Since the Rpn value for natural collagen is 0.13 (38), we concluded that the lipopeptides adopt triple helical conformation, similar to collagen in aqueous solution. The lipopeptides GPO3 and GPO4 showed triple helical conformation in 5 mM phosphate buffer (pH = 8.0). However, the lack of sufficient solubility of the lipopeptides GPO5 in this buffer prevented us from recording and comparing the spectra for all the lipopeptides at pH = 8.0.
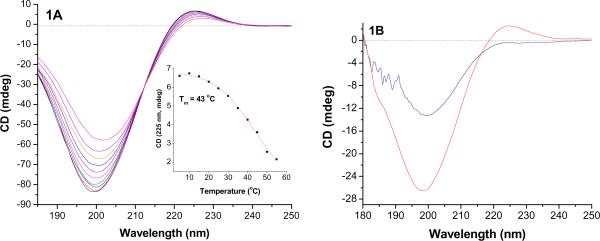
Figure 1.
The temperature dependent CD spectra (5 - 55 °C) of the lipopeptide GPO3 in aqueous solution (1A) and in POPC liposomes prepared in 5 mM phosphate buffer, pH = 8.0 (1B, red trace) are shown. The presence of the positive peak at 225 nm and the large negative peak at 198 nm indicate triple helical structure of this lipopeptide in aqueous solution as well as in liposomes. The inset of 1A shows the melting curve for GPO3 monitored at 225 nm. When the liposomes were incubated with 2.3 μM recombinant MMP-9 for 30 minutes, the triple helicity of the liposome-incorporated GPO3 decreased substantially (1B, blue trace). To improve the signal-to-noise ratio, the spectra shown for the liposome-incorporated GPO3 are the averages of 50 scans. For the last spectrum, the CD spectrum of MMP-9 was subtracted.
Subsequently, we prepared liposomes containing 30 mol% of the synthesized lipopeptides in 25 mM phosphate buffer, pH = 8.0. The major lipid component of the liposomes was varied between POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) and DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine), and the CD spectra were recorded. Highest triple helicities for the liposome-incorporated lipopeptides were observed in the POPC liposomes (Figure 1B, red trace), possibly reflecting the mismatch between the fatty acids of the lipopeptides and POPC and the resultant demixing of the major lipid and the lipopeptides. When we incubated these liposomes with 2.3 μM recombinant MMP-9 for 30 minutes, the triple helicity of the liposome-incorporated lipopeptides was reduced substantially (Figure 1B, blue trace) indicating that the cleavage of the surface-exposed peptide substrates by the enzyme decreases the triple helicity of the lipopeptides.
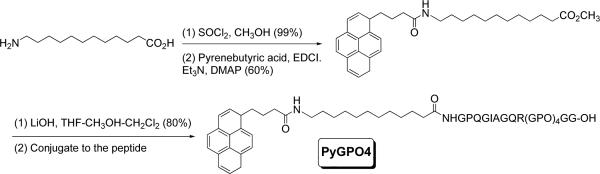
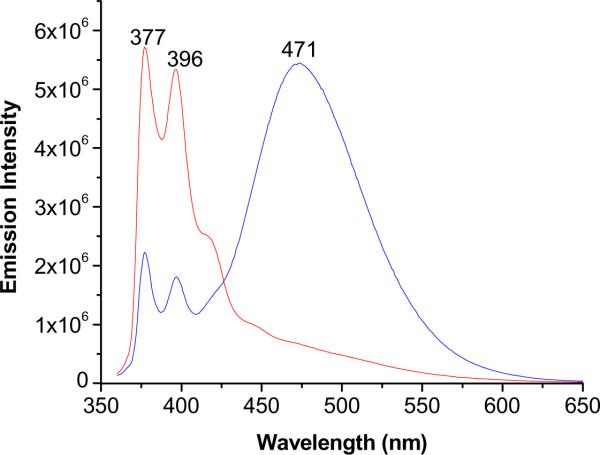
As an independent measure of the triple helicity of the liposome-incorporated lipopeptides, we synthesized a pyrene-containing analog of GPO4 (PyGPO4, Scheme 1, synthetic details are included in Supporting Information). The CD spectrum of this lipopeptide in aqueous solution showed a positive peak at 225 nm and a large negative peak at 198 nm, indicating the formation of triple helix (Tm = 36 °C, Supporting Information). Subsequently, we recorded the fluorescence emission spectra (λex = 340 nm) of POPC liposomes incorporating 30 mol% of PyGPO4 in 25 mM HEPES buffer (pH = 8.0). The presence of a strong excimer peak in the emission spectrum (471 nm) indicated the proximity of the pyrene groups in the hydrophobic fatty acids of the lipopeptides (Figure 2, blue trace). Upon incubation of the liposomes with 3 μM MMP-9 for 15 minutes, we observed that the pyrene excimer emission peak decreased and the monomer emission peaks increased in intensity (Figure 2, red trace). These results indicate that the lipopeptide PyGPO4 is demixed in the lipid bilayer of the liposomes. Hydrolysis of the peptides by MMP-9 leads to the reorganization of the lipids with increased separation between the pyrene groups.
