Abstract
To strengthen active dengue surveillance in Saint Martin and Saint Barthélemy, two French Caribbean islands, we evaluated the epidemiological usefulness of collecting blood samples from NS1-positive dengue patients on filter paper to identify the dengue serotypes circulating in these regions during a 27-month period. This approach allowed dengue serotypes to be identified by reverse transcriptase-polymerase chain reaction in 90.1% of the total set of 666 samples analyzed and, in 95.5% of the samples collected during the acute phase of the disease. This prospective virological surveillance using blood samples absorbed onto filter paper, which were stored at 4°C and shipped at ambient temperature to a specialized laboratory for analysis, allowed us to avoid the logistic and financial costs associated with shipping frozen venous blood samples. This surveillance system offers a low-cost alternative for reinforcing dengue prevention in areas where specialized laboratories do not exist, notably by facilitating the early detection of potentially new dengue serotypes.
Introduction
Dengue fever (DF) and dengue hemorrhagic fever (DHF) are considered among the most important arthropod-borne viral diseases in tropical regions, with estimated annual incidences of 50–100 million cases of DF and more than 500,000 reported cases of DHF and dengue shock syndrome worldwide. This mosquito-borne disease is caused by four dengue virus types (type 1–4), which belong to the Flaviviridae family, genus Flavivirus.1 Moreover, the geographic spread of DF and the simultaneous increase in its mortality (21,000 deaths/year) are worrisome.2,3 To limit and reverse this trend, strengthening and intensifying dengue prevention and control are imperative. Previous exposure to dengue virus, but also viral strain and human host genetic may influence the clinical outcome of dengue infection.2,4,5 Among the preventive and control programs, passive and active DF surveillance systems are essential to provide early warnings of outbreaks and to permit emergency mosquito control measures.6 More specifically, the identification of the circulating dengue serotype and dengue virus strains in a community, may be important indicators of future epidemics.3,7–11
Saint Martin (~37,000 inhabitants in the French part) and Saint Barthélemy (~9,000 inhabitants) are two islands located in the Caribbean subregion of the French West Indies, each one representing an overseas collectivity of France. These two islands are 15 miles apart and are separated geographically by the Saint Barthélemy channel. Since the reemergence of dengue in the Caribbean subregion in the 1970s and the first dengue outbreak identified in Saint Martin in 1977,12 this arboviral disease has become endemic in these islands, with transmissions reported throughout the year and peaks observed mainly during the seasonal heavy rainfalls occurring from May to November.13 However, during the last 4 years epidemics have occurred mainly between October and February.14 To detect and control dengue outbreaks in these islands, an active and passive surveillance system was initiated by public health authorities. It is based on a syndromic approach carried out by a network of sentinel general practitioners (GPs) and biological surveillance of suspected dengue patients carried out by public and private laboratories. Data obtained by this surveillance system made it possible to better monitor and follow the trends of DF in these regions.15 However, the virological surveillance of the dengue serotypes circulating in these populations has not been possible because of the absence of local laboratories able to perform virus isolation and/or genome detection. Moreover, the reference laboratory dedicated to these two islands, and having the specific laboratory capacities, is based in French Guiana. This remote location requires that storage and expensive shipping conditions be used to transport biological samples for analysis.
We showed in previous studies that the collection of blood samples absorbed onto filter paper was an interesting alternative approach for dengue diagnosis.16,17 This sampling procedure allows dengue viral RNA, NS1 viral protein, and specific immunoglobulin M (IgM) to be detected without the logistical and temperature constraints for shipping and storage normally required for biological samples. Dengue viral RNA absorbed onto filter paper remains stable when stored up to 2 months at room temperature.18
We present here the utility of collecting and storing blood sample on filter paper as a tool to strengthen the active dengue surveillance system in the Saint Martin and Saint Barthélemy islands by allowing the dengue serotypes circulating in these islands to be monitored more efficiently.
Material and Methods
Syndromic and biological surveillance of dengue in Saint Martin and Saint Barthélemy.
Dengue surveillance has been structured between different partners since 2004 in Saint Martin and 2005 in Saint Barthélemy. This surveillance is based on three main indicators:
-
1.
The weekly number of dengue-like syndromes (syndromic surveillance). This allows the risk of occurrence or the beginning of an outbreak to be detected early, and to measure the magnitude of outbreaks.
-
2.
The weekly number of biologically confirmed cases of dengue. This is an indicator that contributes to detection and better knowledge of circulating serotypes.
-
3.
The weekly number of hospitalized cases. This figure is used to build the indicators of severity such as i) the hospitalization rate (ratio of the number of hospitalized and biologically confirmed cases to the estimated number of dengue-like syndromes); ii) the severity rate (ratio of the number of severe forms to the estimated number of dengue-like syndromes); and iii) the proportion of severe forms among the hospitalized cases.
Monitoring the trends of these indicators allows monitoring the severity of an epidemic.
