Abstract
This study reviewed all patients diagnosed with imported cutaneous leishmaniasis (CL) at the Hospital for Tropical Diseases in London, United Kingdom, over an 11-year period. Diagnostic and epidemiologic information was collected prospectively for all patients with imported CL to this hospital during 1998–2009. A total of 223 patients were given a diagnosis of CL. Ninety patients were diagnosed with Old World CL, which was caused most commonly by Leishmania donovani complex (n = 20). A total of 71% were tourists to the Mediterranean region, 36% were migrants or visiting friends and relatives, and 17% were soldiers. One hundred thirty-three patients were given a diagnosis of New World CL. The Leishmania subgenus Viannia caused 97 of these cases; 44% of these were in backpackers and 29% were in soldiers. Polymerase chain reaction was more sensitive and faster for detecting Leishmania DNA (86% for Old World CL and 96% for New World CL) than culture. This is the largest study of imported leishmaniasis, and demonstrates that tourists to the Mediterranean and backpackers in Central and South America are at risk for this disease.
Cutaneous leishmaniasis (CL) is the commonest form of leishmaniasis and has an estimated 1.5 million infections annually worldwide.1 This disease predominantly occurs in persons living in disease-endemic countries but also is an increasing problem in travelers. The Hospital for Tropical Diseases in London provides tertiary level clinical care for patients with imported CL, and hosts the United Kingdom Health Protection Agency Reference Laboratory for Parasitology.
Cutaneous leishmaniasis is categorized into Old World CL and New World CL. Differentiation is important because mucosal disease (ML) is caused by New World CL, specifically Leishmania (Viannia) spp. and requires systemic treatment.2 Skin lesions are classically ulcerated, but may be nodular or keratotic.
Endemic CL is caused by L. infantum in Spain and Portugal;3 L. infantum, L. major, and L. tropica in Cyprus and Turkey;4,5 by L. major and L. tropica in Iran, India, and Pakistan;6–8 and by L. aethiopica, L. major, and L. infantum in Ethiopia and Saharan Africa.9–11 L. (Viannia) spp. is the predominant cause of New World throughout Central and South America.12
Non-healing skin lesions in a patient with an appropriate travel history referred to the Hospital for Tropical Diseases undergo a skin biopsy, of which a sample is examined for Leishmania spp. by microscopy, histologic analysis, and culture. The sensitivity of these investigations ranges from 33% to 74%.13,14 The polymerase chain reaction (PCR), which has been used to detect Leishmania DNA, has improved diagnostic sensitivity to 95%.13 The Hospital for Tropical Diseases has used PCR for diagnostic DNA detection in all suspected cases of CL since 1996.
The purpose of this study was to use the large comprehensive Hospital for Tropical Diseases clinical database of all laboratory-proven cases of leishmaniasis over an 11-year period (1998–2009). Our main objective was to report the epidemiology of imported CL, correlations of the type of traveler with the country of travel, and the species of Leishmania acquired. We looked for regional correlations between species known to cause local endemic disease and those species acquired by travelers (Tables 2 and 3).
Table 2.
Epidemiologic distribution of Old World Leishmania spp. causing cutaneous leishmaniasis, Hospital for Tropical Diseases, London, United Kingdom
| Location | L. donovani | L. tropica | L. major | L. aethiopica | Not speciated | Total per country |
|---|---|---|---|---|---|---|
| Mediterranean (European countries bordering the Mediterranean Sea including Cyprus, Greece, Majorca, Malta and Gozo, Portugal, Spain and the Balearic Islands, and Sicily) | 20 | 1 | 0 | 0 | 4 | 25 |
| Near East (Jordan, Turkey, Syria, Yemen, and Israel) | 4 | 3 | 5 | 0 | 5 | 17 |
| Pakistan | 2 | 6 | 1 | 0 | 1 | 10 |
| Afghanistan | 2 | 9 | 2 | 0 | 6 | 19 |
| Africa (Continental Africa including all countries bordering the Sahara) | 0 | 0 | 3 | 1 | 2 | 6 |
| India | 0 | 0 | 0 | 0 | 1 | 1 |
| Iraq | 0 | 0 | 8 | 0 | 4 | 12 |
| Total | 28 | 19 | 19 | 1 | 23 |
Table 3.
