Abstract
Background
Here we provide the most comprehensive study to date on the cranial ossification sequence in Lipotyphla, the group which includes shrews, moles and hedgehogs. This unique group, which encapsulates diverse ecological modes, such as terrestrial, subterranean, and aquatic lifestyles, is used to examine the evolutionary lability of cranial osteogenesis and to investigate the modularity of development.
Results
An acceleration of developmental timing of the vomeronasal complex has occurred in the common ancestor of moles. However, ossification of the nasal bone has shifted late in the more terrestrial shrew mole. Among the lipotyphlans, sequence heterochrony shows no significant association with modules derived from developmental origins (that is, neural crest cells vs. mesoderm derived parts) or with those derived from ossification modes (that is, dermal vs. endochondral ossification).
Conclusions
The drastic acceleration of vomeronasal development in moles is most likely coupled with the increased importance of the rostrum for digging and its use as a specialized tactile surface, both fossorial adaptations. The late development of the nasal in shrew moles, a condition also displayed by hedgehogs and shrews, is suggested to be the result of an ecological reversal to terrestrial lifestyle and reduced functional importance of the rostrum. As an overall pattern in lipotyphlans, our results reject the hypothesis that ossification sequence heterochrony occurs in modular fashion when considering the developmental patterns of the skull. We suggest that shifts in the cranial ossification sequence are not evolutionarily constrained by developmental origins or mode of ossification.
Keywords: skull, heterochrony, Eulipotyphla, embryology, ossification, integration, phylogeny, micro CT
Background
The mammalian skull is one of the most extensively studied anatomical systems among vertebrate structures. One of the major aspects of morphological evolution in the mammalian skull is heterochrony, shifts in the timing and rate of development. The more classic heterochronic studies have focused on quantifying changes in size and shape, whereas the study of 'sequence' heterochrony incorporates changes in the timing of discrete developmental events with many advantages in comparative studies [1-4]. One set of events that has received much attention in recent years is the onset of ossification of individual bones [[5] and references therein]. Sánchez-Villagra et al. [6], presenting the most comprehensive study of sequence heterochrony in the mammalian skull to date, analyzed the ossification sequence of 17 bone elements of the skull among 7 marsupial and 13 species of boreoeutherians (the placental mammals, excluding afrotherians and xenarthrans [7]). A few heterochronies were found to diagnose mammals, marsupials or placentals, but the relative timing of cranial ossification patterns is largely conserved among mammal evolution. Although similarly conserved developmental patterns have been reported for Rodentia [8], further investigation of unexamined species and more phylogenetically inclusive studies are needed to test whether this conservatism is a general pattern of mammals.
Recently, the connection of heterochrony to modularity, another central aspect in the evolution of development, has attracted much attention in vertebrates in general [for example, [9-16]]. Modularity, which is considered to be one of the key concepts to bridge evolutionary biology and developmental biology [17-21], refers to the autonomy of groups of events or structures, as well as the strong associations of developmental events or morphological structures [22]. Independence among anatomical structures is thought to permit unrelated parts to vary and/or evolve separately, while the integration within smaller units maintains functionally necessary relationships among traits [23]. From studies on the genetic, developmental and functional modules across vertebrates, it has been implied that developmental modularity may provide insights into processes of morphological evolution [24-34]. Most studies have focused on the physical relationships among functionally- or developmentally-related structures, and there are only a few studies relating developmental timing to the concept of modularity [9-11,35].
Heterochrony is only considered possible in a modular phenotype, in which some parts are autonomous developmentally from others [10,15,36-38]. It has been suggested that heterochronic change in ossification events of the tetrapod skull occurs among different developmental modules, while maintaining the relative timing of developmental events in each module [11]. Schoch [11] suggested that sets of cranial bones belonging to the same developmental modules could shift ossification timing in unison, although this proposition was not statistically tested. Using the analytical method developed by Poe [39], a study on several therian mammals found that postcranial modules show significant developmental integration of the entire appendicular skeleton in boreoeutherian placentals [10]. However, cranial phenotypic variational modules showed no significant conservation of developmental timing for all sister group comparisons, except within a clade consisting of a mole and a shrew, which displayed integrated shifts of ossification sequence in the facial module [10]. While ossification timing of some skeletal parts was implied to reflect evolutionary modularity in some taxa, it remains unclear whether, and to what degree, heterochrony involves modularity. Although the onset of the ossification sequence of cranial parts in the facial module was indicated to shift together within the mole-shrew clade (Talpa europaea and Cryptotis parva), a detailed and more inclusive study of the diverse group to which these animals belong is needed to strictly test the hypothesis relating sequence heterochrony to modularity. In addition, although a significant relationship between cranial heterochrony and modularity was not found in most taxa examined [10], it is still possible that shifts in the cranial ossification sequence of some taxa are less constrained by developmental modularity, whereas those of other taxa may be strongly constrained by modular pattern. Furthermore, because only the relationship between patterns of sequence heterochrony and modularity of phenotypic variation was compared by Goswami [10], it is yet unclear whether developmental modularity of sequence heterochrony is associated with modules derived from developmental origins (that is, neural crest cells vs. mesoderm derived parts) and/or with those derived from developmental modes (that is, dermal vs. endochondral ossification). The body parts that share developmental origins and functions are suggested to behave as evolutionary modules at various levels [23,26,40]. Therefore, we hypothesize that sequence heterochrony is more likely to occur among different developmental modules, while maintaining the relative timing of developmental events within a single module.
Here we provide the most comprehensive study to date on the ossification sequence of cranial elements in Lipotyphla, the group which includes shrews (Soricidae), moles and 'shrew moles' (Talpidae), hedgehogs (Erinaceidae), and Solenodons (Solenodontidae) [41]. With an extensive series of embryonic specimens from a variety of taxa, we present data on cranial ossification sequences for two species of terrestrial hedgehogs (Erinaceus europaeus and E. amurensis), three species of subterranean mole (Mogera wogura, Condylura cristata, and Scapanus orarius), one terrestrial shrew mole (Urotrichus talpoides), one terrestrial shrew (Suncus murinus), and an aquatic water shrew (Chimarrogale platycephala). In addition, the resolution of sequence data for two moles (Talpa europaea and T. occidentalis) and one shrew species (Cryptotis parva) is much improved compared with the data presented in previous literature [6,42,43]. Using this unique group that encapsulates terrestrial, subterranean and aquatic lifestyles, we examine the evolutionary lability and conservatism of cranial osteogenesis and test the hypothesis that skeletal elements belonging to the same module display coordinated shifts of ossification timing while maintaining the relative sequence within the module.
Materials and methods
Data acquisition
Ossification sequence data of 22 cranial elements in 11 lipotyphlan species were sampled from 238 embryonic specimens held in the collections at the Paleontological Institute and Museum of University of Zürich, Kyoto University Museum, Botanical Gardens Museum of Hokkaido University, Aichigakuin University Dental Science Museum, and Wildlife Laboratory at Tokyo University of Agriculture (Figure 1, Table 1). The jugal and ethmoid were excluded from the analysis because these bones were exceptionally small and difficult to indentify. Data for three boreoeutherian outgroup species, including the greater mouse-eared bat (Myotis myotis), the common treeshrew (Tupaia glis), and the Norway rat (Rattus norvegicus), were obtained from the literature [44-46]. Figure 2 is a composite consensus phylogenetic tree of the studied species based on molecular and morphological analyses [47-52].
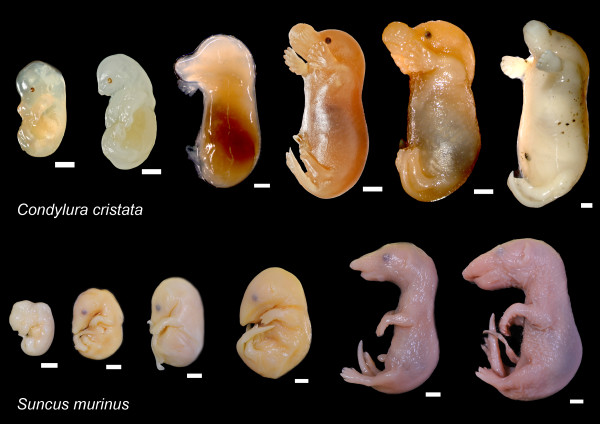
Figure 1.
Ontogenetic series prepared for this study. Upper: Condylura cristata. Lower: Suncus murinus. Scale bar, 2 mm.
Table 1.
Species names, specimen, and stage number
| Species name | Specimen numbers/stages | References |
|---|---|---|
| Erinaceus europaeus | 10/7 | this study |
| Erinaceus amurensis | 21/9 | this study |
| Cryptotis parva | 29/13 | this study |
| Chimarrogale platycephala | 30/8 | this study |
| Suncus murinus | 49/11 | this study |
| Condylura cristata | 10/9 | this study |
| Scapanus orarius | 14/7 | this study |
| Urotrichus talpoides | 13/8 | this study |
| Mogera wogura | 16/8 | this study |
| Talpa europaea | 11/7 | this study |
| Talpa occidentalis | 35/10 | this study |
| Myotis myotis | 26/12 | [28] |
| Tupaia glis | 26/11 | [30] |
| Rattus norvegicus | n.a./7 | [29] |
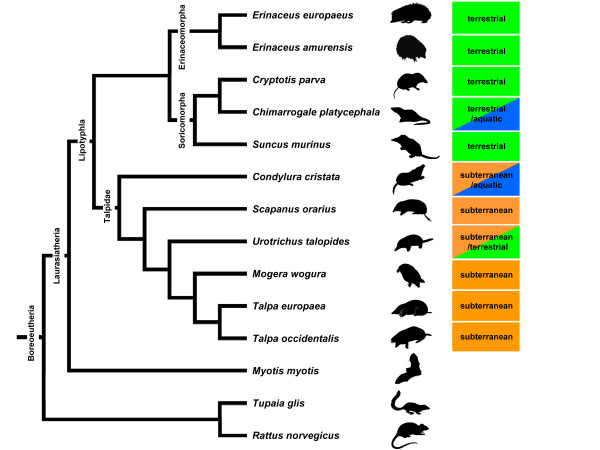
Figure 2.