Data collection and data entry are performed at the Regional Health Agency (Agence Régionale de Santé, ARS). Epidemiological analysis is carried out by the Regional Office of the French Institute for Public Health Surveillance (Institut de Veille Sanitaire, InVS) in the French departments of America. Periodic reports are produced to disseminate information about the local epidemiological situation. Syndromic surveillance data are collected from the GPs of the sentinel network, which was first created in 2004 by local health authorities. However, systematic computer recording began in 2005 in Saint Martin and Saint Barthélemy. The representativeness of the network in terms of GP activity has progressively improved, reaching 48% in Saint Martin and 70% in Saint Barthélemy in 200819; each sentinel GP is called every week to report the number of patients seen for dengue-like syndromes during the previous week. A dengue-like syndrome is defined as a patient with < 7 days of fever (≥ 38.5°C) without evidence of another cause of infection and with at least one sign of pain (headache, retro-orbital pain, myalgia, arthralgia, or back pain). Every week, the total number of dengue cases collected is extrapolated to the whole territory using the ratio of “medical activity of GPs present during the week/medical activity of all the GPs of the concerned territory,” i.e., Saint Martin or Saint Barthélemy.
Biological data (positive NS1 antigen detection and/or positive anti-dengue IgM detection) are collected from local laboratories, which report all biological results related to dengue to the Regional Health Agency. Because these islands cover a small population, there are only two private laboratories in Saint Martin (one of them is the service provider of the only local hospital) and one in Saint Barthélemy (also the service provider of the local hospital). Although serological tests for anti-dengue IgM detection have been used since the beginning of surveillance, the use of commercial NS1 detection has been more recently introduced into the detection routines of these laboratories. Three tests are currently used: the Platelia Dengue NS1 Ag test, an enzyme-linked immunosorbent assay from Bio-Rad Laboratories (Marnes La Coquette, France), and two immunochromatographic tests, the Dengue NS1 Ag STRIP (Bio-Rad) and the SD Dengue Duo rapid test (Standard Diagnostics, Kyonggi-do, Korea).20,21
Hospital wards transmit the data related to biologically confirm hospitalized cases. Each case is classified according to World Health Organization (WHO) criteria, based on the clinical and biological information collected.3 Local indicators were established using the maximum expected values obtained by modeling the data (Serfling method), so as to detect changes in the epidemiological situation (Figures 1 and 2).22,23 Thus, a pre-epidemic alert is determined by a combination of various criteria such as: if the estimated number of dengue-like syndromes exceeds the maximum expected value for the period during three consecutive weeks, and the number of biologically confirmed cases exceeds the maximum expected value during two consecutive weeks. Other qualitative and/or quantitative criteria may also be taken into account in the interpretation of results such as the emergence of a new serotype, an increasing positivity rate, the number of hospitalized cases, the number of severe cases, and the slope of the increase. The next phase (epidemic alert) is determined by an additional 2-week period of values exceeding the expected maximum levels for both indicators (estimated dengue-like syndrome and laboratory-confirmed cases). The end of the epidemic is declared when the estimated number of dengue-like syndromes and the number of biologically confirmed cases are below the maximum expected values for two consecutive weeks.
Figure 1.
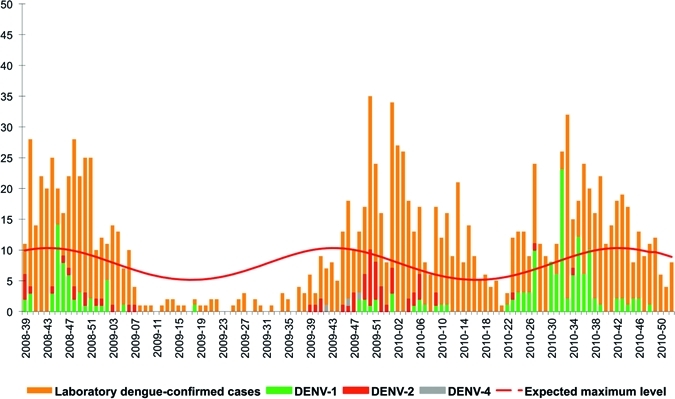
Weekly number of biologically confirmed cases of dengue and weekly distribution of serotype, Saint Martin, Epi-week 2008-39–2010-52.
Figure 2.
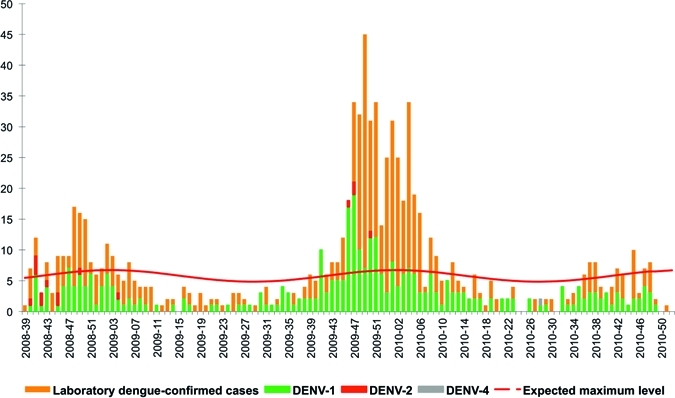
Weekly number of biologically confirmed cases of dengue and weekly distribution of serotype, Saint Barthélemy, Epi-week 2008-39–2010-52.