Epidemiologic distribution of New World Leishmania spp. causing cutaneous leishmaniasis, Hospital for Tropical Diseases, London, United Kingdom
| Location | L. (Viannia) spp. | L. mexicana | L. amazonensis | Total per country |
|---|---|---|---|---|
| Belize | 38 | 18 | 0 | 56 |
| Bolivia | 19 | 1 | 0 | 20 |
| Peru | 11 | 0 | 0 | 11 |
| Central America (excluding Belize and the northern border of Mexico to the northern border of Colombia) | 18 | 7 | 0 | 25 |
| South America (excluding Bolivia or Peru) | 11 | 1 | 3 | 15 |
| Total | 97 | 27 | 3 | 127 |
Clinical details were obtained from case notes when the laboratory recorded a positive result for Leishmania spp. Data obtained included type of traveler, country of travel, time to referral, diagnostic tests performed, and Leishmania species.
The laboratory received fresh and fixed specimens. A dab preparation was made from the fresh specimens and stained with Giemsa or modified Rapid Field stain and inspected microscopically for amastigotes. Fresh material was also inoculated into modified Novy-Nicolle-McNeil medium and growth was checked weekly for 14–21 days. The time from the sample being received by the laboratory to confirming the diagnosis of infection with Leishmania spp. by any modality was recorded. Leishmania DNA was extracted, and amplification was attempted by using PCR on all samples from cases of suspected leishmaniasis.
All extractions were performed by using the Qiagen Tissue and Blood Mini Kit (Qiagen, Crawley, United Kingdom). Histologic sections were de-waxed by using xylene and absolute ethanol before extraction. During the study, primer sets were changed and the repertoire increased, as shown in the algorithm in Figure 1, because more species-specific primers became available.
Figure 1.
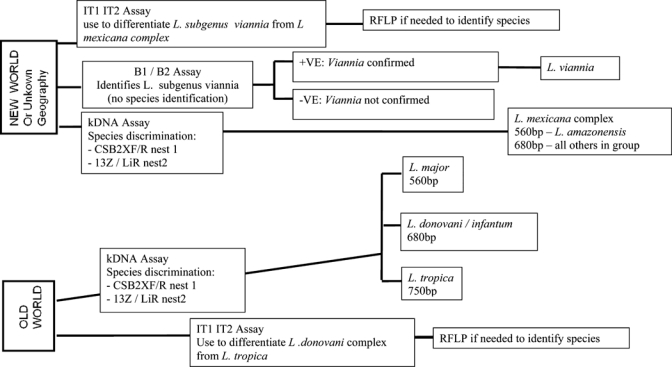
Algorithm for diagnosis of Leishmania infection by polymerase chain reaction, Hospital for Tropical Diseases, London, United Kingdom.
The most sensitive PCR (nested) used in species identification for most cases uses kinetoplast minicircle DNA.15,16 The PCR products were subjected to electrophoresis on agarose gels containing ethidium bromide, and diagnosis and species identification was performed by comparison with a molecular mass standard and amplified DNA from control strains.
A total of 223 patients were given a diagnosis of CL during 1998–2009. Of these patients, 133 were given a diagnosis of New World CL and 90 were given a diagnosis of Old World CL (Table 1). More males than females were infected (67% with Old World CL versus 67% with New World CL 70%), and the median age was 26.5 years. The median time from lesion onset to definitive diagnosis was five months for patients with Old World CL (range = < 1–35 months) and two months for patients with New World CL (range = < 1–24 months). Tourists (32%) and migrants either entering the United Kingdom or visiting friends and relatives in a disease-endemic country (36%) predominated in the Old World CL group. Twenty-five tourists acquired Old World CL in the Mediterranean basin, particularly in Spain (11 of 25, 44%). All military personnel acquired their disease in Iraq or Afghanistan.