Scheme 1.
The synthesis of the lipopeptide PyGPO4.
Figure 2.
The emission spectra of POPC liposomes incorporating 30 mol% of the lipopeptide PyGPO4 (blue trace) in 25 mM HEPES buffer, pH = 8.0 containing 100 mM NaCl, 10 mM CaCl2 and 10 mM ZnCl2. The presence of the intense emission at 471 nm indicated the pyrene excimer formation with the lipid bilayer of the liposomes. The emission spectrum of the liposomes after incubation with 2.3 μM MMP-9 for 15 minutes is shown with the red trace.
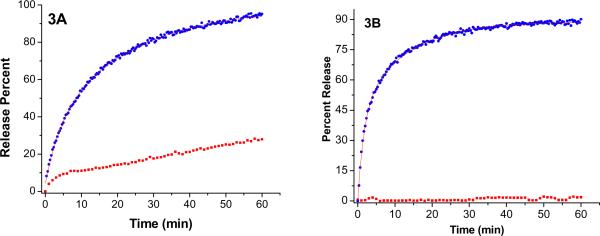
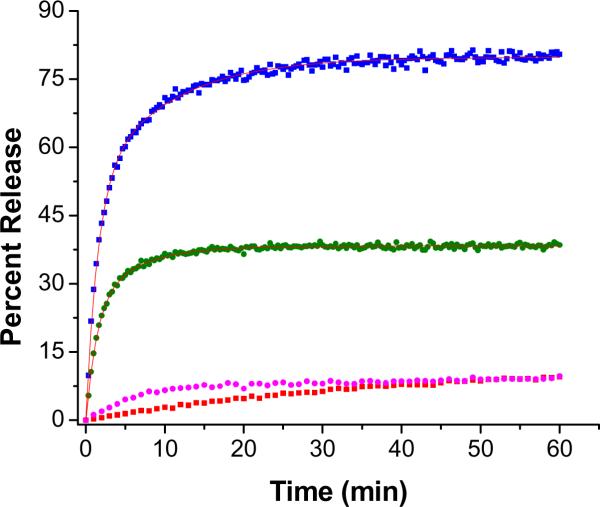
Next, we proceeded to determine the optimal liposomal formulation for the release methodology. We varied the amounts, the triple helicity of the lipopeptides and the major lipid component of the liposomes. First, the amount of lipopeptide in the liposomes (30 mol% of GPO3) and the concentration of MMP-9 (2.3 μM) were kept constant, and the major lipid component was varied (DOPC, POPC and DSPC). The rates of release of encapsulated carboxyfluorescein from these liposomal formulations were monitored (λex = 480 nm; λem = 518 nm, Figure 3). When DOPC was the major lipid, we observed complete release of the encapsulated dye within 30 minutes (Figure 3A). However, these liposomes released about 25% of the contents during the same time in the absence of any enzyme (Figure 3A). For the POPC liposomes, 90% of the encapsulated dye was released by MMP-9 in 30 minutes (Figure 3B). We were pleased to note that for these liposomes, no dye released in the absence of MMP-9 (Figure 3B). When DSPC was the major lipid, no significant dye release from the liposomes was observed either in the presence and or in absence of MMP-9 (data now shown). As discussed before, incorporation of the lipopeptides in the liposomes results in lipid demixing. The degree of this demixing depends on the mismatch between the hydrophobic fatty acid moieties of the lipopeptides and the major lipid component of the liposomes. For the synthesized lipopeptides, the saturated stearoyl moiety does not mix well with the unsaturated oleoyl groups of the lipids POPC and DOPC in the liposomes. However, when DSPC is the major component, the lipopeptides are well mixed within the lipid bilayer of the liposomes.
Figure 3.
The release profiles for encapsulated carboxyfluorescein (λex = 480 nm; λem = 518 nm) for the liposomes incorporating 30 mol% of the lipopeptide GPO3 in the presence and absence of MMP-9 in 25 mM HEPES buffer, pH = 8.0 containing 100 mM NaCl, 10 mM CaCl2 and 10 mM ZnCl2. For DOPC liposomes (3A), complete dye release was observed (blue circles) in the presence of 2.3 μM MMP-9 and about 25% release was observed in the absence of the enzyme (red squares). For POPC liposomes (3B), 90% of the encapsulated dye was released with 2.3 μM MMP-9 (blue circles); in the absence of the enzyme, no dye release was detected (red squares). The lines through the release profiles are the fitted curved using a biexponential rate equation.