Design of dengue virological surveillance.
A prospective virological surveillance based on the identification of dengue serotype was set up in October 2008 to reinforce the dengue epidemiological surveillance in Saint Martin and Saint Barthélemy. Two private laboratories (one in each island) that contributed to biological surveillance of dengue voluntarily participated to the evaluation of this new approach. Biological samples from patients clinically suspected of dengue infection and positive for NS1 antigen were selected for dengue serotype identification by the reference laboratory based at the Institut Pasteur de la Guyane (IPG), in French Guiana. Once a sample was detected positive for NS1 in either the Saint Martin or Saint Barthélemy laboratory, three drops of whole blood (20 µL for each drop) were individually absorbed onto filter paper (Schleicher and Schuell, Germany, -Whatmann 3M). One drop was dedicated for identification of the dengue serotype, whereas the two others were kept for NS1 and/or IgM detection on specific request from the private laboratory. Filter papers containing dried blood drops were stored at 4°C. Every eight or 15 days (depending of the epidemiological situation of dengue transmission), filter papers were sent by post at ambient temperature (20–25°C and 85–90% relative humidity) to IPG for identification of dengue serotype by reverse transcriptase-polymerase chain reaction (RT-PCR). An information sheet included with each specimen recorded the origin of samples, the date of illness onset, the date of sample collection and finally, the NS1 results and the commercial test used. A maximum of 10 NS1-positive samples per week and per laboratory were randomly selected during the epidemic period and were sent to IPG for identification of dengue serotype. This quantity of samples was considered manageable by the IPG and sufficient for surveillance purposes. During the inter-epidemic period, the protocol advocated that all NS1-positive samples detected by the two laboratories be sent to the IPG.
Identification of dengue serotype.
For each sample tested, one drop of dried whole blood on filter paper was cut into strips and placed in a 1.8 mL tube. Viral RNA was extracted from the filter paper of each patient and from 10 µL of a positive dengue serum control using TRIzol (Invitrogen Life Technologies, Paisley, Refrewshire, UK), according to the manufacturer's recommendations. The positive control used usually belongs to a DENV serotype that is not currently detected during a studied period. The extracts were precipitated with isopropanol and 1 µL of glycogen (5 µg/µL; Roche Diagnostics, Mannheim, Germany). Air-dried RNA pellets were suspended in 20 µL of water. Next, 5 µL of RNA was reverse transcribed into cDNA using random hexamer oligonucleotides (Roche) and moloney murine leukemia virus reverse transcriptase (Invitrogen Life Technologies). The detection and typing of dengue viruses were carried out according to Lanciotti and others.24
Results
Biological samples received by the reference laboratory.
Between October 2008 (week 39-2008) and December 2010 (week 52-2010), the IPG reference laboratory received 740 dried venous blood samples on filter paper for molecular analysis. Seventy-four samples (10% of the total; 50 from Saint Martin and 24 from Saint Barthélemy) that were collected mainly during the first months of the study were not analyzed for lack of sufficient sample information, such as date of illness onset, date of sample collection, or NS1 results.
Detection of dengue serotype according to time of disease.
A total of 666 samples was analyzed by RT-PCR among which 288 (43.2%) were from Saint Martin and 378 (56.8%) were from Saint Barthélemy (Table 1). Five hundred thirty-two samples (79.9%) were obtained during the acute phase of the disease (from Day 0 to Day 4 after fever onset, where Day 0 represents the first day of disease), 46 samples (6.9%) were obtained during the early-convalescent phase of disease (Day 5 to Day 11), and 88 samples (13.2%) were collected at an undetermined disease phase. This prospective virological surveillance allowed dengue serotypes to be identified for 90.1% (600 of 666) of all NS1-positive specimens, for 86.1% (248 of 288) of those from Saint Martin, and 93.1% (352 of 378) of those from Saint Barthélemy (P = 0.003, Fisher test) (Table 1).
Table 1.
Percentage of positive reverse transcriptase-polymerase chain reaction (RT-PCR) obtained from blood samples absorbed onto filter paper of NS1-positive samples according to the time of collection
| Days after onset | Saint Martin | Saint Barthélemy | P* | Total |
|---|---|---|---|---|
| Day 0 | 90.0% (9/10) | 100.0% (6/6) | P = 1.0 | 93.8% (15/16) |
| Day 1 | 92.7% (38/41) | 97.5% (116/119) | P = 0.2 | 96.3% (154/160) |
| Day 2 | 98.2% (56/57) | 99.0% (102/103) | P = 1.0 | 98.8% (158/160) |
| Day 3 | 96.4% (54/56) | 95.2% (59/62) | P = 1.0 | 95.8% (113/118) |
| Day 4 | 88.6% (39/44) | 85.3% (29/34) | P = 0.7 | 87.2% (68/78) |
| Acute phase | 94.2% (196/208) | 96.3% (312/324) | P = 0.3 | 95.5% (508/532) |
| Day 5 | 28.6% (4/14) | 81.8% (9/11) | P = 0.008 | 52.0% (13/25) |
| Day 6 to Day 11 | 41.7% (5/12) | 33.3% (3/9) | P = 1.0 | 38.1% (8/21) |
| Early convalescent phase | 34.6% (9/26) | 60.0% (12/20) | P = 0.09 | 45.7% (21/46) |
| Unknown | 79.6% (43/54) | 82.4% (28/34) | P = 0.8 | 80.7% (71/88) |
| Total | 86.1% (248/288) | 93.1% (352/378) | P = 0.003 | 90.1% (600/666) |
P, Fisher test, P ≤ 0.05 was considered statistically significant (comparison between the percentages of serotype detection by site, at each sampling day).