Phylogenetic relationships among the lipotyphlan species and outgroups included in this study. The phylogenetic framework on which the data were examined is a composite of several sources [47,51,52].
We employed an enzymatic clearing and staining method and a high-resolution tomography (μCT) technique for detecting ossification. Some specimens were cleared and stained by a modified method of a standard enzymatic procedure [43], and the earliest appearance of ossifications was recorded based on uptake of alizarin red (Figure 3) [6]. As other species were historical museum specimens, the appearance of bones was also assessed noninvasively by acquiring shadow images taken by μCT at the University Museum, University of Tokyo (TXS225-ACTIS, TESCO, Tokyo) and at the Anthropological Institute, University of Zurich (μCT80, Scano Medical, Bassersdorf, Switzerland). Three dimensional visualization and analysis of shadow images were conducted in Avizo 6.1 (Visualization Sciences Group, Burlington, MA, USA) (Figure 4). Since using different visualization techniques to obtain ossification sequence is reported not to represent a confounding issue and that differences in detection thresholds do not yield erroneous sequences [53,54], we consider minimum error will be involved with employing both clear staining and μCT methods.
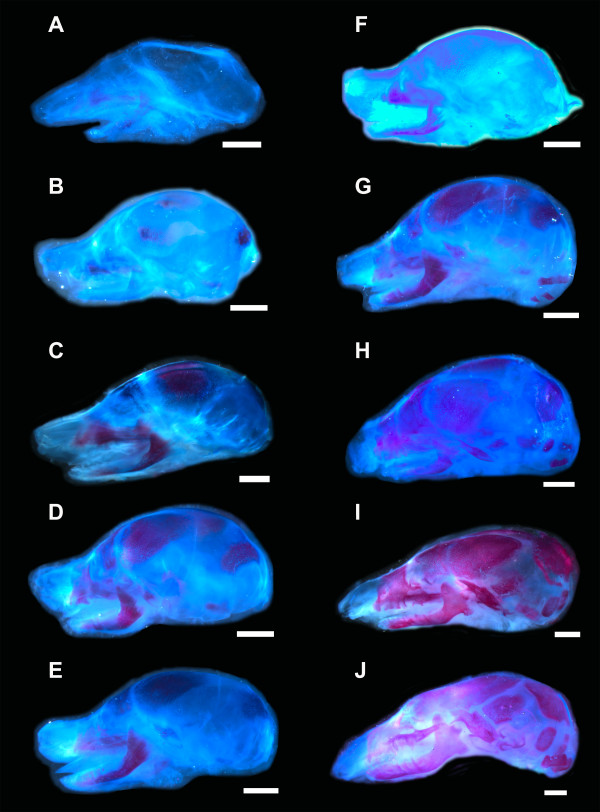
Figure 3.
Cleared and stained specimens of Talpa occidentalis. Calcified structures are stained in red, cartilages in blue, and connective tissue in light blue. Scale bar, 2 mm.
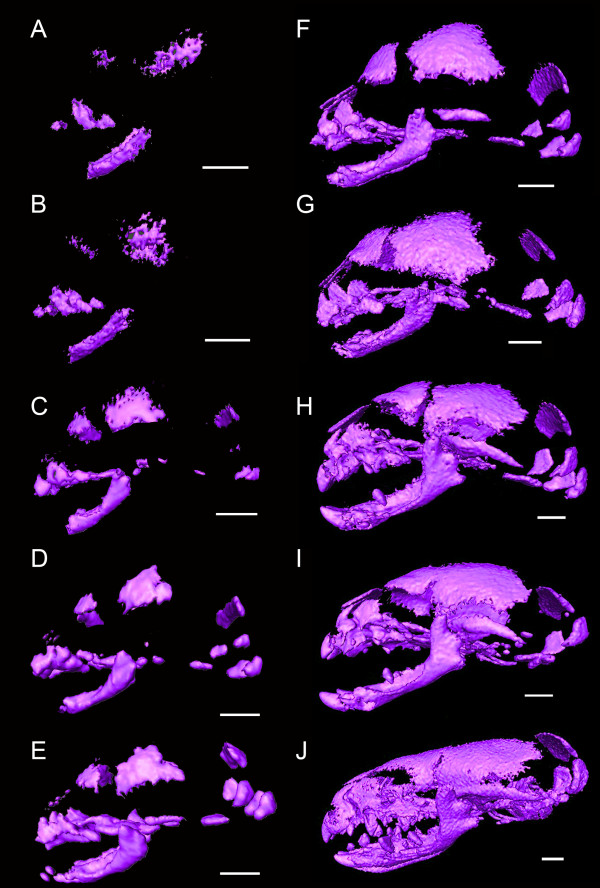
Figure 4.
Three-dimensional reconstructions of μCT scans of Suncus murinus. Scale bar, 1 mm.
Analysis of variation in ossification sequence
To examine the rank variation in sequence of a particular ossification event, we scaled the rank of each ossification event as:
in which r is the absolute rank of a given ossification event, and rmax is the total number of ranks for each species. Therefore, the relative ranks of each species are distributed between 0 and 1. This allowed us to remove the differences of maximum rank between species resulting from differing levels of sampling resolution between species. A similar approach as standardizing the absolute rank r by the maximum number of ranks (rmax) has been applied in previous sequence heterochrony studies [for example, [6,8,53]]. As the ranks are distributed between 1/rmax and 1 with this method, the relative ranks of the earliest bone to ossify can vary, depending on rmax. However, the method used here circumvents this problem because the relative ranks of the earliest event will always be scaled to zero. Nevertheless, some noise remains because species with higher rmax have a lower influence on the variance [53]. The range in rank variation across species was assessed to examine the variability of a particular element in the ossification sequence. The frequency distribution of ranks was also calculated to examine the distribution of ossification events within the rank sequence. In order to assess the variability of ossification timing between modules, rank variation between hypothetical modules was also compared.
Event pairing and PARSIMOV analysis
To identify heterochronies within the ossification sequence, the timing of each ossification event of 22 bones was compared with every other ossification event, that is, the ossification rank of one bone was compared with that of another bone within the species (Table 2). This resulted in 231 event pairs for each species, which were treated as 'characters' [55]. Three character states, before (score 0), simultaneous (score 1), or after (score 2), were given respectively to reflect the relative timing of one ossification event relative to another. For example, if the premaxilla is earlier than the nasal in a certain species, then this event pair is scored as 0. In this way, differences in sequence resolution can be overcome and event pairs can be compared among species. Simultaneous events are usually the result of low resolution of sampling, because the onset of ossification of two bones is unlikely to occur exactly at the same time [6,56]. After constructing event pairs, the character state at each node of the phylogeny can be inferred by a parsimonious approach (but see [57]). Parsimov [55] was employed to map event pairs onto the given phylogeny (Figure 2) and to analyze the evolutionary change of developmental timing. Outgroup species (M. myotis, T. glis, and R. norvegicus) were used to polarize the characters. This parsimony-based phylogenetic method investigates all possible sets of event timing changes on each branch of the phylogeny. Then it computes the minimal number of heterochronic events that account for every event-pair change and yields a consensus that contains all hypotheses of movement that must necessarily form part of any equally parsimonious solution to the observed event-pair changes [55]. This analysis provides a conservative estimate of change compared to the simple mapping method or the more subjective cracking method [6,58]. As recommended by Jeffery et al. [55], optimizations were performed using both ACCTRAN and DELTRAN options, and the consensus of the two was accepted as the most conservative estimate of heterochronic shifts. It should be noted that Parsimov is a highly conservative method and that the consensus of ACCTRAN and DELTRAN is an estimate of minimal heterochronic changes [59].
Table 2.
Cranial events ranked according to relative timing of onset of ossification
| E. europaeus | E. amurensis | Cr. parva | Ch. platycephala | Su. murinus | Co. cristata | Sc. orarius | U. talpoides | M. wogura | T. europaea | T. occidentalis | Myotis | Tupaia | Rattus | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Premaxilla | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 |
| Maxilla | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 |
| Palatine | 3 | 3 | 2 | 2 | 3 | 1 | 1 | 1 | 2 | 1 | 1 | 6 | 3 | 2 |
| Dentary | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Frontal | 1 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 4 | 1 |
| Parietal | 4 | 1 | 3 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 4 | 2 |
| Squamosal | 4 | 3 | 5 | 2 | 4 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 4 | 2 |
| Basioccipital | 5 | 3 | 6 | 3 | 3 | 1 | 5 | 3 | 3 | 2 | 1 | 8 | 6 | 2 |
| Nasal | 4 | 3 | 4 | 2 | 5 | 1 | 1 | 6 | 2 | 2 | 2 | 7 | 7 | 3 |
| Pterygoid | 1 | 3 | 3 | 2 | 4 | 1 | 1 | 1 | 2 | 2 | 4 | 3 | 5 | 2 |
| Exoccipital | 5 | 3 | 6 | 2 | 4 | 1 | 2 | 4 | 4 | 3 | 1 | 10 | 7 | 2 |
| Basisphenoid | 4 | 3 | 8 | 5 | 6 | 4 | 3 | 5 | 7 | 4 | 6 | 9 | 9 | 4 |
| Lacrimal | 6 | 5 | 6 | 6 | 5 | 6 | 1 | 8 | 4 | 4 | 3 | ? | 5 | 3 |
| Alisphenoid | 5 | 3 | 9 | 6 | 8 | 8 | 7 | 8 | 8 | 7 | 8 | 9 | 7 | 4 |
| Orbitosphenoid | 5 | 4 | 10 | 6 | 9 | 7 | 6 | 7 | 6 | 6 | 7 | 12 | 9 | 7 |
| Petrosal | 7 | 8 | 11 | 7 | 10 | 9 | 7 | 8 | 8 | 7 | 9 | 11 | 11 | 6 |
| Mastoid | 7 | 9 | 12 | 7 | 11 | 5 | 5 | 6 | 5 | 4 | 3 | 11 | 11 | 7 |
| Vomer | 2 | 3 | 2 | 2 | 5 | 1 | 1 | 1 | 2 | 1 | 1 | 8 | 5 | 2 |
| Presphenoid | 7 | 9 | 13 | 8 | 11 | 9 | 7 | 8 | 8 | 7 | 10 | ? | 10 | 6 |
| Supraoccipital | 5 | 3 | 5 | 2 | 3 | 1 | 2 | 5 | 2 | 2 | 1 | 9 | 8 | 5 |
| Ectotympanic | 6 | 6 | 6 | 4 | 7 | 3 | 2 | 8 | 6 | 5 | 5 | 5 | 6 | 3 |
| Goniale | 6 | 7 | 7 | 6 | 8 | 6 | 4 | 8 | 2 | 5 | 7 | 4 | 6 | ? |
Data for three boreoeutherian outgroup species, the greater mouse-eared bat (Myotis myotis), the common treeshrew (Tupaia glis), and the Norway rat (Rattus norvegicus) were compiled from the literature [44-46]. Simultaneous events are usually the result of low resolution of sampling, since the onset of ossification of two bones is unlikely to occur exactly at the same time. Therefore, the resolutions for oral bones in Co. cristata, Sc. Orarius, U. talpoides, and T. europaea are limited in this study.