Previously, we have observed that the dye release profiles for liposomes incorporating 30 mol% of the lipopeptide GPO4 were single exponential in nature (25). The dye release profiles shown in Figure 3 can be fitted best with a biexponential rate equation: F (release) = F0 − (A1*e−k1t + A2*e−k2t) to obtain the rate constants k1 and k2 for the dye release and the amplitudes A1 and A2. The calculated rate constants for the dye release from the DOPC and POPC liposomes are shown in Table 2. For the POPC liposomes, both rate constants were faster compared to those for the DOPC liposomes (Table 2). When we prepared liposomes incorporating 30 mol% of the lipopeptide GPO5 with POPC as the major lipid, encapsulating sufficient concentration of carboxyfluorescein was found to be very difficult. It should be noted that the triple helices are formed on the outside as well as in the inside of the liposomes. The increased molecular volume of the lipopeptide of GPO5 possibly hinders the efficient encapsulation of carboxyfluorescein inside the liposomes. When we decreased the amount of GPO5 to 20 mol% in the POPC liposomes, the dye was encapsulated efficiently. However, the release from these liposomes was inefficient (< 40%, Supporting Information) in the presence of 2.3 μM MMP-9. The stability of the triple helices increases with the number of GPO repeat units (Table 1, 44). Increased thermal stability of the triple helices leads to decreased cleavage rates by the MMPs (44).
Table 2.
Calculated rate constants (k1, k2) and amplitudes for the release of encapsulated carboxyfluorescein from DOPC and POPC liposomes containing 30 mol% of GPO3 in the presence of 2.3 μM MMP-9 (errors: 3 – 6%).
| Major lipid | k1 (sec−1) | Amplitude (%) | k2 (sec−1) | Amplitude (%) |
|---|---|---|---|---|
| DOPC | 7.0 × 10−4 | 67 | 3.8 × 10−3 | 33 |
| POPC | 1.3 × 10−3 | 51 | 9.9 × 10−3 | 49 |
Based on these results, we selected POPC as the major lipid component of the liposomes. Our next objective was to determine the amount of lipopeptide needed in the liposomes for optimal release of encapsulated carboxyfluorescein. We have previously observed that 30 mol% of lipopeptide GPO4 in POPC liposomes exhibited the high amount of contents release (> 95%) in the presence of MMP-9 and low amount of background release (< 10%) in the absence of the enzyme (25). When 30 mol% of the lipopeptide GPO3 was incorporated in POPC liposomes, 90% of the encapsulated carboxyfluorescein was released in 30 minutes in the presence of 2.3 μM MMP-9 (Figure 3B). No release of the dye from the liposomes was observed in the absence of the enzyme (Figure 3B). When we reduced the amount of GPO3 to 20 mol% in the POPC liposomes, 80% of the contents were released in the presence of 2.3 μM MMP-9 (Figure 4, blue squares). For these liposomes, we observed that 10% of the encapsulated dye was released in the absence of any enzyme (Figure 4, red squares). If the liposomes contained 10 mol% of the lipopeptide GPO3, only 38% of the contents was released by MMP-9 (2.3 μM) in 1 hour (Figure 4, green circles) with a background release of 10% in the absence of the enzyme (Figure 4, magenta circles). Increasing the amount to GPO3 to 40 mol% in the liposomes led to a drastic reduction of the release efficiency (< 10%, data not shown) in the presence of 2.3 μM of MMP-9. This is not due to insufficient peptide cleavage due to shortage of MMP-9 as addition of more enzyme did not improve the release efficiency. Variation of the amount of GPO5 in the POPC liposomes did not improve the efficiency of the contents release in the presence of the enzyme (Supporting Information).
Figure 4.
The release profiles for liposome-encapsulated carboxyfluorescein (λex = 480 nm; λem = 518 nm) in the presence and absence of 2.3 μM MMP-9 in 25 mM HEPES buffer, pH = 8.0 containing 100 mM NaCl, 10 mM CaCl2 and 10 mM ZnCl2 for POPC liposomes incorporating 20 mol% (blue squares) and 10 mol% of lipopeptide GPO3 (green circles). The background releases of the dye in the absence of the enzyme are also shown (red squares: 20 mol% GPO3; magenta circles: 10 mol% GPO3). The lines through the release profiles are the fitted curved using a biexponential rate equation.