Dengue serotypes were detected in 95.5% (508 of 532) of all samples obtained during the acute phase of disease (Table 1). No significant difference was observed between RT-PCR results obtained from Saint Martin specimens (94.2%; 196 of 208) and those obtained from Saint Barthélemy specimens (96.3%; 312 of 324) (P = 0.3, Fisher test). As expected, dengue serotypes were detected in fewer of the NS1-positive blood samples collected during the early-convalescent phase of disease than in those collected during the acute phase, with serotypes identified in only 45.7% (21 of 46) of early-convalescent samples. At Day 5 of disease, a significant difference in RT-PCR detection was observed between samples received from Saint Martin (28.6%; 4 of 14) and those from Saint Barthélemy (81.8%; 9 of 11), respectively (P = 0.008, Fisher test). Finally, 80.7% (71 of 88) of the specimens collected from patients for whom the date of disease was undetermined were positive by RT-PCR (Table 1).
Dengue virological surveillance in Saint Martin.
Among 288 NS1-positive dengue blood samples received during the period of the study, a dengue serotype was identified in 86.1% of them (248 of 288). Among these samples, 71.8% (178 of 248) were positive for the DENV-1 serotype, 26.6% (66 of 248) were positive for the DENV-2 serotype, and 1.6% were positive (4 of 248) for the DENV-4 serotype (Table 2).
Table 2.
Trends of dengue virological surveillance in Saint Martin between September 2008 and December 2010
| Epidemic transmission | Inter-epidemic transmission | Epidemic transmission | Inter-epidemic transmission | Epidemic transmission | Total | |
|---|---|---|---|---|---|---|
| Period (week no./year) | 39-2008–07-2009 | 08-2009–48-2009 | 49-2009–18-2010 | 19-2010–23-2010 | 24-2010–52-2010 | |
| Duration (number of weeks) | 21 | 41 | 23 | 5 | 27 | 119 |
| Number of biological confirmed cases† | 361 | 128 | 371 | 25 | 409 | 1294 |
| Number of analyzed samples | 70 | 27 | 62 | 5 | 124 | 288 |
| Number of positive RT-PCR* (%) | 70 (100%) | 15 (55.6%) | 53 (85.5%) | 4 (80.0%) | 106 (85.5%) | 248 (86.1%) |
| DENV-1 | 53 (75.7%) | 3 (20.0%) | 15 (28.3%) | 3 (75.0%) | 104 (98.2%) | 178 (71.8%) |
| DENV-2 | 17 (24.3%) | 9 (60.0%) | 38 (71.7%) | 1 (25.0%) | 1 (0.9%) | 66 (26.6%) |
| DENV-3 | – | – | – | – | – | – |
| DENV-4 | – | 3 (20.0%) | – | – | 1 (0.9%) | 4 (1.6%) |
RT-PCR = reverse transcriptase-polymerase chain reaction.
Laboratory-confirmed dengue cases based on positive NS1 or/and IgM patients reported by the laboratories.
Three successive epidemic phases occurred in Saint Martin during the period of the study (Table 2 and Figure 1). The first began in September 2008 (Week 39), corresponding to the period when the evaluation system was being setup. During this outbreak, the biological surveillance network reported 361 dengue cases on the basis of positive NS1 and/or specific IgM results. Seventy of the specimens (19.4%; 70 of 361) were selected for RT-PCR analysis, which revealed that two dengue serotypes were co-circulating during this outbreak; 75.7% (53 of 70) contained DENV-1 and 24.3% (17 of 70) contained DENV-2. This epidemic was followed by an inter-epidemic phase of 41 weeks, during which three dengue serotypes were detected, DENV-1 (20%; 3 of 15), DENV-2 (60%; 9 of 15), and DENV-4 (20%; 3 of 15).
A new recrudescence of dengue cases was then observed from December 2009 to May 2010 with 371 laboratory-confirmed dengue cases, among which 53 were found to be positive (Table 2 and Figure 1). This outbreak was characterized by a reversal in the proportions of the two dengue serotypes circulating in the previous outbreak (September 2008): 71.7% (38 of 53) of the samples contained DENV-2 and 28.3% (15 of 53) contained DENV-1 (Table 2).