Table 1.
Demographic characteristics of study population, Hospital for Tropical Diseases, London, United Kingdom*
| Characteristic | Old World CL† | Range or % | New World CL† | Range or % |
|---|---|---|---|---|
| Total | 90 | 133 | ||
| Male sex | 60 (67) | 93 (70) | ||
| Median age at presentation, years | 27 | 3–86 | 26 | 2–80 |
| HIV infected‡ | 0/11 | 0/21 | ||
| Median time to presentation, months | 5 | < 1–35 | 2 | < 1–24 |
| Mucosal involvement | 0/90 | 11/133 | ||
| Type of traveler | ||||
| Visiting friends and relatives/new entrant | 33 | 36 | 2 | 1.5 |
| Tourist | 29 | 32 | 7 | 5 |
| Soldier/military | 15 | 17 | 39 | 29 |
| Backpacker/NGO worker | 3 | 3 | 59 | 44 |
| Other (business, missionary) | 8 | 9 | 24 | 18 |
CL = cutaneous leishmaniasis; HIV = human immunodeficiency virus; NGO = non-government organization.
Values are no. (%) unless otherwise indicated.
Thirty-two persons were tested for HIV.
New World CL occurred mainly in travelers on backpacking trips (44%) and in soldiers (29%) performing jungle training in Belize. There were 11 cases of ML in patients with New World CL; all were caused by L. (Viannia) spp. (Table 1).
Countries of acquisition of Old World CL are shown in Table 2. The commonest region was the Mediterranean; this region had 20 cases caused by the L. donovani complex (the Hospital for Tropical Diseases PCR cannot distinguish between the DNA of L. donovani and L. infantum). Leishmania was the predominant species and caused Old World CL in Afghanistan, Pakistan, and Iraq (n = 18). Leishmania major caused Old World CL in Pakistan, Iraq, Afghanistan, and Africa (n = 19).
Countries of acquisition for New World CL are shown in Table 3. Of 133 cases, country of acquisition was available for 127 travelers. Eighty-one had traveled to Central America and 46 to South America. Leushmania (Viannia) spp. caused 97 (86%) cases of New World CL and was associated with travel to Central America (n = 56) and South America (n = 41). Leishmania mexicana was detected in 26 (32%) cases associated with Central America, particularly Belize; only one case was acquired in South America. There were three cases of infection with L. amazonensis, all of which were acquired in South America.
For Old World CL, sensitivity was 86% for the PCR, 52% for culture, and 62% for microscopy. The sensitivity of tests for NWCL was 96% for PCR, 68% for culture and 77% for microscopy (Table 4). Because positive results for histologic samples were not fully recorded in the database, these data are not reported. The mean time to a positive diagnosis by PCR was 8 days and to any result either positive or negative by culture was 11.6 days (n = 134) (P < 0.005, by paired two-tailed t-test).
Table 4.
Sensitivity of diagnostic tests used to identify Leishmania spp. causing cutaneous leishmaniasis, Hospital for Tropical Diseases, London, United Kingdom*
| Test | Old World CL, no. positive/no. tested (%) | New World CL, no. positive/no. tested (%) |
|---|---|---|
| Microscopy | 38/61 (62) | 90/117 (77) |
| Culture | 33/63 (52) | 78/114 (68) |
| PCR | 67/78 (86) | 124/129 (96) |
CL = cutaneous leishmaniasis; PCR = polymerase chain reaction. Sensitivities reported are for the Hospital for Tropical Diseases PCR overall for New World CL and Old World CL. The primer set increased during the study (Figure 1), but individual sensitivities for each primer set have not been calculated. There were no patients from whom samples were culture positive and PCR negative.
This study is the largest case series of imported CL from a tertiary referral center in Europe. Data clearly show that different Old World CL Leishmania species in travelers are found in distinct geographic areas. Endemic disease patterns correlate closely with travel-acquired CL, and identical species cause disease in each region.