Interestingly, these release data also exhibited biexponential rate profiles. The release of the liposomal contents involves several intermediate steps: binding of the enzyme to the exposed peptides, hydrolysis of the peptides, creation of lipid “defects” and lipid reorganization, release of encapsulated carboxyfluorescein, and repairing of the “defects” created in the lipid bilayer. Fitting the observed release data by a biexponential rate equation is clearly an over simplification of the complex set of steps involved in the triggered release process. Notably, when we decreased the amount of the lipopeptide GPO3 in the liposomes, the magnitudes of the rate constants for the release remained nearly the same but the amplitude of the faster rate process progressively increased and the amplitude for the slower step decreased (Table 3). It is possible that the faster of the two steps is the repair of the “defects” created in the lipid bilayer as MMP-9 hydrolyzes the exposed peptides. With less amount of the lipopeptide incorporated in the liposomes, it is expected that less amount of “defects” are created in the bilayers and hence, they are repaired quickly. Based on these observations, we selected 30 mol% as the optimal amount of lipopeptide in our liposomal formulations.
Table 3.
Calculated rate constants (k1, k2) and amplitudes for the release of encapsulated carboxyfluorescein from POPC liposomes containing varying amounts of the lipopeptide GPO3 in the presence of 2.3 μM MMP-9 (errors: 3 – 6%).
| Amount of GPO3 (mol%) | k1 (sec−1) | Amplitude (%) | k2 (sec−1) | Amplitude (%) |
|---|---|---|---|---|
| 30 | 1.3 × 10−3 | 51 | 9.9 × 10−3 | 49 |
| 20 | 1.7 × 10−3 | 37 | 11.1 × 10−3 | 63 |
| 10 | 1.9 × 10−3 | 18 | 11.8 × 10−3 | 72 |
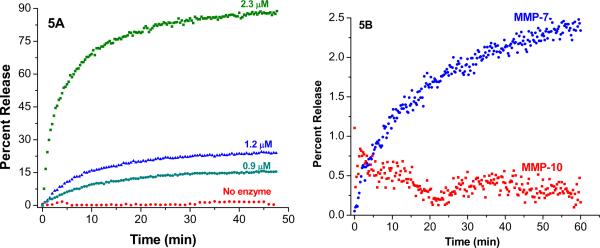
After optimizing the major lipid component and the amount of lipopeptides in the liposomes, we turned our attention to the response of the release methodology to MMP-9 concentration. It is reported that in normal tissues, the concentration of active form of MMP-9 is about 10 nM (39). However, for lung cancer patients, the concentration of this enzyme in the bronchial lavage fluid can be as high as 200 nM (40). We have previously observed that for the POPC liposomes containing 30 mol% of the lipopeptide GPO4, the rates and the amounts of contents release progressively decreased with decreasing concentrations of MMP-9 (25). While 2.3 μM MMP-9 completely released the contents within 30 minutes, 50% of the encapsulated dye release was observed in 1 hour in the presence of 200 nM enzyme (25). In order to determine if such a desirable property exists for the POPC liposomes containing 30 mol% of the lipopeptide GPO3, we added varying concentrations of recombinant human MMP-9 to these liposomes and monitored the release of encapsulated carboxyfluorescein (Figure 5A).
Figure 5.
The release profiles for POPC liposome-encapsulated carboxyfluorescein (λex = 480 nm; λem = 518 nm) containing 30 mol% of lipopeptide GPO3 in the presence 2.3 μM (green squares), 1.2 μM (blue triangles) and 900 nM (cyan circles) of MMP-9 in 25 mM HEPES buffer, pH = 8.0 (5A) containing 100 mM NaCl, 10 mM CaCl2 and 10 mM ZnCl2. The background release in the absence of any MMP-9 is shown in red circles (5A). Figure 5B shows the release of the encapsulated dye from these liposomes in the presence of 2.3 μM MMP-7 (blue circles) and 2.3 μM MMP-10 (red squares) under the same conditions.
When the concentration of the enzyme was decreased from 2.3 μM to 1.2 μM, we observed a significant reduction in the amount of dye release from the liposomes (Figure 5A). In the presence of 900 nM of MMP-9, only 15% of the encapsulated dye was released. The difference in release profiles between the lipopeptides GPO4 and GPO3 may be related to their triple helicity. The Rpn value for GPO4 is 0.11 while that for GPO3 is 0.08 (Table 1). Thus, lesser fraction of the lipopeptide GPO3 exist in triple helical conformation compared to GPO4. Upon peptide hydrolysis by MMP-9, the structural perturbation within the lipid bilayer of the liposomes will be more pronounced in the case of the lipopeptide GPO4 compared to GPO3. The liposomes did not release any significant amount of the encapsulated dye in the presence of other MMPs which cannot hydrolyze triple helical peptides (e.g., MMP-7 and -10; Figure 5B) or in the presence of trypsin (data not shown). Taking into consideration all of these results, we decided to proceed to the cell-based studies employing liposomal formulation incorporating 30 mol% of the lipopepide GPO3 or GPO4 with POPC as the major lipid component.