After a short period in which the number of laboratory-confirmed dengue cases decreased, a third dengue epidemic was observed. The biological surveillance system reported 409 dengue cases based on positive NS1 and/or IgM results. Among the 124 (30.3%; 124 of 409) blood samples analyzed by RT-PCR, DENV-1 was the serotype the most extensively detected (98.2%; 104 of 106) (Table 2).
Dengue virological surveillance in Saint Barthélemy.
Two successive dengue epidemics occurred in Saint Barthélemy between September 2008 and December 2010. During this prospective virological surveillance, 378 blood samples were collected and sent to the IPG. Molecular analysis of 352 of these samples revealed the serotype proportions to be 95.4% for DENV-1 (336 of 352), 4.3% for DENV-2 (15 of 352), and 0.3% for DENV-4 (1 of 352) (Table 3).
Table 3.
Trends of dengue virological surveillance in Saint Barthélemy between September 2008 and December 2010
| Inter-epidemic transmission | Epidemic transmission | Inter-epidemic transmission | Epidemic transmission | Inter-epidemic transmission | Total | |
|---|---|---|---|---|---|---|
| Period (week no./year) | 39-2008–42-2008 | 43-2008–07-2009 | 08-2009–46-2009 | 47-2009–09-2010 | 10-2010–52-2010 | |
| Duration (number of weeks) | 4 | 17 | 39 | 16 | 43 | 119 |
| Number of biological confirmed cases† | 23 | 152 | 131 | 379 | 150 | 835 |
| Number of analyzed samples | 14 | 62 | 94 | 126 | 82 | 378 |
| Number of positive RT-PCR* (%) | 14 (100%) | 62 (100%) | 83 (88.3%) | 117 (92.9%) | 76 (92.7%) | 352 (93.1%) |
| DENV-1 | 8 (57.1%) | 57 (91.9%) | 82 (98.8%) | 114 (97.4%) | 75 (98.7%) | 336 (95.4%) |
| DENV-2 | 6 (42.9%) | 5 (8.1%) | 1 (1.2%) | 3 (2.6%) | – | 15 (4.3%) |
| DENV-3 | – | – | – | – | – | – |
| DENV-4 | – | – | – | – | 1 (1.3%) | 1 (0.3%) |
RT-PCR = reverse transcriptase-polymerase chain reaction.
Laboratory-confirmed dengue cases based on positive NS1 or/and IgM patients reported by laboratories.
The first epidemic reported in Saint Barthélemy began at the end of October 2008 (Week 43), within 3 weeks after the dengue epidemic alert was issued for Saint Martin (Table 3and Figure 2). The biological surveillance network reported 152 cases, of which 62 of them were analyzed by RT-PCR (Table 3). This epidemic was similar to the one observed in neighboring Saint Martin in two characteristics: i) DENV-1 was the predominant serotype (91.9%; 57 of 62), and ii) the epidemic ended in February 2010 (Week 07). This dengue outbreak was followed by a 39-week long inter-epidemic period of sporadic transmission.
In mid-November 2009, another dengue outbreak took place in Saint Barthélemy. Although the two epidemics had similar durations (17 weeks in 2008–2009 versus 16 weeks in 2009–2010), 2-fold more laboratory-confirmed dengue cases were reported for the latter Saint Barthélemy outbreak (N = 379) than for the previous one (N = 152). One hundred seventeen NS1-positive blood samples were subtyped by RT-PCR: 114 were DENV-1 (97.4%) and 3 were DENV-2 (2.6%). Although the DENV-1 serotype predominated during this period in Saint Barthélemy, the DENV-2 serotype was found to predominate among specimens provided from the same period in Saint Martin.
Discussion
Dengue has become a major public health burden in tropical and sub-tropical regions of the world, partly because of the increased number of reported cases, particularly severe clinical forms, and the increased number of deaths attributable to this disease. With no specific treatments available for patient management and no vaccine yet available to protect vulnerable populations, prevention of this arboviral disease plays a crucial role in its control. Among the different approaches implemented, an active surveillance system was advocated in the 1980s notably to detect dengue outbreaks early and to monitor the introduction of new dengue subtypes.6,25–27
We reported here the epidemiological usefulness of collecting venous blood samples on filter paper to monitor the dengue serotypes in circulation in two French territories in the Caribbean region: the islands of Saint Martin and Saint Barthélemy. This prospective approach to the virological surveillance of dengue serotypes was evaluated during a 27-month period. It allows detecting dengue serotypes using RT-PCR in 90.1% of overall blood samples provided from NS1-positive dengue cases (Table 1).
For samples collected during the acute phase of the disease, the percentage in which dengue virus RNA was detected by RT-PCR analysis was 95.5%. First, these results show the relevance of using, as a first line, such highly specific diagnostic tools for acute dengue diagnosis in areas where techniques for dengue serotype identification are not available. Although the use of some commercial NS1 tests is still debated because of their unequal sensitivities, most of the commercial NS1 tests exhibit specificities over 95%.20,21,28–30 The high specificity of these tests limits the number of false positive NS1 results and thus ensures that dengue serotypes will be identified in high percentages of the samples analyzed by RT-PCR. Second, these results confirm the good stability of dengue viral RNA absorbed into filter paper stored at 4°C and shipped at ambient temperature.