The large numbers of L. donovani complex cases acquired in Spain and the Balearic Islands is probably associated with increasing numbers of British tourists traveling to Spain. A study from our institute (n = 42 over 3 years) showed that 40% of patients with CL were tourists.17 Other centers have reported similar trends for Old World CL and New World CL.18,19 Lawn and others at the Hospital for Tropical Diseases reported increasing numbers of imported cases of New World CL during 1999–2003 (79 CL and 6 ML),20 which reflected increasing numbers of travelers at risk for acquiring CL rather than an overall increase in the disease. Infections acquired in the Near East/Middle East and Africa were in military personnel and persons visiting friends and relatives. Soldiers have been in Iraq and Afghanistan since 2001. The increase in cases in persons visiting friends and relatives may be caused by increasing numbers of migrants to the United Kingdom; soldiers caused the predominance of males in the Old World CL group.
The large number of case of New World CL seen in travelers was unexpected. A study in the Netherlands reported 78 patients seen over 10 years, of whom 78% had New World CL, but did not give detailed reasons for travel.19 In our study, most cases of New World CL from this region were military personnel, (from the United Kingdom Government's jungle training facility in Belize) and adventure tourists. Only two (1.5%) of our cases of New World CL were in immigrants/ persons visiting friends and relatives compared with 36% in the Old World CL group.
As shown in other studies,5,15,21 detection of Leishmania DNA by using PCR is highly sensitive for the diagnosis of CL and identifies species. Specificity of PCR is also high. Previous work from our laboratory on smaller numbers of samples has shown a sensitivity of »100% for detection of L. (Viannia) spp. DNA by using PCR.17,20,22 This larger data set showed a definitive sensitivity of PCR at the Hospital for Tropical Diseases of 86% for Old World CL and 96% for New World CL. Diagnosis by using PCR significantly reduces the time to obtain a definitive diagnosis and is the only technique yielding a species that can be used on fixed specimens.
Only 32 patients were tested for infection with human immunodeficiency virus; all had negative results. This finding contrasts with that for visceral leishmaniasis, for which infection with this virus is an established risk factor. Our study was restricted to demographic details that had been recorded in the case notes for most cases. Therefore, we were unable to obtain detailed information for some patients.
Polymerase chain reaction was performed for all specimens, but the date at which a negative PCR result is final was not routinely recorded. Therefore, we could only calculate the diagnosis times for cases with positive results, which led to potential bias. We also only included patients seen in our own unit. Therefore, our data are likely not to be completely representative of all cases of CL in the United Kingdom.
Our study is the largest case series of imported CL reported in the literature. We have identified a range of different Leishmania species in travelers, associated with particular demographic groups, who all acquired locally endemic disease. The use of PCR to detect Leishmania DNA has improved the sensitivity and specificity of laboratory diagnosis. Use of this technique to identify species enables the treating clinician to select the most appropriate therapy and significantly reduces time to definitive diagnosis.
ACKNOWLEDGMENTS
We thank Katherine Bowers and Claire Dance (Department of Clinical Parasitology, Hospital for Tropical Diseases) for assistance with CL specimens, Dr. Debbie Nolder (London School of Tropical Medicine and Hygiene) for technical contributions to the manuscript and use of her unpublished data, Professor Francisco Vega-Lopez for contributing patient information to the database, Lt. Col. Mark Bailey for ensuring that military personnel with CL are referred to the Hospital for Tropical Diseases and for technical contributions to the manuscript, and Dr. Gavin Dreyer for statistical support.
Footnotes
Financial support: This study was supported by the Special Trustees of the Hospital for Tropical Diseases. Margaret Armstrong is supported by The Special Trustees of the Hospital for Tropical Diseases. All authors are supported by the University College London Hospitals Comprehensive Biomedical Research Centre Infection Theme. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure: None of the authors have any conflicts of interests.