After optimizing the liposomal formulations for contents release triggered by recombinant human MMP-9, we proceeded to determine the release property in the presence of cell-secreted enzyme. We noted, a priori, that our recombinant MMP-9 contains the catalytic and the fibronectin domains, but the cell-secreted enzyme has the additional hemopexin domain (41). For the cell-based studies, the metastatic breast adenocarcinoma cells MCF7 was selected as this cell line is known to secrete high amounts of MMP-9 and -2 in the extracellular matrix (42). The human colorectal adenocarcinoma cells HT-29 was used as the control, since these cells do not secrete high levels of MMP-9 and -2 in the extracellular matrix (43). All cells were obtained and cultured following the protocols from American Type Culture Collection (Manassas, VA, USA). When the cells achieved 70% of confluency, the old culture media was replaced with fresh culture medium without phenol red. The cells were further incubated for 24 h, collected, and centrifuged. The supernatants were collected for the release experiments from the liposomes.
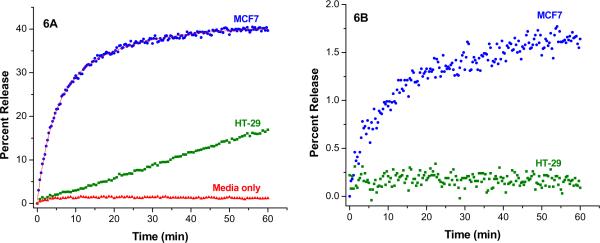
For the POPC liposomes containing 30 mol% of the lipopeptide GPO4, the conditioned media collected from the cultured MCF7 cells rapidly released 40% of the encapsulated dye within 30 minutes (Figure 6A). The release data can be fitted with a biexponential rate equation, with k1 = 1.0 × 10−3 s−1 (44% amplitude) and k2 = 2.5 × 10−3 s−1 (56% amplitude). These liposomes in the presence of the conditioned media collected from the HT-29 cells showed a very different profile (linear), slowly releasing less that 10% of the contents in 30 minutes. The cell culture media (for both cells) did not release any liposome encapsulated dye during the same period of time (Figure 6A). However, liposomes incorporating 30 mol% lipopeptide GPO3 failed to release any encapsulated contents in the presence of conditioned media from the MCF7 or the HT-29 cells (Figure 6B).
Figure 6.
The release profiles for POPC liposome-encapsulated carboxyfluorescein (λex = 480 nm; λem = 518 nm) containing 30 mol% of lipopeptide GPO4 in the presence of cell secreted MMP-9 in 25 mM HEPES buffer, pH = 8.0 containing 100 mM NaCl, 10 mM CaCl2 and 10 mM ZnCl2 (6A). The conditioned media from MCF7 cells released 40% of the encapsulated dye (blue circles) in 30 minutes; the conditioned media from the HT-29 cells released less than 10% of the contents in 30 minutes (green squares) and the culture media did not release any contents from the liposomes (red triangles). The line through the blue circles represents the fitted curve using the biexponential rate equation. The liposomes containing 30 mol% of lipopeptide GPO3 failed to release significant amount of the encapsulated dye under identical experimental conditions with conditioned media from MCF7 (6B, blue circles) and HT-29 (6B, green squares).
The concentration of active MMP-9 and -2 was estimated to be 150 nM in the conditioned media from the MCF7 cells and 25 – 30 nM for the conditioned media from the HT-29 cells (by employing a fluorogenic MMP-9 activity assay kit from Biomol International). Previously we have observed that POPC liposomes with 30 mol% of lipopeptide GPO4 released 50% of the encapsulated contents in the presence of 200 nM recombinant, two-domain MMP-9 (25). Since the additional hemopexin domain present in the cell-secreted MMP-9 does not participate either in unwinding or in catalysis of small triple helical peptide substrates (44), the results obtained with the recombinant MMP-9 corroborate the results with cell-secreted MMP-9. The substrate selectivity for MMP-9 and -2 are very similar (34), and it is expected that the MMP-2 present in the conditioned media will also hydrolyze the triple-helical peptides on the liposome surface and contribute to the release of the encapsulated dye.
The release profile in Figure 6A in the presence of the conditioned media from the MCF7 cells showed an initial linear phase followed by a slowly decreasing phase. The first step in the enzymatic release of the dye from the liposomes is the cleavage of the lipopeptide GPO4 by the cell-secreted MMP-2 and -9. We estimated that the concentration of GPO4 on the outer lipid layer of the liposomes to be about 100 nM and the concentration of MMP-2 and -9 in the conditioned media to be about 150 nM. Initially, the lipopeptides are rapidly cleaved by MMP-2 and -9 in the conditioned media, leading to the rapid release of the encapsulated carboxyfluorescein. As the concentration of GPO4 decreases, the rate of cleavage of this substrate by MMP-2 and -9 is reduced, leading to the slow release of the liposomal contents. On the other hand, the concentration of MMP-2 and -9 in the conditioned media from the HT-29 cells is about 30 nM. Since the concentration of GPO4 in the liposomes are higher compared to the MMP concentration, the substrate cleavage by the enzymes proceeds slowly. This is reflected in the slow linear release of the liposome-encapsulated dye in the presence of the conditioned media from the HT-29 cells (Figure 6A).