Although event-pair data are unsuitable for phylogenetic analysis due to non-independence of event pairs [60], and although the main objective of this study is not to infer phylogenetic relationships among lipotyphlans, parsimony analysis can be applied to explore the phylogenetic signal within the data. A parsimony analysis of event-pair data was conducted using PAUP∗ version 4.0b10 [61].
Modularity analysis
Using the data on the cranial ossification sequence, we tested whether the cranial elements which belong to the same hypothetical module exhibit coordinated shifts in ossification timing [39]. To test for coordinated shifts in ossification timing, theoretical developmental and phenotypic variational modules were first constructed. Theoretical modules are composed of sets of elements that are predicted to exhibit coordinated timing of the first ossification. In this study, three independent hypothetical divisions were tested. Cranial elements were divided into two developmental origin modules, the mesoderm module and neural crest cell module, according to their reported developmental origin in the mouse [62]. Alternatively, cranial elements were classified into another independent developmental division according to the mode of ossification, that is, the endochondral bone module and dermal bone module [63]. Phenotypic variational module division was based on the morphometric analyses, which have identified five sub-divisions in the adult mammalian skull: oral, zygomatic, nasal, cranial base, and cranial vault [34,64,65]. Hypothetical module associations of bone elements used in the analysis are listed in Table 3.
Table 3.
Hypothetical module associations of bone elements
| Developmental origin module | Ossification mode module | Phenotypic module | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Neural crest cells | Mesoderm | Endochondral | Dermal | Oral | Nasal | Zygomatic | Vault | Basicranium | |
| Premaxilla | ◯ | ◯ | ◯ | ◯ | |||||
| Maxilla | ◯ | ◯ | ◯ | ◯ | ◯ | ||||
| Palatine | ◯ | ◯ | ◯ | ◯ | ◯ | ||||
| Dentary | ◯ | ◯ | ◯ | ||||||
| Frontal | ◯ | ◯ | ◯ | ◯ | |||||
| Parietal | ◯ | ◯ | ◯ | ||||||
| Squamosal | ◯ | ◯ | ◯ | ◯ | |||||
| Basiocciptial | ◯ | ◯ | ◯ | ||||||
| Nasal | ◯ | ◯ | ◯ | ||||||
| Pterygoid | ◯ | ◯ | ◯ | ◯ | ◯ | ||||
| Exoccipital | ◯ | ◯ | ◯ | ◯ | |||||
| Basisphenoid | ◯ | ◯ | ◯ | ◯ | |||||
| Lacrimal | ◯ | ◯ | ◯ | ◯ | |||||
| Alisphenoid | ◯ | ◯ | ◯ | ◯ | |||||
| Orbitosphenoid | ◯ | ◯ | ◯ | ◯ | |||||
| Petrosal | ◯ | ◯ | ◯ | ||||||
| Mastoid | ◯ | ◯ | ◯ | ◯ | |||||
| Vomer | ◯ | ◯ | ◯ | ◯ | |||||
| Presphenoid | ◯ | ◯ | ◯ | ||||||
| Supraoccipital | ◯ | ◯ | ◯ | ||||||
| Ectotympanic | ◯ | ◯ | ◯ | ||||||
| Goniale | ◯ | ◯ | ◯ | ||||||
Integration of developmental timing was analyzed by testing for the conservation of rank orders within sets of traits across phylogeny [39]. If the order of development of a set of modular events is evolutionarily conserved, then the correlation of developmental sequence between taxa is expected to be high. This we examine statistically with the correlation coefficient for a given hypothesized events [39]. Ossification sequences in pairs of sister taxa were compared using Kendall's τ, which measures the degree of similarity of their sequences. Here, the correlation coefficient Kendall's τ is calculated as:
in which nc is the number of concordant pairs of ranks, nd is the number of discordant pairs, nx is the number of tied events in the first taxon (that is, the number of simultaneous ossification events within the taxon), and ny is the number of tied events in the other taxon. We must note that the Kendall's τ correlation coefficient can be biased in cases of many tied events, but our dataset has enough resolution to calculate the coefficient.
The significance of the observed Kendall's τ between sister taxa was assessed for each module by comparing it within the null distribution of similar numbered sets of developmental events. We randomly sampled sets of a comparable number of developmental events from the whole events (iteration 1,000 times) and obtained the correlation value (that is, Kendall's τ) between taxa. For instance, the vault module contains nine bones. In this case, the rank correlation of nine vault bones between two sister taxa is first computed. Next, nine random events are sampled from the whole events, and then the correlation of nine randomly sampled events between the sister groups is computed. The null distribution for correlation values is obtained by conducting this procedure 1,000 times. If the vault module is significantly shifting as a whole, observed correlation value (that is, Kendall's τ) between the two taxa shall be ranked high among the null distribution of randomly obtained correlation values.
This test is simple if only two taxa are compared (that is, when comparing between terminal nodes), but comparisons on multiple species may involve statistical non-independence, which necessitates an approach that incorporates information on phylogeny [39]. In these cases, a version of Felsenstein's independent contrast [66] can circumvent the issue of non-independence of multiple species comparison [39]. First, the hypothetical ancestral developmental sequence is computed for each node. Then independent comparisons of sequences are conducted between pairs of independent nodes of the phylogeny [39]. Here the test statistic is the average Kendall's τ value for these node-pairs. The null distribution is obtained by calculating average correlation values of random samples (iteration 1,000 times). The sequence of ancestral nodes was reconstructed by averaging the sequences of sister taxa joined at a particular node [39]. The averaged sequence was considered as the hypothetical ancestral sequence and then used for a pairwise significance test of Kendall's τ. It should be noted that since the actual (extinct) ancestral sequence may not be simply the average of descendent taxa, this technique potentially produces biases. However, averaging is widely adopted in methods such as independent contrasts [9,66]. If heterochronies in ossification sequences have a strong modular pattern, such a pattern would be detectable with this approach [9,39]. In this study, a 0.05 significance level was adopted, and this significance level was corrected using Bonferroni threshold (P = 0.05/99). A random sampling procedure was done with Poptools (Pest Animal Control Co-operative Research Centre, Canberra, Australia) [67] and statistical analyses were conducted in PAST [68].
Results
Variation in ossification sequence
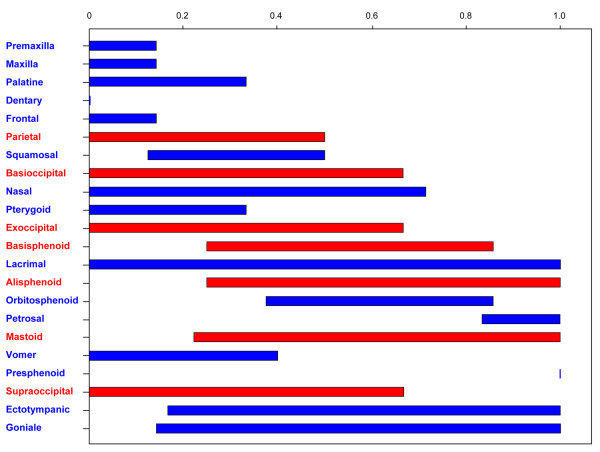
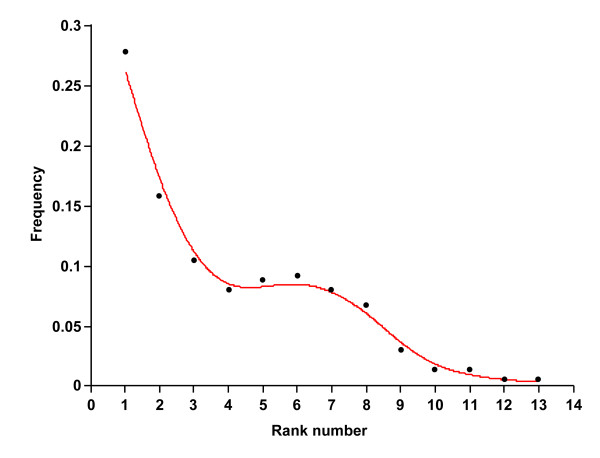
Ossification sequences of each species are listed in Table 2, and the frequency distribution of each stage is given in Additional file 1. The rank variation of each cranial element across lipotyphlan species is summarized in Figure 5. The distributions of the elements show that an ossification event occurs predominantly in the early stages of the whole ossification sequence (Figure 6). No intraspecific variation was found in ossification sequence for all species. The most interspecifically variable elements in ossification sequence were lacrimal, goniale, ectotympanic and mastoid. Presphenoid, dentary, frontal, maxilla, premaxilla and petrosal were among the least variable elements. The mean rank range for each phenotypic and developmental module is listed in Table 4. Among the five phenotypic modules, the vault module was the most variable followed by the basicranial and zygomatic modules. When developmental origin modules were compared, the mesoderm module was more variable than the neural crest cell module. Among the ossification mode modules, the cartilaginous bone module was more variable than the dermal bone module.