Authors' addresses: Emma C. Wall and Margaret Armstrong, Hospital for Tropical Diseases, Mortimer Market Centre, Capper Street, London WC1E 6JB, United Kingdom, E-mails: emma.wall@doctors.org.uk and Margaret.Armstrong@uclh.nhs.uk. Julie Watson, Department of Clinical Parasitology, Hospital for Tropical Diseases, Mortimer Market Centre, Capper Street, London WC1E 6JB, United Kingdom, E-mail: Julie.Watson@uclh.nhs.uk. Peter L. Chiodini, Hospital for Tropical Diseases, Mortimer Market Centre, Capper Street, London WC1E 6JB, United Kingdom, Department of Clinical Parasitology, Hospital for Tropical Diseases, Mortimer Market Centre, Capper Street, London WC1E 6JB, United Kingdfom, and London School of Hygiene and Tropical Medicine, Keppel Street, London WC1E 7HT, United Kingdom, E-mail: Peter.Chiodini@uclh.nhs.uk. Diana N. Lockwood, Hospital for Tropical Diseases, Mortimer Market Centre, Capper Street, London WC1E 6JB, United Kingdom and London School of Hygiene and Tropical Medicine, Keppel Street, London WC1E 7HT, United Kingdom, E-mail: Diana.Lockwood@lshtm.ac.uk.
References
- 1.Desjeux P. Leishmaniasis. Nat Rev Microbiol. 2004;2:692. doi: 10.1038/nrmicro981. [DOI] [PubMed] [Google Scholar]
- 2.Bailey M, Lockwood DN. Cutaneous leishmaniasis. Clin Dermatol. 2007;25:203–211. doi: 10.1016/j.clindermatol.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Campino L, Pratlong F, Abranches P, Rioux JA, Santos-Gomes G, Alves-Pires C, Cortes S, Ramada J, Cristovão JM, Afonso MO, Dedet JP. Leishmaniasis in Portugal: enzyme polymorphism of Leishmania infantum based on the identification of 213 strains. Trop Med Int Health. 2006;11:1708–1714. doi: 10.1111/j.1365-3156.2006.01728.x. [DOI] [PubMed] [Google Scholar]
- 4.Antoniou M, Haralambous C, Mazeris A, Pratlong F, Dedet JP, Soteriadou K. Leishmania donovani leishmaniasis in Cyprus. Lancet Infect Dis. 2009;9:76–77. doi: 10.1016/S1473-3099(09)70004-0. [DOI] [PubMed] [Google Scholar]
- 5.Akkafa F, Dilmec F, Alpua Z. Identification of Leishmania parasites in clinical samples obtained from cutaneous leishmaniasis patients using PCR-RFLP technique in endemic region, Sanliurfa Province, in Turkey. Parasitol Res. 2008;103:583–586. doi: 10.1007/s00436-008-1013-5. [DOI] [PubMed] [Google Scholar]
- 6.Al-Hucheimi SN, Sultan BA, Al-Dhalimi MA. A comparative study of the diagnosis of Old World cutaneous leishmaniasis in Iraq by polymerase chain reaction and microbiologic and histopathologic methods. Int J Dermatol. 2009;48:404–408. doi: 10.1111/j.1365-4632.2009.03903.x. [DOI] [PubMed] [Google Scholar]
- 7.Bhutto AM, Soomro FR, Baloch JH, Matsumoto J, Uezato H, Hashiguchi Y, Katakura K. Cutaneous leishmaniasis caused by Leishmania (L.) major infection in Sindh province, Pakistan. Acta Trop. 2009;111:295–298. doi: 10.1016/j.actatropica.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Katakura K. Molecular epidemiology of leishmaniasis in Asia (focus on cutaneous infections) Curr Opin Infect Dis. 2009;22:126–130. doi: 10.1097/QCO.0b013e3283229ff2. [DOI] [PubMed] [Google Scholar]