The percentages of samples collected during the acute phase of disease that were determined to be positive by RT-PCR were similar for the specimens collected in Saint Martin (94.2%) and those collected in Saint Barthélemy (96.3%). However, for those sampled during the early convalescent phase, significant difference was only observed with samples collected at Day 5 after onset of fever: 81.8% of those from Saint Barthélemy were positive, whereas only 28.6% of those from Saint Martin were positive (Table 1). Although this observation remains unexplained, a plausible hypothesis could be the usual practice of the Saint Martin laboratory to load filter papers with slightly less than the necessary 20 µL volume of blood. Alternatively, it is also possible that the precise phase of the disease in which these Saint Barthélemy samples were collected is erroneous. In other words, some samples apparently collected on Day 5 of the disease could have been in fact collected earlier, resulting in samples having higher levels of virus, which yielded higher apparent RT-PCR sensitivity. However, we did not have the means to investigate these possible differences in recording the dates of onset to validate this hypothesis.
This approach enabled us to initiate an efficient surveillance system for dengue serotypes. With the ability to monitor the distribution of the circulating serotypes in these two islands, we could verify if any differences in terms of severity occurred according to the distributions observed. No differences in either hospitalization or severity rates were observed in Saint Martin between the second epidemic, which predominantly consisted of the DENV-2 serotype, and either of the two other DENV-1 serotype epidemics (data not shown). The introduction of DENV-2 serotype did not generate an increase in disease severity as might have been expected, because this serotype has been reported to be associated with a higher frequency of severe dengue forms.31–33
The reason why the dengue subtypes circulating in endemic regions are not widely monitored is often because of the lack of local specialized laboratories and the lack of financial resources to allow blood samples to be shipped in cold conditions to reference laboratories. In Saint Barthélemy and in Saint Martin, similar limitations impaired the implementation of an efficient surveillance system of the circulating serotypes, until this new approach was introduced. Indeed, it offers the possibility to detect dengue subtypes in blood samples stored at room temperature and thus allows circumventing costly logistical problems associated with shipment of frozen samples.
In the future this virological surveillance will have to be strengthened, particularly during the inter-epidemic transmission phase. Indeed, only 21% (27 of 128) and 20% (5 of 25) of positive dengue cases occurring in the two inter-epidemic phases at Saint Martin were investigated by RT-PCR. The collection of additional data during these periods would allow the proportion of circulating dengue serotypes to be more precisely assessed, which is useful for detecting the precocious introduction of a new serotype. It could also be beneficial to improve this virological surveillance system by including the sequencing analyses of the dengue serotypes detected in these two islands. This additional data would allow monitoring of the potential introduction of new dengue viral genotype, which could be associated with risk of severity and the study of DENV gene flow throughout the French territories of America and the surrounding Caribbean islands. Moreover, autochthonous dengue cases in the South-East of France mainland were detected recently in August 2010 and were found to be mostly related to concomitant epidemics in the French West Indies (Martinique and Guadeloupe) that occurred from the beginning of 2010.34 Dengue genotype surveillance should identify whether viral spread is a result of cultural and economic ties between French territories of America and mainland France.35 In conclusion, virological surveillance of dengue serotypes based on genome detection of laboratory-confirmed dengue cases, using blood samples absorbed onto filter paper, is a good alternative approach to advocate in regions lacking local reference laboratories able to perform genome detection.
ACKNOWLEDGMENTS
We are grateful to all the staff of the two laboratories (Laboratoire Lepers and Laboratoire Winicki) for their help and collaboration in this study.
Disclaimer: We declare that we have no conflicts of interest.
Footnotes
Financial support: This study was supported by the Conseil Régional de la Guyane (agreement no. 60/2007/CR). The laboratory of virology, as National Reference Center, receives financial support from Institut Pasteur and Institut de Veille Sanitaire.
Authors' addresses: Séverine Matheus, Bhetty Labeau, Laetitia Bremand, and Philippe Dussart, Laboratoire de Virologie, Centre National de Référence des Arbovirus, Institut Pasteur de la Guyane, 97306 Cayenne Cedex, Guyane Française, E-mails: smatheus@pasteur-cayenne.fr, blabeau@pasteur-cayenne.fr, lbremand@pasteur-cayenne.fr, and pdussart@pasteur-cayenne.fr. Jean-Loup Chappert, Sylvie Cassadou, and Philippe Quenel, Cellule Interrégionale d'Epidémiologie Antilles-Guyane, Fort-de-France, Martinique, E-mails: Jean-Loup.CHAPPERT@ars.sante.fr, Sylvie.CASSADOU@ars.sante.fr, and Philippe.QUENEL@ars.sante.fr. Franck Berger, Unité d'Epidémiologie, Institut Pasteur de la Guyane, 97306 Cayenne Cedex, Guyane française. E-mail: fberger@pasteur-cayenne.fr. Alain Winicki, Laboratoire d'Analyses Médicales, Saint Barthélemy, La pointe Gustavia, 97133 Saint Barthélemy, E-mail: awinicki@wanadoo.fr. Patricia Huc-Anais, Laboratoire d'Analyses Médicales, Marigot, Saint Martin. E-mail: Laboratoire.Lepers@wanadoo.fr.