Figure 5.
Adjusted rank ranges of single bones across species. Ranks range are from above 0 (ossifying first) to 1 (ossifying last). Dentary was always the first bone to ossify, and presphenoid was always the last bone to ossify in all species. Bones derived from the neural crest cells are colored in blue, and bones derived from the mesoderm are illustrated in red.
Figure 6.
Frequency variation plot of cranial ossifying events in this study.
Table 4.
Adjusted rank range for each bone set
| Rank range | ||
|---|---|---|
| Developmental origin | Neural crest cells | 0.41 |
| Mesoderm | 0.60 | |
| Ossification mode | Endochondral | 0.55 |
| Dermal | 0.38 | |
| Phenotypic module | Oral | 0.23 |
| Nasal | 0.31 | |
| Zygomatic | 0.50 | |
| Vault | 0.60 | |
| Basicranial | 0.55 | |
| Origin + ossification mode | Mesoderm (endochondral) | 0.61 |
| Mesoderm (dermal) | 0.50 | |
| Neural crest cells (endochondral) | 0.48 | |
| Neural crest cells (dermal) | 0.36 |
Event paring and PARSIMOV analyses
A total of 231 event-pairs revealed 124 parsimony-informative characters (53.7% of total), with 67 characters constant (29.0%), and 40 variable pairs parsimony-uninformative (17.3%). Table 5 contains the consensus list of the movements in the timing of ossification of cranial elements detected by Parsimov. One potentially autapomorphic heterochronic pattern that characterizes lipotyphlans was found: the late onset of ossification of the lacrimal bone relative to the pterygoid. No heterochrony was found for nodes of Laurasiatheria, hedgehogs and shrews, respectively. On the other hand, moles were characterized by early ossifications of palatine (with respect to premaxilla and frontal), nasal (with respect to frontal and pterygoid), pterygoid (with respect to premaxilla, frontal and parietal), and vomer (with respect to premaxilla, palatine, frontal and parietal). The clade consisting of Scapanus, Urotrichus, Mogera, and Talpa was distinguished by a late onset of ossification of alisphenoid in relation to petrosal and presphenoid. The aquatic shrew Ch. pltaycephala was characterized by late shifts of maxilla and premaxilla (with respect to palatine, dentary and frontal), and lacrimal (with respect to basisphenoid and goniale) and early shifts of squamosal (with respect to palatine, dentary and frontal), nasal (with respect to palatine, dentary, frontal and pterygoid), pterygoid (with respect to palatine and frontal), exoccipital (with respect to palatine, frontal, parietal and supraoccipital), supraoccipital (with respect to frontal, parietal, nasal and pterygoid). The aquatic/fossorial mole Co. cristata was characterized by early development of basioccipital (with respect to premaxilla, maxilla, palatine, dentary, frontal and parietal, squamosal, pterygoid and supraoccipital), nasal (with respect to basiocciptial), exoccipital (with respect to premaxilla, maxilla, palatine, dentary, frontal, parietal, squamosal, basiocciptial, pterygoid and vomer), and supraoccipital (with respect to premaxilla, maxilla, palatine, dentary, frontal, parietal, pterygoid and vomer) and a late shift of lacrimal. The terrestrial mole U. talpoides was characterized by late shifts of nasal (with respect to frontal, pterygoid, exoccipital, basisphenoid, mastoid and supraoccipital), lacrimal (with respect to basisphenoid, alisphenoid, orbitosphenoid, petrosal, mastoid, presphenoid, ectotympanic and goniale), supraoccipital (with respect to squamosal, basiocciptial and basisphenoid), ectotympanic (with respect to basisphenoid, alisphenoid, orbitosphenoid, petrosal and presphenoid), and goniale (with respect to alisphenoid, orbitosphenoid, petrosal and presphenoid).
Table 5.
List of the heterochronic movements reconstructed by the Parsimov method (consensus of ACCTRAN and DELTRAN)
| Clade | event | movemet | ...in relation to... |
|---|---|---|---|
| Laurasiatheria | No movement | ||
| Chiroptera | Palatine | late | Parietal, Squamosal, Pterygoid, Ectotympanic |
| Basioccipital | late | Nasal, Ectotympanic | |
| Exoccipital | late | Nasal, Alisphenoid | |
| Vomer | late | Nasal, Pterygoid, Ectotympanic | |
| Rodentia | Premaxilla | late | Palatine, Squamosal |
| Palatine | late | Parietal, Squamosal | |
| Exoccipital | early | Basiocciptial, Nasal, Pterygoid, Alisphenoid, Vomer | |
| Orbitosphenoid | late | Mastoid, Presphenoid | |
| Petrosal | early | Mastoid, Presphenoid | |
| Lipotyphla | Lacrimal | late | Pterygoid |
| Erinaceidae | No movement | ||
| Erinaceus europaeus | Frontal | early | Premaxilla, Maxilla, Dentary |
| Parital | late | Premaxilla, Maxilla | |
| Pterygoid | early | Premaxilla, Maxilla, Dentary, Frontal, Vomer | |
| Erinaceus amurensis | No movement | ||
| Soricidae | No movement | ||
| Soricinae | No movement | ||
| Cryptosis parva | Parital | late | Premaxilla, Maxilla, Frontal |
| Exoccipital | late | Squamosal, Nasal | |
| Vomer | early | Nasal, Pterygoid | |
| Chimarrogale platycephala | Premaxilla | late | Palatine, Dentary, Frontal |
| Maxilla | late | Palatine, Dentary, Frontal | |
| Squamosal | early | Palatine, Frontal, Parietal | |
| Nasal | early | Palatine, Frontal, Parietal, Pterygoid | |
| Pterygoid | early | Palatine, Frontal | |
| Exoccipital | early | Palatine, Frontal, Parietal, Supraoccipital | |
| Lacrimal | late | Basisphenoid, Goniale | |
| Supraoccipital | early | Frontal, Parietal, Nasal, Pterygoid | |
| Suncus murinus | Parital | early | Palatine, Dentary, Frontal |
| Basioccipital | early | Palatine, Squamosal | |
| Vomer | late | Pterygoid, Lacrimal | |
| Talpidae | Palatine | early | Premaxilla, Frontal |
| Nasal | early | Frontal, Pterygoid | |
| Pterygoid | early | Premaxilla, Frontal, Parietal | |
| Vomer | early | Premaxilla, Palatine, Frontal, Parietal | |
| Condylura cristata | Basioccipital | early | Premaxilla, Maxilla, Palatine, Dentary, Frontal, Parietal, |
| Squamosal, Pterygoid, Supraoccipital | |||
| Nasal | early | Basiocciptial | |
| Exoccipital | early | Premaxilla, Maxilla, Palatine, Dentary, Frontal, Parietal, | |
| Squamosal, Basiocciptial, Pterygoid, Vomer | |||
| Lacrimal | late | Basisphenoid, Ectotympanic, Goniale | |
| Supraoccipital | early | Premaxilla, Maxilla, Palatine, Dentary, Frontal, Parietal, | |
| Pterygoid, Vomer | |||
| Scapanus + Urotrichus + Mogera + Talpa | Alisphenoid | late | Petrosal, Presphenoid |
| Scapanus orarius | Basioccipital | late | Basisphenoid, Mastoid |
| Lacrimal | early | Premaxilla, Maxilla, Palatine, Dentary, Frontal, Parietal | |
| Nasal, Pterygoid, Vomer | |||
| Urotrichus + Mogera + Talpa | No movement | ||
| Urotrichus talpoides | Nasal | late | Frontal, Pterygoid, Exoccipital, Basisphenoid, Mastoid, |
| Supraoccipital | |||
| Lacrimal | late | Basisphenoid, Alisphenoid, Orbitosphenoid, Petrosal, | |
| Mastoid, Presphenoid, Ectotympanic, Goniale | |||
| Supraoccipital | late | Squamosal, Basiocciptial, Basisphenoid | |
| Ectotympanic | late | Basisphenoid, Alisphenoid, Orbitosphenoid, Petrosal, | |
| Presphenoid | |||
| Goniale | late | Alisphenoid, Orbitosphenoid, Petrosal, Presphenoid | |
| Mogera + Talpa | Pterygoid | late | Premaxilla, Maxilla, Dentary, Parietal, Squamosal |
| Mogera wogura | Palatine | late | Premaxilla, Maxilla, Dentary, Parietal, Squamosal |
| Frontal | late | Squamosal, Supraoccipital | |
| Orbitosphenoid | early | Basisphenoid, Ectotympanic | |
| Vomer | late | Premaxilla, Maxilla, Dentary, Parietal, Squamosal | |
| Goniale | early | Squamosal, Nasal, Pterygoid, Basisphenoid, Lacrimal, | |
| Supraoccipital, Ectotympanic | |||
| Talpa | Pterygoid | late | Palatine, Frontal |
| Talpa europaea | Frontal | early | Nasal |
| Basioccipital | early | Squamosal | |
| Basisphenoid | early | Lacrimal, Mastoid | |
| Talpa occidentalis | Frontal | late | Palatine, Vomer |
| Squamosal | late | Nasal, Lacrimal, Mastoid | |
| Basioccipital | early | Premaxilla, Maxilla, Palatine, Dentary, Parietal, Vomer | |
| Pterygoid | late | Squamosal, Nasal, Lacrimal, Mastoid | |
| Exoccipital | early | Premaxilla, Maxilla, Palatine, Dentary, Parietal, | |
| Basiocciptial, Nasal, Vomer, Supraoccipital | |||
| Supraoccipital | early | Premaxilla, Maxilla, Dentary, Parietal, Nasal |
The results from the single ACCTRAN and DELTRAN analyses are also provided in Additional file 2. The separate results obtained from ACCTRAN and DELTRAN showed that Laurasiatheria is characterized by late shifts of premaxilla (with respect to parietal and squamosal), alisphenoid (with respect to nasal and basisphenoid), and orbitosphenoid (with respect to basisphenoid and petrosal). An early shift of parietal (with respect to maxilla and palatine) and late shifts of palatine (with respect to parietal and pterygoid), lacrimal (with respect to basioccipital, exoccipital, pterygoid and vomer), alisphenod (with respect to exoccipital and basisphenoid), presphenoid, ectotympanic and goniale were detected for Lipotyphla. The clade consisting of hedgehogs and shrews was characterized by early shifts of nasal (with respect to lacrimal and vomer) and supraoccipital (with respect to basioccipital, exoccipital and lacrimal) and a late shift of goniale (with respect to basioccipital and ectotympanic).