CONCLUSION
We have demonstrated that by optimizing the structure and the amount of the lipopeptide substrate and the major lipid component, liposomal contents can be rapidly released in the presence of the extracellular pathogenic enzyme MMP-9. The amount of contents released depends on the mismatch between the fatty acid moieties of the lipopeptide and the major lipid of the liposomal formulations. By utilizing the optimized liposomal formulations, we also demonstrated that while cancer cells (HT-29) which secrete low levels of MMP-9 failed to release significant amounts of the liposomal contents, metastatic cancer cells (MCF7) secreting high amounts of the enzyme rapidly released the encapsulated contents from the liposomes. We are currently optimizing the liposomal formulations by incorporating 10 mol% of a polyethyleneglycol containing lipid to impart long circulating property to the liposomes. These studies and release studies using a mouse model of metastatic breast adenocarcinoma will be reported in the future.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the NIH grants 1R01 CA113746, 1 R01 CA 132034 and NSF DMR-0705767 to SM and DKS. JS thanks the support of the proteomics core facility by NIH Grant P20 RR016741 from the INBRE Program of the NCRR.
Footnotes
Supporting Information Available. Synthetic details for the synthesis of the pyrene containing fatty acid, liposomal release studies with the lipopeptide GPO5. This material is available free of charge via the internet at http://pubs.acs.org.
LITERATURE CITED
- (1).Jesorka A, Orwar O. Liposomes: Technologies and Analytical Applications. Annu. Rev. Anal. Chem. 2008;1:801–832. doi: 10.1146/annurev.anchem.1.031207.112747. [DOI] [PubMed] [Google Scholar]
- (2).Bangham AD, Standish MM, Watkens JC. Diffusion of univalent ions across lamellae of swollen phospholipids. J. Mol. Biol. 1965;13:238–252. doi: 10.1016/s0022-2836(65)80093-6. [DOI] [PubMed] [Google Scholar]
- (3).Torchilin VP. Recent advances with liposomes as pharmaceutical carriers. Nat. Rev. Drug Discov. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- (4).Samad A, Sultana Y, Aqil M. Liposomal Drug Delivery Systems: An Update review. Current Drug Delivery. 2007;4:297–305. doi: 10.2174/156720107782151269. [DOI] [PubMed] [Google Scholar]
- (5).Torchilin VP. Targeted Pharmaceutical Nanocarriers for cancer Therapy and Imaging. The AAPS Journal. 2007;9:128–147. doi: 10.1208/aapsj0902015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gomez-Hens A, Fernandez-Romero J. The role of liposomes in analytical processes. Trends in Analytical Chemistry. 2005;24:9–19. [Google Scholar]
- (7).Allen TM, Hansen C, Rutledge J. Liposomes with prolonged circulation times: factors affecting uptake by reticuloendothelial and other tissues. Biochim. Biophys. Acta. 1989;981:27–35. doi: 10.1016/0005-2736(89)90078-3. 1989. [DOI] [PubMed] [Google Scholar]
- (8).O' Shaughnessy JA. Pegylated liposomal doxorubicin in the treatment of breast cancer. Clin. Breast Cancer. 2003;4:318–328. doi: 10.3816/cbc.2003.n.037. [DOI] [PubMed] [Google Scholar]
- (9).Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotechnology. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- (10).ElBayoumi TA, Torchilin VP. Tumor-specific anti-nucleosome antibody improves therapeutic efficacy of doxorubicin-loaded long-circulating liposomes against primary and metastatic tumor in mice. Mol. Pharmaceutics. 2009;6:246–254. doi: 10.1021/mp8001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Garg A, Tisdale AW, Haidari E, Kokkoli E. Targeting colon cancer cells using PEGylated liposomes modified with a fibronectin-mimetic peptide. Int. J. Pharm. 2009;366:201–210. doi: 10.1016/j.ijpharm.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kawano K, Onose E, Hattori Y, Maitani Y. Higher liposomal membrane fluidity enhances the in vitro antitumor activity of folate - targeted liposomal mitoxantrone. Mol. Pharmaceutics. 2009;6:98–104. doi: 10.1021/mp800069c. [DOI] [PubMed] [Google Scholar]
- (13).Boman NL, Bally MB, Cullis PR, Mayer LD, Webb MS. Encapsulation of vincristine in liposomes reduces toxicity and improves antitumor efficacy. J. Liposome Res. 1995;5:523–541. [Google Scholar]
- (14). [(accessed Feb 9, 2009)];For a list of FDA-approved liposomal drugs to treat cancer. see http://www.icare.org/fda.htm.