References
- 1.Monath TP. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzman MG, Kouri G. Dengue: an update. Lancet Infect Dis. 2002;2:33–42. doi: 10.1016/s1473-3099(01)00171-2. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization . Dengue Haemorrhagic Fever: Diagnosis, Treatment, Prevention and Control. New edition. Geneva: World Health Organization; 2009. [Google Scholar]
- 4.Rico-Hesse R. Dengue virus evolution and virulence models. Clin Infect Dis. 2007;44:1462–1466. doi: 10.1086/517587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakuntabhai A, Turbpaiboon C, Casademont I, Chuansumrit A, Lowhnoo T, Kajaste-Rudnitski A, Kalayanarooj SM, Tangnararatchakit K, Tangthawornchaikul N, Vasanawathana S, Chaiyaratana W, Yenchitsomanus PT, Suriyaphol P, Avirutnan P, Chokephaibulkit K, Matsuda F, Yoksan S, Jacob Y, Lathrop GM, Malasit P, Desprès P, Julier C. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37:507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gubler DJ, Castra-Valez A. A program for prevention and control of epidemic dengue and dengue hemorrhagic fever in Puerto-Rico and the U.S. Virgin Islands. Bull Pan Am Health Organ. 1991;25:237–247. [PubMed] [Google Scholar]
- 7.Gubler DJ. Surveillance for dengue and dengue hemorrhagic fever. Bull Pan Am Health Organ. 1989;23:397–404. [PubMed] [Google Scholar]
- 8.Lam SK. Two decades of dengue in Malaysia. Trop Med. 1993;35:195–200. [Google Scholar]
- 9.Sithiprasasna R, Patpoparn S, Attatippaholkun W, Suvannadabba S, Srisuphanunt M. The geographic information system as an epidemiological tool in the surveillance of dengue virus-infected Aedes mosquitoes. Southeast Asian J Trop Med Public Health. 2004;35:918–926. [PubMed] [Google Scholar]
- 10.Beatty ME, Stone A, Fitzsimons DW, Hanna JN, Lam SK, Vong S, Guzman MG, Mendez-Galvan JF, Halstead SB, Letson GW, Kuritsky J, Mahoney R, Margolis HS. Best practices in dengue surveillance: a report from the Asia-Pacific and Americas Dengue Prevention Boards. PLoS Negl Trop Dis. 2010;16:e890. doi: 10.1371/journal.pntd.0000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halstead SB. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 12.Van der Sar A, Woodall JP, Temmer LE. Dengue in the Caribbean, 1977. Washington, DC: Pan American Health Organization; 1979. pp. 55–59. (The dengue epidemic in the Leeward Islands of the Netherlands Antilles: Saba, St. Eustatius, and St. Martin, 1977). [Google Scholar]
- 13.Peyrefitte CN, Pastorino BA, Bessaud M, Gravier P, Tock F, Couissinier-Paris P, Martial J, Huc-Anais P, Césaire R, Grandadam M, Tolou HJ. Dengue type 3 virus, Saint Martin, 2003–2004. EID. 2005;11:757–761. doi: 10.3201/eid1105.040959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anonymous Bulletin d'Alerte et de Surveillance Antilles Guyane No. 6. 2008. http://www.invs.sante.fr/Actualites/Publications/basag/Basag2008_6.pdf Available at. Accessed June 2008.
- 15.Anonymous Bulletin d'Alerte et de Surveillance Antilles Guyane No. 4. 2008. http://www.invs.sante.fr/publications/basag/basag2008_4.pdf Available at. Accessed April 2008.
- 16.Matheus S, Meynard JB, Lacoste V, Morvan J, Deparis X. Use of capillary blood samples as a new approach for diagnosis of Dengue virus infection. J Clin Microbiol. 2007;45:887–890. doi: 10.1128/JCM.02063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matheus S, Meynard JB, Lavergne A, Girod R, Moua D, Labeau B, Dussart P, Lacoste V, Deparis X. Dengue-3 outbreak in Paraguay: investigations using capillary blood samples on filter paper. Am J Trop Med Hyg. 2008;79:685–687. [PubMed] [Google Scholar]
- 18.Prado I, Rosario D, Bernardo L, Alvarez M, Rodríguez R, Vázquez S, Guzmán MG. PCR detection of dengue virus using dried whole blood spotted on filter paper. J Virol Methods. 2005;125:75–81. doi: 10.1016/j.jviromet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Anonymous Bulletin d'Alerte et de Surveillance Antilles Guyane No. 10. 2008. http://www.invs.sante.fr/publications/basag/basag2008_10 Available at. Accessed October 2008.