The consensus tree of two equally parsimonious trees resulting from the parsimony analysis of the event-pairing data (Additional file 3) was mostly incongruent with the phylogeny taken as reference (Figure 2). The terrestrial outgroup species E. amurensis, S. murinus, and R. norvegicus were clustered together. Although all six mole species were clustered together, this cluster was paraphyletic, given the position in it of C. platycephala's position.
Modularity analysis
Results of Poe's modularity test [39] are summarized in Table 6. No sets showed significant correlation for the hypothetical modules. For the analysis of phenotypic variational modules, one comparison showed a P-value of 0.027 (oral module in T. europaea vs. T. occidentalis), but this was not statistically significant under the Bonferroni threshold (0.05/99). Among the developmental origin modules, although three sister group comparisons (Erinaceidae-Soricidae node vs. Talpidae, Erinaceidae vs. Soricidae and T. europaea vs. T. occidentalis) showed P-values lower than 0.05 for the neural crest cell module, these values did not reach the Bonferroni corrected significance level. No significant correlation was found for the mesoderm module in any sister group comparison.
Table 6.
Results of modularity analysis in pairs of studied taxa.
| Developmental origin module | Ossification mode module | Phenotypic module | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Neural crest cells | Mesoderm | Endochondral | Dermal | Oral | Nasal | Zygomatic | Vault | Basicranial | |||
| E. europaeus | vs. | E. amurensis | 0.877 (0.167) | 0.843 (0.444) | 0.825 (0.512) | 0.608 (0.931) | 0.667 (0.916) | 0.783 (0.620) | 0.754 (0.678) | 0.814 (0.667) | 0.867 (0.342) |
| Cr.parva | vs. | Ch. platycephala | 0.803 (0.610) | 0.904 (0.176) | 0.888 (0.097) | 0.613 (0.958) | 0.405 (0.972) | - | 0.737 (0.769) | 0.787 (0.680) | 0.866 (0.277) |
| Cr.parva+Ch. platycephala | vs. | Su. murinus | 0.829 (0.310) | 0.907 (0.166) | 0.854 (0.250) | 0.613 (0.941) | 0.563 (0.978) | - | 0.860 (0.454) | 0.851 (0.472) | 0.833 (0.494) |
| Erinaceidae | vs. | Soricidae | 0.867 (0.043) | 0.799 (0.639) | 0.780 (0.708) | 0.671 (0.944) | 0.668 (0.917) | 0.826 (0.550) | 0.809 (0.620) | 0.818 (0.500) | 0.823 (0.486) |
| T. europaea | vs. | T. occidentalis | 0.894 (0.041) | 0.853 (0.333) | 0.786 (0.597) | 0.808 (0.487) | 1.000 (0.027) | 0.632 (0.981) | 0.863 (0.110) | 0.699 (0.875) | 0.747 (0.750) |
| Talpa | vs. | Mogera | 0.830 (0.148) | 0.908 (0.111) | 0.704 (0.903) | 0.760 (0.666) | 0.724 (0.708) | 0.552 (0.982) | 0.855 (0.269) | 0.783 (0.625) | 0.631 (0.944) |
| Talpa+Mogera | vs. | Urotrichus | 0.732 (0.611) | 0.852 (0.231) | 0.608 (0.981) | 0.716 (0.569) | - | 0.546 (0.972) | 0.771 (0.519) | 0.805 (0.292) | 0.562 (0.958) |
| Talpa+Mogera+Urotrichus | vs. | Scapanus | 0.684 (0.630) | 0.692 (0.657) | 0.613 (0.875) | - | - | - | 0.710 (0.611) | 0.706 (0.486) | 0.522 (0.917) |
| Talpa+Mogera+Urotrichus+Scapanus | vs. | Cristatus | 0.731 (0.361) | 0.753 (0.491) | 0.722 (0.528) | - | - | - | 0.797 (0.324) | 0.700 (0.583) | 0.630 (0.722) |
| Erinaceidae+Soricidae | vs. | Talpidae | 0.839 (0.037) | 0.764 (0.731) | 0.730 (0.931) | 0.711 (0.902) | - | - | 0.799 (0.639) | 0.695 (0.917) | 0.795 (0.431) |
| Lipotyphla | vs. | Myotis | 0.711 (0.370) | 0.702 (0.602) | 0.555 (0.944) | 0.802 (0.111) | 0.725 (0.403) | 0.814 (0.278) | 0.760 (0.380) | 0.648 (0.667) | 0.544 (0.875) |
Kendall's τ is listed for each comparison and P-values are given in parentheses. Significance level was set as P < 0.05/99 (that is, P < 0.0005) by Bonferroni threshold. Values are not given for those sets in which Kendall's τ was indeterminate owing to low element numbers and/or high conservation of developmental sequence, denoted "-".
Discussion
Sequence heterochrony
It has recently been proposed that the sequence of cranial ossification is quite conservative and that few heterochronic shifts have occurred in mammalian evolution based on the study of 17 boreoeutherians [6]. This previous study, and other similar ones for other groups, used the fairly conservative approach of Parsimov [69] to detect heterochrony. To make our study comparable, we also used Parsimov. Although we confirmed the general vertebrate pattern (such as the early ossification of jaw bones with respect to the neurocranial bones [11,70]) in lipotyphlans, our analysis suggests that some heterochronies have occurred during the evolution of this group. For example, the late shifting of the lacrimal in respect to pterygoid characterizes Lipotyphla. In addition, moles exhibit considerably derived features, such as the accelerated development of the palatine, nasal, pterygoid and vomer. This indicates that the relatively early development of the vomeronasal complex is an autapomorphic feature of moles. In many mammals, bones in the vomeronasal region are known to ossify later than the anterior dermal bones (premaxilla, maxilla, palatine and frontal) [8,46,71]. However, it appears that the development of these bones occurs virtually simultaneously with the above mentioned anterior dermal bones in moles.
It is known that rostrum morphology is relatively robust and box-shaped in moles compared to that of shrews and hedgehogs. A robust snout is often used for boring through the soil and to push away encountered objects [72,73]. Mole snouts are equipped with highly touch-sensitive, domed mechanosensory organs called Eimer's organs, which are not found in other lipotyphlans, such as shrews, hedgehogs and solenodons [74,75]. Moles have poor eyesight and gather much of their information by probing the environment with the glabrous tip of their nose (rhinarium) [74,76]. Each Eimer's organ functions as a tactile receptor that contains a number of separate elements, including sensory receptors and supporting epidermal tissue in a specific configuration [74]. The drastic early shift of the vomeronasal complex may be related to the increased importance of the rostrum for fossorial lifestyle in moles, since the number of Eimer's organs reflects the surface area of the rhinarium [77], which is relatively large in moles.
In contrast, the shrew mole U. talpoides, which is more terrestrial [51,78], shows a secondary late shift of the nasal. This indicates that, whereas all fossorial moles develop the vomeronasal region early in embryogenesis, the terrestrial shrew-like U. talpoides is an exception. It is reported that U. talpoides possesses 1,310 Eimer's organs, which is a considerably small number compared to other true moles (Mogera, 2,200; Talpa, 2,200; Scapanus, 2,470; Condylura, 26,000; Catania 2,000). It is also noteworthy that relative size and robustness of the vomeronasal complex is reduced in U. talpoides compared to that of other moles and that its pointed gracile vomernasal organ resembles that of shrews [74,79,80]. In contrast to other fossorial moles, the more terrestrial U. talpoides has functional eyes and a pointed gracile rostrum used for sniffing as in shrews, whereas the role of the rostrum as a tactile sense and boring appendage is reduced [Hisashi Abe, personal communication]. It is conceivable that while the developmental timing of the vomeronasal complex shifted earlier in the common ancestor of moles, since the role of the snout was emphasized for subterranean lifestyle, the nasal reverted to a later development through secondary terrestrial adaptations in U. talpoides. This scenario results from the phylogenetic framework taken as reference, but it should be noted that the position of the shrew moles in talpid phylogeny is contested [47,51,52]
Another shift of the onset of ossification timing in U. talpoides is found for the supraoccipital. Although the supraoccipital has moved early in the common ancestor of moles, this bone reverts to a later development in U. talpoides. It is known that moles have undergone considerable morphological transformations associated with digging [81-83]. For example, the supraoccipital region is relatively increased in moles compared to other lipotyphlans [84] and provides the space to attach the enlarged musculus rhomboideus that functions as the major digging muscle [83]. Whereas the m. rhomboideus capitis is generally small in mammals, this muscle is considerably enlarged in moles [81,83]. Given that the enlarged supraoccipital provides the attachment site for the m. rhomboideus capitis, the early onset of the supraoccipital may be functionally coupled to the enlargement of this bone in moles. At the same time, the reduced occipital size in U. talpoides may reflect the reduced importance of digging muscles.