- (15). [accessed Feb 9, 2009];There are currently 357 clinical trials in progress on the use of liposomes to treat various diseases, including cancer. ( http://www.clinicaltrials.gov;)
- (16).Chandra B, Mallik S, Srivastava DK. Design of photocleavable lipids and their application in liposomal “uncorking”. J. Chem. Soc. Chem. Commun. 2005:3021–3023. doi: 10.1039/b503423j. [DOI] [PubMed] [Google Scholar]
- (17).Dromi S, Frenkel V, Luk A, Traughber B, Angstadt M, Bur M, Poff J, Xie J, Libutti SK, Li KCP, Wood BJ. Pulsed-high intensity focused ultrasound and low temperature-sensitive liposomes for enhanced targeted drug delivery and antitumor effect. J. Clin. Cancer Res. 2007;13:2722–2727. doi: 10.1158/1078-0432.CCR-06-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sawant RM, Hurley JP, Salmaso S, Kale A, Tolcheva E, Levchenko TS, Torchilin VP. “SMART” drug delivery systems: double-targeted pH-responsive pharmaceutical nanocarriers. Bioconjugate Chem. 2006;17:943–949. doi: 10.1021/bc060080h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wu G, Mikhailovsky A, Khant HA, Fu C, Chiu W, Zasadzinski JA. Remotely triggered liposome release by near-infrared light absorption via hollow gold nanoshells. J. Am. Chem. Soc. 2008;130:8175–8177. doi: 10.1021/ja802656d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Huang S-L, MacDonald RC. Acoustically active liposomes for drug encapsulation and ultrasound – triggered release. Biochim. Biophys. Acta. 2004;1665:134–141. doi: 10.1016/j.bbamem.2004.07.003. [DOI] [PubMed] [Google Scholar]
- (21).Davis SC, Szoka FC., Jr. Cholesterol phosphate derivatives: synthesis and incorporation into a phosphatase and calcium-sensitive triggered release liposome. Bioconjugate Chem. 1998:783–792. doi: 10.1021/bc980047y. [DOI] [PubMed] [Google Scholar]
- (22).Foged C, Nielsen HM, Frokjaer S. Phospholipase A2 sensitive liposomes for delivery of small interfering RNA (siRNA) J. Liposome Res. 2007;17:191–196. doi: 10.1080/08982100701530373. [DOI] [PubMed] [Google Scholar]
- (23).Romberg B, Flesch FM, Hennink WE, Storm G. Enzyme-induced shedding of a poly (amino acid)-coating triggers contents release from dioleoyl phosphatidylethanolamine liposomes. Int. J. Pharm. 2008;355:108–113. doi: 10.1016/j.ijpharm.2007.11.055. [DOI] [PubMed] [Google Scholar]
- (24).Sarkar N, Banerjee J, Hanson AJ, Elegbede AI, Rosendahl T, Krueger AB, Banerjee AL, Tobwala S, Wang R, Lu X, Mallik S, Srivastava DK. Matrix Metalloproteinase-assisted triggered release of liposomal contents. Bioconjugate Chem. 2008;19:57–64. doi: 10.1021/bc070081p. [DOI] [PubMed] [Google Scholar]
- (25).Elegbede AI, Banerjee J, Hanson AJ, Tobwala S, Ganguli B, Wang R, Lu X, Srivastava DK, Mallik S. Mechanistic studies of the triggered release of liposomal contents by matrix metalloproteinase-9. J. Am. Chem. Soc. 2008;130:10633–10642. doi: 10.1021/ja801548g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Nagase H, Woessner J, Frederick Matrix metalloproteinases. J. Biol Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- (27).Abraham D, Ponticos M, Nagase H. Connective tissue remodeling: cross-talk between endothelins and matrix metalloproteinases. Current Vascular Pharmacology. 2005;3:369–379. doi: 10.2174/157016105774329480. [DOI] [PubMed] [Google Scholar]
- (28).McCawley LJ, Matrisian LM. Matrix metalloproteinases: multifunctional contributors to tumor progression. Mol. Med. Today. 2000;6:149–156. doi: 10.1016/s1357-4310(00)01686-5. [DOI] [PubMed] [Google Scholar]
- (29).Murphy G, Nagase H. Progress in matrix metalloproteinase research. Mol Aspects Med. 2008;29:290–308. doi: 10.1016/j.mam.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Curran S, Murray GI. Matrix metalloproteinases. Molecular aspects of their roles in tumor invasion and metastasis. Eur. J. Cancer. 2000;36:1621–1630. doi: 10.1016/s0959-8049(00)00156-8. [DOI] [PubMed] [Google Scholar]