- 20.Dussart P, Petit L, Labeau B, Bremand L, Leduc A, Moua D, Matheus S, Baril L. Evaluation of two new commercial tests for the diagnosis of acute dengue virus infection using NS1 antigen detection in human serum. PLoS Negl Trop Dis. 2008;20:e280. doi: 10.1371/journal.pntd.0000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tricou V, Vu HT, Quynh NV, Nguyen CV, Tran HT, Farrar J, Wills B, Simmons CP. Comparison of two dengue NS1 rapid tests for sensitivity, specificity and relationship to viraemia and antibody responses. BMC Infect Dis. 2010;28:142–150. doi: 10.1186/1471-2334-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serfling RE. Methods for current statistical analysis of excess pneumonia-influenza deaths. Public Health Rep. 1963;78:494–506. [PMC free article] [PubMed] [Google Scholar]
- 23.Costagliola D, Flahault A, Galinec D, Garnerin P, Menares J, Valleron AJ. A routine tool for detection and assessment of epidemics of influenza-like syndromes in France. Am J Public Health. 1991;81:97–99. doi: 10.2105/ajph.81.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–551. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gubler DJ, Suharyono W, Sumarmo Wulur H, Jahja E, Sulianto Saroso J. Virological surveillance for dengue hemorrhagic fever in Indonesia using the mosquito inoculation technique. Bull World Health Organ. 1979;57:931–936. [PMC free article] [PubMed] [Google Scholar]
- 26.Rigau-Perez JG, Gubler DJ. In: Dengue and Dengue Hemorrhagic Fever. Gubler DJ, Kuno G, editors. Oxford: CAB International; 1997. pp. 405–424. (Surveillance for dengue and dengue hemorrhagic fever). [Google Scholar]
- 27.Ooi EE, Gubler DJ. Dengue in Southeast Asia: epidemiological characteristics and strategic challenges in disease prevention. Cad Saude Publica. 2009;25:S115–S124. doi: 10.1590/s0102-311x2009001300011. [DOI] [PubMed] [Google Scholar]
- 28.Kumarasamy V, Wahab AH, Chua SK, Hassan Z, Chem YK, Mohamad M, Chua KB. Evaluation of a commercial dengue NS1 antigen-capture ELISA for laboratory diagnosis of acute dengue virus infection. J Virol Methods. 2007;140:75–79. doi: 10.1016/j.jviromet.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Lima Mda R, Nogueira RM, Schatzmayr HG, dos Santos FB. Comparison of three commercially available dengue NS1 antigen capture assays for acute diagnosis of dengue in Brazil. PLoS Negl Trop Dis. 2010;4:e738. doi: 10.1371/journal.pntd.0000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang SM, Sekaran SD. Early diagnosis of Dengue infection using a commercial Dengue Duo rapid test kit for the detection of NS1, IGM, and IGG. Am J Trop Med Hyg. 2010;83:690–695. doi: 10.4269/ajtmh.2010.10-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas L, Verlaeten O, Cabié A, Kaidomar S, Moravie V, Martial J, Najioullah F, Plumelle Y, Fonteau C, Dussart P, Césaire R. Influence of the dengue serotype, previous dengue infection, and plasma viral load on clinical presentation and outcome during a dengue-2 and dengue-4 co-epidemic. Am J Trop Med Hyg. 2008;78:990–998. [PubMed] [Google Scholar]
- 32.Guzmán MG, Kourí G, Valdés L, Bravo J, Vázquez S, Halstead SB. Enhanced severity of secondary dengue-2 infections: death rates in 1981 and 1997 Cuban outbreaks. Rev Panam Salud Publica. 2002;11:223–227. doi: 10.1590/s1020-49892002000400003. [DOI] [PubMed] [Google Scholar]
- 33.Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa MT, Nogueira RN, da Rosa AT. Origins of dengue type 2 viruses associated with increased pathogenesis in the Americas. Virology. 1997;230:244–251. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- 34.La Ruche G, Souarès Y, Armengaud A, Peloux-Petiot F, Delaunay P, Desprès P, Lenglet A, Jourdain F, Leparc-Goffart I, Charlet F, Ollier L, Mantey K, Mollet T, Fournier JP, Torrents R, Leitmeyer K, Hilairet P, Zeller H, Van Bortel W, Dejour-Salamanca D, Grandadam M, Gastellu-Etchegorry M. First two autochthonous dengue virus infections in metropolitan France. Euro Surveill. 2010;5:19676. [PubMed] [Google Scholar]
- 35.Domingo C, Niedrig M, Gascon J, Palacios G, Reyes N, Malo MJ, Wichmann O, Ruiz J, Schultze D, Schunk M, Puente S, Vinner L, Van Esbroeck M, Schuffenecker I, Grandadam M, Lopez-Velez R, Tenorio A. Molecular surveillance of circulating dengue genotypes through European travelers. J Trop Med. 2011;18:183–190. doi: 10.1111/j.1708-8305.2011.00501.x. [DOI] [PubMed] [Google Scholar]