Another notable heterochrony found among lipotyphlans is that the lacrimal (with respect to basisphenoid and goniale), exoccipital (with respect to palatine, frontal and parietal), and supraoccipital (with respect to frontal and parietal) shift early in Ch. platycephala and in Co. cristata, respectively. These two phylogenetically distant species (Figure 2) are unique in comparison to other lipotyphlans studied here in having independently acquired a semi-aquatic lifestyle [85-87]. Although it is difficult to sort out the functional significance of early development of these bones, it may be related to adaptations for an aquatic lifestyle and swimming ability.
The phylogenetic relationships among shrews, hedgehogs and moles have been highly controversial [88-90]. While anatomical studies have conventionally suggested a shrew + mole clade to the excursion of hedgehogs [90], recent molecular studies supports the monophyly of shrews and hedgehogs [49,88,89]. Although event pairs are generally unsuitable for phylogenetic analysis due to the non-independence of data [6,11,91], it is worth noting that our exploratory parsimony based analysis resulted in a clustering of moles with the inclusion of Ch. platycephala (Additional file 3).
Rank variability
Sánchez-Villagra et al. [6], whose study included 20 mammalian species, reported that the most interspecifically variable cranial elements in terms of relative ossification timing are the basioccipital, basisphenoid, jugal, parietal, pterygoid and squamosal. In contrast, the lacrimal, goniale, ectotympanic, mastoid, alisphenoid and nasal bones are found as the most variable elements during the onset of ossification in lipotyphlans (Figure 5). In a more inclusive study on Rodentia, it is reported that the parietal, alisphenoid, squamosal, jugal, pterygoid and basisphenoid are the most labile bones in terms of relative ossification timing [8]. Although it appears that alisphenoid is similarly variable in rodents and lipotyphlans, this bone is not reported to be highly variable across the major clades in mammals [6]. Presphenoid, dentary, frontal, premaxilla, maxilla and petrosal are the most conservative elements in lipotyphlans. Consistent with our results, petrosal, dentary and premaxilla are reported to be the least variable cranial elements in rodents [8]. Similarly, Sánchez-Villagra et al. [6] reported that the least variable elements include anterior facial bones, such as the dentary, maxilla, frontal and premaxilla. This suggests that facial elements, which generally ossify at early stages of bone development (Figure 5), are less variable in terms of ossification timing.
Overall, the amount of variability in timing of ossification onset in lipotyphlan cranial bones was as follows, in order of highest to lowest variability: mesoderm derived endochondral bones, mesoderm derived dermal bones, neural crest cell derived endochondral bones, and neural crest cell derived dermal bones (Table 4). Thus, the cartilaginous bones are more variable as a whole than dermal bones among the mesoderm derived bones, whereas endochondral bones are similarly more variable than dermal bones within the neural crest cell derived bones (Table 4). We also found that mesoderm derived bones tend to be more variable than neural crest cell derived bones. Sánchez-Villagra et al. [6] noted that the relative ossification timing of neurocranial traits, especially the basicranial bones, are more labile than the facial elements. The basicranial bones are also apparently highly variable as a whole within lipotyphlans (Figure 5, Table 4). However, the cranial base also includes less variable bones, such as presphenoid, petrosal and pterygoid. Instead, both developmental origin (mesoderm or neural crest cells) and ossification mode (dermal or endochondral) are coupled with the evolutionary lability of ossification timing.
Modularity of ossification heterochrony
On the whole there was little integration across all the modules tested. No developmental module showed significant correlation for any sister group comparison. This indicates that lipotyphlan ossification heterochrony is not constrained by differences in developmental origins (mesoderm or neural crest cells) or ossification mode (dermal or endochondral). Although developmental origin and ossification mode of bones are considered to be highly influential to morphological evolution in vertebrates [62,92-94], it appears that ossification heterochrony is an exception to this pattern. It is possible that the independence of ossification timing of each bone may facilitate the labile alteration of ossification sequence and, thus, ease consequent evolutionary changes in phenotypes. In addition, the lack of clear integration in ossification heterochrony among phenotypic modules indicates that sequence heterochrony is virtually independent from morphometrically derived phenotypic modules of the cranium, which is consistent with the results reported by Goswami and colleagues [9,10]. Although adult phenotypic (metric) traits of the mammalian skull are reported to exhibit a highly modular pattern [for example, [33,34,95]], our results suggest that modularity in adult phenotypic traits is not strongly linked to integration in ossification sequence heterochrony. Functionally- or developmentally-integrated structures have often been suggested to exhibit coordinated shifts in developmental timing [10,11,13,96]. However, very few studies have explicitly examined the relationship between modularity and sequence heterochrony [10,95]. Our results presented here suggest that the hypothesis raised by Schoch [11] and others [12,13,96] - that there is a modular pattern in sequence heterochrony - is not supported for the case of lipotyphlans. Either the lipotyphlan case is an exception, or the sampling or methods used in previous analyses are in need of expansion or revision.
Although the integration of ossification heterochrony for a priori hypothetical modules was tested in this study, we did not attempt to detect heterochronic modules from our ossification dataset. We must admit that the modularity test and the following Bonferroni correction adopted in this study are rather conservative to detect modularity in sequence heterochrony. Development of analytical tools to detect modularity directly from sequence heterochrony data shall provide new avenues to understand the role of modularity in the evolution of vertebrate cranial diversity.
Conclusion
The extensive examination of lipotyphlan craniogenesis reveals notable heterochronies in the onset of ossification. An early shift of developmental timing of the vomeronasal complex has occurred in the common ancestor of moles. This shift is most likely coupled with the robust nature of the rostrum for boring and tactile sensing in subterranean life. On the other hand, the nasal has reverted to a more typical later development in the more terrestrial shrew mole. This secondary shift in developmental timing can be linked to the secondary shift (concluded from phylogeny) to a terrestrial lifestyle and corresponding reduction of shrew mole digging and sensory requirements. Finally, we find no significant conservation of ossification timing within modules constructed depending on developmental origins or ossification modes. We reject the hypothesis that ossification sequence heterochrony is more likely to occur among different developmental modules. Although developmental origin and ossification mode of bones are considered to be highly influential to morphological evolution in vertebrates; in general, it is suggestible that lipotyphlan ossification heterochrony is not evolutionarily constrained by differences in developmental origins (mesoderm or neural crest cells) or ossification mode (dermal or endochondral).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DBK and MRSV designed the study. DBK, HE, CM, SO, KK, MA and MRSV collected samples. DBK, GS and CPEZ conducted analysis of CT data. DBK analyzed the data and wrote the paper. All authors read, discussed and approved the final manuscript.
Supplementary Material
Frequency distribution of specimens for each stage. Ossification events are skewed toward earlier stages of sequences, suggesting a greater concentration of ossification events and/or less resolution early in the sequence.
Heterochronies reported by the ACCTRAN and DELTRAN. All the heterochronic events detected by the ACCTRAN and DELTRAN are summarized.
Phenogram obtained from parsimony analysis of event-pair characters. The consensus tree is obtained from parsimony analysis of event-paring scores. Although all six mole species were clustered together, the phenogram was mostly incongruent with the commonly accepted phylogenetic relationships.
Contributor Information
Daisuke Koyabu, Email: dsk8evoluxion@gmail.com.
Hideki Endo, Email: hendo@um.u-tokyo.ac.jp.
Christian Mitgutsch, Email: christian.mitgutsch@gmail.com.
Gen Suwa, Email: suwa@um.u-tokyo.ac.jp.
Kenneth C Catania, Email: ken.catania@vanderbilt.edu.
Christoph PE Zollikofer, Email: zolli@aim.uzh.ch.
Sen-ichi Oda, Email: oda@zool.ous.ac.jp.
Kazuhiko Koyasu, Email: k0yasu@dpc.agu.ac.jp.
Motokazu Ando, Email: mando@nodai.ac.jp.
Marcelo R Sánchez-Villagra, Email: m.sanchez@pim.uzh.ch.
Acknowledgements
Ingmar Werneburg and Laura A. B. Wilson critically read and commented on a previous version of the manuscript. The authors are thankful to Laura A. B. Wilson for giving us statistical advice. Two reviewers provided very important suggestions and criticisms and one of them led us to reconsider some fundamental assumptions of our initial conclusions. To this anonymous reviewer, we express our sincere thanks for her/his intellectual contribution during the collegiate review-process. We appreciate discussions with Hisashi Abe, Shin-ichiro Kawada, Osamu Kondo, Shigeru Kuratani, Akio Shinohara, Masaharu Motokawa, Satoshi Suzuki, Hisayoshi Nozaki, Kinya G. Ota, Masaki Takechi and Naoki Irie. We thank Ryusuke Fujimoto, Yoori Furuta, Misato Hosojima, Hideo Ichikawa, Masaru Kato, Masaharu Motokawa, Torsten Scheyer and Fumihito Takaya for access to specimens and various kinds of help. We are grateful to Naoki Morimoto for assistance with micro CT scans. This study was supported by the Japan Society for Promotion of Science Research Fellowship for Young Scientists (no. 234832) to DBK, by the Japan Society for Promotion of Science Grant-in-Aids for Scientific Research (no. 22405002) to HE, and by the Swiss National Fund (3100A0-133032/1) to MRSV.