- (31).Tai KY, Shieh YS, Lee CS, Shiah SG, Wu CW. Axl promotes cell invasion by inducing MMP – 9 activity through activation of NF-κ B and Brg-1. Oncogene. 2008;27:4044–4055. doi: 10.1038/onc.2008.57. [DOI] [PubMed] [Google Scholar]
- (32).Owen JL, Torroella-Kouri M, Iragavarapu-Charyulu V. Molecular events involved in the increased expression of matrix metalloproteinase- 9 by T lymphocytes of mammary tumor-bearing mice. Int. J. Mo.l Med. 2008;21:125–134. [PubMed] [Google Scholar]
- (33).Cauwe B, Martens E, Van den Steen PE, Proost P, Van Aelst I, Blockmans D, Opdenakker G. Adenylyl cyclase-associated protein-1/CAP1 as a biological target substrate of gelatinase B/ MMP - 9. Exp. Cell Res. 2008;314:2739–2749. doi: 10.1016/j.yexcr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- (34).Lauer-Fields JL, Sritharan T, Stack MS, Nagase H, Fields GB. Selective Hydrolysis of Triple-helical Substrates by Matrix Metalloproteinase-2 and -9. J. Biol. Chem. 2003;278:18140–18145. doi: 10.1074/jbc.M211330200. [DOI] [PubMed] [Google Scholar]
- (35).Cejas MA, Kinney WA, Chen C, Vinter JG, Almond HR, Jr., Balss KM, Maryanoff CA, Schmidt U, Breslav M, Mahan A, Lacy E, Maryanoff BE. Thrombogenic collagen-mimetic peptides: self-assembly of triple helix-based fibrils driven by hydrophobic interactions. Proc. Natl. Acad. Sci. USA. 2008;105:8513–8518. doi: 10.1073/pnas.0800291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Sarkar NR, Rosendahl T, Krueger AB, Banerjee AL, Benton K, Mallik S, Srivastava DK. “Uncorking” of liposomes by matrix metalloproteinase-9. J. Chem. Soc. Chem. Commun. 2005:999–1001. doi: 10.1039/b416827e. [DOI] [PubMed] [Google Scholar]
- (37).Sabatino G, Papini AM. Advances in automatic, manual and microwave - assisted solid - phase peptide synthesis. Current Opinion in Drug Discovery Development. 2008;11:762–770. [PubMed] [Google Scholar]
- (38).Feng Y, Melacini G, Taulane JP, Goodman M. Acetyl-terminated and template-assembled collagen-based polypeptides Composed of Gly-Pro-Hyp sequences. 1. Synthesis and conformational analysis by circular dichroism, ultraviolet absorbance, and optical rotation. J. Am. Chem. Soc. 1996;118:10351–10358. [Google Scholar]
- (39).Iizasa T, Fujisawa T, Suzuki M, Motohashi S-I, Yasufuku K, Yasukawa T, Baba M, Shiba M. Elevated levels of circulating plasma matrix metalloproteinase 9 in non-small cell lung cancer patients. Clin. Cancer Res. 1999;5:149–153. [PubMed] [Google Scholar]
- (40).Turpeenniemi-Hujanen T. Gelatinases (MMP-2 and -9) and their natural inhibitors as prognostic indicators in solid cancers. Biochimie. 2005;87:287–297. doi: 10.1016/j.biochi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- (41).Ezhilarasan R, Jadhav U, Mohanam I, Rao JS, Gujrati M, Mohanam S. The hemopexin domain of MMP - 9 inhibits angiogenesis and retards the growth of intracranial glioblastoma xenograft in nude mice. Int. J. Cancer. 2009;124:306–315. doi: 10.1002/ijc.23951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Cortes-Reynosa P, Robledo T, Macias-Silva M, Wu SV, Salazar EP. Src kinase regulates metalloproteinase- 9 secretion induced by type IV collagen in MCF - 7 human breast cancer cells. Matr. Biol. 2008;27:220–231. doi: 10.1016/j.matbio.2007.11.003. [DOI] [PubMed] [Google Scholar]
- (43).Svetlana AT, Moriggl R, Corvinus FM, Wiederanders B, Schutz A, Kovacic B, Friedrich K. Signal transducer and activator of transcription 3 activation promotes invasive growth of colon carcinomas through matrix metalloproteinase induction. Neoplasia. 2007;9:279–291. doi: 10.1593/neo.06820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, Visse R, Nagase H, Fields GB. The roles of substrate thermal stability and the P2 and P1' subsite identity on matrix metalloproteinase triple-helical peptidase activity and collagen specificity. J. Biol. Chem. 2006;281:28302–38313. doi: 10.1074/jbc.M606004200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.