- 9.Gadisa E, Genetu A, Kuru T, Jirata D, Dagne K, Aseffa A, Gedamu L. Leishmania (Kinetoplastida): species typing with isoenzyme and PCR-RFLP from cutaneous leishmaniasis patients in Ethiopia. Exp Parasitol. 2007;115:339–343. doi: 10.1016/j.exppara.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Rhajaoui M, Nasereddin A, Fellah H, Azmi K, Amarir F, Al-Jawabreh A, Ereqat S, Planer J, Abdeen Z. New clinico-epidemiologic profile of cutaneous leishmaniasis, Morocco. Emerg Infect Dis. 2007;13:1358–1360. doi: 10.3201/eid1309.070946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benmously-Mlika R, Fenniche S, Kerkeni N, Aoun K, Khedim A, Mokhtar I. Primary Leishmania infantum MON-80 endonasal leishmaniasis in Tunisia [in French] Ann Dermatol Venereol. 2008;135:389–392. doi: 10.1016/j.annder.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Gontijo B, de Carvalho ML. American cutaneous leishmaniasis [in Portuguese] Rev Soc Bras Med Trop. 2003;36:71–80. doi: 10.1590/s0037-86822003000100011. [DOI] [PubMed] [Google Scholar]
- 13.Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S. Cutaneous leishmaniasis. Lancet Infect Dis. 2007;7:581–596. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- 14.Reithinger R, Dujardin JC. Molecular diagnosis of leishmaniasis: current status and future applications. J Clin Microbiol. 2007;45:21–25. doi: 10.1128/JCM.02029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noyes HA, Reyburn H, Bailey JW, Smith D. A nested-PCR-based schizodeme method for identifying Leishmania kinetoplast minicircle classes directly from clinical samples and its application to the study of the epidemiology of Leishmania tropica in Pakistan. J Clin Microbiol. 1998;36:2877–2881. doi: 10.1128/jcm.36.10.2877-2881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bruijn MH, Barker DC. Diagnosis of New World leishmaniasis: specific detection of species of the Leishmania braziliensis complex by amplification of kinetoplast DNA. Acta Trop. 1992;52:45–58. doi: 10.1016/0001-706x(92)90006-j. [DOI] [PubMed] [Google Scholar]
- 17.Scarisbrick JJ, Chiodini PL, Watson J, Moody A, Armstrong M, Lockwood D, Bryceson A, Vega-López F. Clinical features and diagnosis of 42 travellers with cutaneous leishmaniasis. Travel Med Infect Dis. 2006;4:14–21. doi: 10.1016/j.tmaid.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Faber WR, Becht M, van Ginkel CJ, van der Kaay HJ, Vermeer BJ, Kager PA. Cutaneous leishmaniasis in 49 patients in The Netherlands [in Dutch] Ned Tijdschr Geneeskd. 1991;135:229–233. [PubMed] [Google Scholar]
- 19.Zeegelaar JE, Steketee WH, van Thiel PP, Wetsteyn JC, Kager PA, Faber WR. Changing pattern of imported cutaneous leishmaniasis in the Netherlands. Clin Exp Dermatol. 2005;30:1–5. doi: 10.1111/j.1365-2230.2004.01677.x. [DOI] [PubMed] [Google Scholar]
- 20.Lawn SD, Whetham J, Chiodini PL, Kanagalingam J, Watson J, Behrens RH, Lockwood DN. New world mucosal and cutaneous leishmaniasis: an emerging health problem among British travellers. QJM. 2004;97:781–788. doi: 10.1093/qjmed/hch127. [DOI] [PubMed] [Google Scholar]
- 21.Faber WR, Oskam L, van Gool T, Kroon NC, Knegt-Junk KJ, Hofwegen H, van der Wal AC, Kager PA. Value of diagnostic techniques for cutaneous leishmaniasis. J Am Acad Dermatol. 2003;49:70–74. doi: 10.1067/mjd.2003.492. [DOI] [PubMed] [Google Scholar]
- 22.Seaton RA, Morrison J, Man I, Watson J, Nathwani D. Out-patient parenteral antimicrobial therapy: a viable option for the management of cutaneous leishmaniasis. QJM. 1999;92:659–667. doi: 10.1093/qjmed/92.11.659. [DOI] [PubMed] [Google Scholar]