References
- Richardson MK, Jeffery JE, Coates MI, Bininda-Emonds ORP. Comparative methods in developmental biology. Zoology. 2001;104:278–283. doi: 10.1078/0944-2006-00033. [DOI] [PubMed] [Google Scholar]
- Maxwell EE, Harrison LB. Methods for the analysis of developmental sequence data. Evol Dev. 2009;11:109–119. doi: 10.1111/j.1525-142X.2008.00307.x. [DOI] [PubMed] [Google Scholar]
- de Jong IML, Colbert MW, Witte F, Richardson MK. Polymorphism in developmental timing: intraspecific heterochrony in a Lake Victoria cichlid. Evol Dev. 2009;11:625–635. doi: 10.1111/j.1525-142X.2009.00370.x. [DOI] [PubMed] [Google Scholar]
- de Jong IML, Witte F, Richardson MK. Developmental stages until hatching of the Lake Victoria cichlid Haplochromis piceatus (Teleostei: Cichlidae) J Morphol. 2009;270:519–535. doi: 10.1002/jmor.10716. [DOI] [PubMed] [Google Scholar]
- Mitgutsch C, Wimmer C, Sánchez-Villagra MR, Hahnloser R, Schneider RA. Timing of ossification in duck, quail, and zebra finch: intraspecific variation, heterochronies, and life histgory evolution. Zoolog Sci. 2011;28:491–500. doi: 10.2108/zsj.28.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Villagra MR, Goswami A, Weisbecker V, Mock O, Kuratani S. Conserved relative timing of cranial ossification patterns in early mammalian evolution. Evol Dev. 2008;10:519–530. doi: 10.1111/j.1525-142X.2008.00267.x. [DOI] [PubMed] [Google Scholar]
- Springer MS, de Jong WW. Which mammalian supertree to bark up? Science. 2001;291:1709. doi: 10.1126/science.1059434. [DOI] [PubMed] [Google Scholar]
- Wilson LAB, Schradin C, Mitgutsch C, Galliari FC, Mess A, Sánchez-Villagra MR. Skeletogenesis and sequence heterochrony in rodent evolution, with particular emphasis on the African striped mouse, Rhabdomys pumilio (Mammalia) Org Divers Evol. 2010;10:243–258. doi: 10.1007/s13127-010-0020-4. [DOI] [Google Scholar]
- Goswami A. Cranial modularity and sequence heterochrony in mammals. Evol Dev. 2007;9:290–298. doi: 10.1111/j.1525-142X.2007.00161.x. [DOI] [PubMed] [Google Scholar]
- Goswami A, Weisbecker V, Sánchez-Villagra MR. Developmental modularity and the marsupial-placental dichotomy. J Exp Zool B Mol Dev Evol. 2009;312:186–195. doi: 10.1002/jez.b.21283. [DOI] [PubMed] [Google Scholar]
- Schoch RR. Skull ontogeny: developmental patterns of fishes conserved across major tetrapod clades. Evol Dev. 2006;8:524–536. doi: 10.1111/j.1525-142X.2006.00125.x. [DOI] [PubMed] [Google Scholar]
- Shubin NH, Davis MC. In: Modularity in Development and Evolution. Schlosser G, Wagner GP, editor. Chicago, IL: University of Chicago Press; 2004. Modularity in the evolution of vertebrate appendages; pp. 429–440. [Google Scholar]
- Harrison LB, Larsson HCE. Estimating evolution of temporal sequence changes: a practical approach to inferring ancestral developmental sequences and sequence heterochrony. Syst Biol. 2008;57:378–387. doi: 10.1080/10635150802164421. [DOI] [PubMed] [Google Scholar]
- Blomquist GE. Methods of sequence heterochrony for describing modular developmental changes in human evolution. Am J Phys Anthropol. 2008;138:231–238. doi: 10.1002/ajpa.20963. [DOI] [PubMed] [Google Scholar]
- Olson ME, Rosell JA. Using heterochrony to detect modularity in the evolution of stem diversity in the plant family Moringaceae. Evolution. 2006;60:724–734. [PubMed] [Google Scholar]
- Schmidt K, Starck JM. Developmental variability during early embryonic development of zebra fish, Danio rerio. J Exp Zool B Mol Dev Evol. 2004;302:446–457. doi: 10.1002/jez.b.21010. [DOI] [PubMed] [Google Scholar]
- Hendrikse JL, Parsons TE, Hallgrímsson B. Evolvability as the proper focus of evolutionary developmental biology. Evol Dev. 2007;9:393–401. doi: 10.1111/j.1525-142X.2007.00176.x. [DOI] [PubMed] [Google Scholar]
- Müller GB. Evo-devo: extending the evolutionary synthesis. Nat Rev Genet. 2007;8:943–949. doi: 10.1038/nrg2219. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Pavlicev M, Cheverud JM. The road to modularity. Nat Rev Genet. 2007;8:921–931. doi: 10.1038/nrg2267. [DOI] [PubMed] [Google Scholar]
- Hallgrímsson B, Lieberman DE. Mouse models and the evolutionary developmental biology of the skull. Integr Comp Biol. 2008;48:373. doi: 10.1093/icb/icn076. [DOI] [PubMed] [Google Scholar]
- Pigliucci M. Is evolvability evolvable? Nat Rev Genet. 2008;9:75–82. doi: 10.1038/nrg2278. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. In: Variation: A Central Concept in Biology. Hallgrímsson B, Hall BK, editor. Amsterdam, Netherlands: Elsevier; 2005. Developmental constraints, modules and evolvability; pp. 219–247. [Google Scholar]
- Wagner GP. Homologues, natural kinds and the evolution of modularity. Am Zool. 1996;36:36–43. [Google Scholar]
- Badyaev AV, Foresman KR, Young RL. Evolution of morphological integration: developmental accommodation of stress-induced variation. Am Nat. 2005;166:382–395. doi: 10.1086/432559. [DOI] [PubMed] [Google Scholar]
- Cheverud JM. Phenotypic, genetic, and environmental morphological integration in the cranium. Evolution. 1982;36:499–516. doi: 10.2307/2408096. [DOI] [PubMed] [Google Scholar]
- Cheverud JM. Developmental integration and the evolution of pleiotropy. Am Zool. 1996;36:44–50. [Google Scholar]
- Cheverud JM, Ehrich TH, Vaughn TT, Koreishi SF, Linsey RB, Pletscher LS. Pleiotropic effects on mandibular morphology II: differential epistasis and genetic variation in morphological integration. J Exp Zool B Mol Dev Evol. 2004;302:424–435. doi: 10.1002/jez.b.21008. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Rutledge JJ, Atchley WR. Quantitative genetics of development: genetic correlations among age-specific trait values and the evolution of ontogeny. Evolution. 1983;37:895–905. doi: 10.2307/2408405. [DOI] [PubMed] [Google Scholar]
- Zelditch ML. Ontogenetic variation in patterns of developmental and functional integration in the laboratory rat. Evolution. 1988;42:28–41. doi: 10.2307/2409113. [DOI] [PubMed] [Google Scholar]
- Zelditch ML, Moscarella RA. In: Phenotypic Integration. Pigliucci M, Preston K, editor. Oxford, United Kingdom: Oxford University Press; 2004. Form, function and life history: spatial and temporal dynamics of integration; pp. 274–301. [Google Scholar]
- Klingenberg CP, Leamy LJ, Cheverud JM. Integration and modularity of quantitative trait locus effects on geometric shape in the mandible. Genetics. 2003;166:1909–1921. doi: 10.1534/genetics.166.4.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP, Mebus K, Auffray JC. Developmental integration in a complex morphological structure: how distinct are the modules in the mouse mandible? Evol Dev. 2003;5:522–531. doi: 10.1046/j.1525-142X.2003.03057.x. [DOI] [PubMed] [Google Scholar]
- Marroig G, Shirai LT, Porto A, de Oliveira FB, De Conto V. The evolution of modularity in the mammalian skull II: evolutionary consequences. Evol Biol. 2009;36:136–148. doi: 10.1007/s11692-009-9051-1. [DOI] [Google Scholar]
- Porto A, de Oliveira FB, Shirai LT, De Conto V, Marroig G. The evolution of modularity in the mammalian skull I: morphological integration patterns and magnitudes. Evol Biol. 2009;36:118–135. doi: 10.1007/s11692-008-9038-3. [DOI] [Google Scholar]
- Kaji T, Tsukagoshi A. Heterochrony and modularity in the degeneration of maxillopodan nauplius eyes. Biol J Linn Soc. 2010;99:521–529. doi: 10.1111/j.1095-8312.2009.01382.x. [DOI] [Google Scholar]
- de Beer GR. The Development of the Vertebrate Skull. London: Oxford University Press; 1937. [Google Scholar]
- Gould SJ. Ontogeny and Phylogeny. Cambridge, MA: Harvard University Press; 1977. [Google Scholar]
- Raff EC, Raff RA. Dissociability, modularity, evolvability. Evol Dev. 2000;2:235–237. doi: 10.1046/j.1525-142x.2000.00069.x. [DOI] [PubMed] [Google Scholar]
- Poe S. A test for patterns of modularity in sequences of developmental events. Evolution. 2004;58:1852–1855. doi: 10.1111/j.0014-3820.2004.tb00468.x. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. In: Phenotypic Integration: Studying the Ecology and Evolution of Complex Phenotypes. Pigliucci M, Preston K, editor. Oxford, United Kingdom: Oxford University Press; 2004. Integration, modules and development: molecules to morphology to evolution; pp. 213–230. [Google Scholar]
- Asher RJ, Helgen KM. Nomenclature and placental mammal phylogeny. BMC Evol Biol. 2010;10:102. doi: 10.1186/1471-2148-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami A, Prochel J. Ontogenetic morphology and allometry of the cranium in the common european mole (Talpa europaea) J Mammal. 2007;88:667–677. doi: 10.1644/06-MAMM-A-315R.1. [DOI] [Google Scholar]
- Prochel J, Goswami A, David Carmona F, Jimenéz R. Ossification sequence in the mole Talpa occidentalis (Eulipotyphla, Talpidae) and comparison with other mammals. Mammal Biol. 2008;73:399–403. doi: 10.1016/j.mambio.2007.05.014. [DOI] [Google Scholar]
- Frick H. Die Entwicklung und Morphologie des Chondrokraniums von Myotis Kaup. Stuttgart, Germany: Georg Thieme Verlag; 1954. [Google Scholar]
- Strong RM. The order, time, and rate of ossification of the albino rat (Mus norvegicus albinus) skeleton. Am J Anat. 1925;36:313–355. doi: 10.1002/aja.1000360206. [DOI] [Google Scholar]
- Zeller U. In: Mammalia Depicta. Kuhn HX, Zeller U, editor. Hamburg/Berlin, Germany: Verlag Paul Parey; 1987. Morphogenesis of the mammalian skull with special reference to Tupaia; pp. 17–50. [Google Scholar]
- Sánchez-Villagra MR, Horovitz I, Motokawa M. A comprehensive morphological analysis of talpid moles (Mammalia) phylogenetic relationships. Cladistics. 2006;22:59–88. doi: 10.1111/j.1096-0031.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- Amrine-Madsen H, Koepfli KP, Wayne RK, Springer MS. A new phylogenetic marker, apolipoprotein B, provides compelling evidence for eutherian relationships. Mol Phylogenet Evol. 2003;28:225–240. doi: 10.1016/S1055-7903(03)00118-0. [DOI] [PubMed] [Google Scholar]
- Dubey S, Salamin N, Ohdachi SD, Barrière P, Vogel P. Molecular phylogenetics of shrews (Mammalia: Soricidae) reveal timing of transcontinental colonizations. Mol Phylogenet Evol. 2007;44:126–137. doi: 10.1016/j.ympev.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Ohdachi SD, Hasegawa M, Iwasa MA, Vogel P, Oshida T, Lin LK, Abe H. Molecular phylogenetics of soricid shrews (Mammalia) based on mitochondrial cytochrome b gene sequences: with special reference to the Soricinae. J Zool. 2006;270:177–191. [Google Scholar]
- Shinohara A, Campbell KL, Suzuki H. Molecular phylogenetic relationships of moles, shrew moles, and desmans from the new and old worlds. Mol Phylogenet Evol. 2003;27:247–258. doi: 10.1016/S1055-7903(02)00416-5. [DOI] [PubMed] [Google Scholar]
- Shinohara A. PhD thesis. Hokkaido University Graduate School of Environmental Earth Science; 2008. Molecular phylogeny and phylogeography of the family Talpidae (Eulipotyphla Mammalia) [Google Scholar]
- Weisbecker V, Goswami A, Wroe S, Sánchez-Villagra MR. Ossification heterochrony in the therian postcranial skeleton and the marsupial-placental dichotomy. Evolution. 2008;62:2027–2041. doi: 10.1111/j.1558-5646.2008.00424.x. [DOI] [PubMed] [Google Scholar]
- Hautier L, Weisbecker V, Sánchez-Villagra MR, Goswami A, Asher RJ. Skeletal development in sloths and the evolution of mammalian vertebral patterning. Proc Natl Acad Sci USA. 2010;107:18903–18908. doi: 10.1073/pnas.1010335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery JE, Bininda-Emonds ORP, Coates MI, Richardson MK. A new technique for identifying sequence heterochrony. Syst Biol. 2005;54:230. doi: 10.1080/10635150590923227. [DOI] [PubMed] [Google Scholar]
- Nunn CL, Smith KK. Statistical analyses of developmental sequences: the craniofacial region in marsupial and placental mammals. Am Nat. 1998;152:82–101. doi: 10.1086/286151. [DOI] [PubMed] [Google Scholar]
- Schulmeister S, Wheeler WC. Comparative and phylogenetic analysis of developmental sequences. Evol Dev. 2004;6:50–57. doi: 10.1111/j.1525-142X.2004.04005.x. [DOI] [PubMed] [Google Scholar]
- Jeffery JE, Bininda-Emonds ORP, Coates MI, Richardson MK. Analyzing evolutionary patterns in amniote embryonic development. Evol Dev. 2002;4:292–302. doi: 10.1046/j.1525-142X.2002.02018.x. [DOI] [PubMed] [Google Scholar]
- Hautier L, Weisbecker V, Goswami A, Knight F, Kardjilov N, Asher RJ. Skeletal ossification and sequence heterochrony in xenarthran evolution. Evol Dev. in press . [DOI] [PubMed]
- Bininda-Emonds ORP, Jeffrey JE, Richardson MK. Is sequence heterochrony an important evolutionary mechanism in mammals? J Mamm Evol. 2003;10:335–361. [Google Scholar]
- Swofford DL. PAUP: Phylogenetic Analysis Using Parsimony, version 4.0 b10. Sunderland. 2001.
- Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BK. Bones and Cartilage: Developmental and Evolutionary Skeletal Biology. New York: Academic Press; 2005. [Google Scholar]
- Cheverud JM. Morphological integration in the saddle-back tamarin (Saguinus fuscicollis) cranium. Am Nat. 1995;145:63–89. doi: 10.1086/285728. [DOI] [Google Scholar]
- Marroig G, Cheverud JM. A comparison of phenotypic variation and covariation patterns and the role of phylogeny ecology and ontogeny during cranial evolution of New World monkeys. Evolution. 2001;55:2576–2600. doi: 10.1111/j.0014-3820.2001.tb00770.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. doi: 10.1086/284325. [DOI] [Google Scholar]
- Hood G. PopTools. Canberra: Pest Animal Control CRC; 2000. [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electronica. 2001;4:9. [Google Scholar]
- Weisbecker V, Mitgutsch C. A large-scale survey of heterochrony in anuran cranial ossification patterns. J Zool Syst Evol Res. 2010;48:332–347. doi: 10.1111/j.1439-0469.2010.00570.x. [DOI] [Google Scholar]
- Smith KK. Comparative patterns of craniofacial development in eutherian and metatherian mammals. Evolution. 1997;51:1663–1678. doi: 10.2307/2411218. [DOI] [PubMed] [Google Scholar]
- Novacek MJ. In: The Skull: Patterns of Structural and Systematic Diversity. Hanken J, Hall BK, editor. Vol. 2. Chicago, IL: University of Chicago Press; 1993. Patterns of diversity in the mammalian skull; pp. 438–545. [Google Scholar]
- Hornaday WT. The American Natural History: A Foundation of Useful Knowledge of the Higher Animals of North America. New York, NY: Charles Scribner's Sons; 1904. [Google Scholar]
- Yalden DW. The anatomy of mole locomotion. J Zool. 1966;149:55–64. [Google Scholar]
- Catania KC. Epidermal sensory organs of moles, shrew moles, and desmans: a study of the family talpidae with comments on the function and evolution of Eimer's organ. Brain Behav Evol. 2000;56:146–174. doi: 10.1159/000047201. [DOI] [PubMed] [Google Scholar]
- Shibanai S. Ultrastructure of the Eimer's organs of the Japanese shrew mole, Urotrichus talpoides (Insectivora, Mammalia) and their changes following infraorbital axotomy. Anat Anz. 1988;165:105–129. [PubMed] [Google Scholar]
- Catania KC. A comparison of the Eimer's organs of three north american moles: the hairy-tailed mole (Parascalops breweri), the star-nosed mole (Condylura cristata), and the eastern mole (Scalopus aquaticus) J Comp Neurol. 1995;354:150–160. doi: 10.1002/cne.903540110. [DOI] [PubMed] [Google Scholar]
- Catania KC, Kaas JH. Somatosensory fovea in the star-nosed mole: behavioral use of the star in relation to innervation patterns and cortical representation. J Comp Neurol. 1997;387:215–233. doi: 10.1002/(SICI)1096-9861(19971020)387:2<215::AID-CNE4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Abe H. In: The Planet of Animals. Abe H, Imafuku M, editor. Vol. 39. Tokyo, Japan: Asahi Shinbun; 1992. Insectivores and xenathrans; pp. 66–96. [Google Scholar]
- Abe H. Illustrated Skulls of Japanese Mammals. Sapporo: Hokkaido University Press; 2000. [Google Scholar]
- Ishii N. In: The Wild Mammals of Japan. Ohdachi SD, Ishibashi Y, Iwasa MA, Saitoh T, editor. Kyoto, Japan: Shoukadoh Kyoto; 2009. Urotrichus talpoides Temminck, 1841. [Google Scholar]
- Whidden HP. Comparative myology of moles and the phylogeny of the Talpidae (Mammalia, Lipotyphla) Am Mus Nov. 2000;3294:1–53. [Google Scholar]
- Mitgutsch C, Richardson MK, Jiménez R, Martin JE, Kondrashov P, de Bakker MAG, Sánchez-Villagra MR. Circumventing the polydactyly 'constraint': the mole's 'thumb'. Biol Lett. in press . [DOI] [PMC free article] [PubMed]
- Edwards LF. Morphology of the forelimb of the mole (Scalops aquaticus, L.) in relation to its fossorial habits. Ohio J Sci. 1937;37:20–41. [Google Scholar]
- Starck D. Lehrbuch der speziellen Zoologie. Wirbeltiere. Teil 5: Säugetiere. Jena, Germany: Gustav Fischer; 1995. [Google Scholar]
- Catania KC. Olfaction: underwater 'sniffing' by semi-aquatic mammals. Nature. 2006;444:1024–1025. doi: 10.1038/4441024a. [DOI] [PubMed] [Google Scholar]
- McIntyre IW, Campbell KL, MacArthur RA. Body oxygen stores, aerobic dive limits and diving behaviour of the star-nosed mole (Condylura cristata) and comparisons with non-aquatic talpids. J Exp Biol. 2002;205:45–54. doi: 10.1242/jeb.205.1.45. [DOI] [PubMed] [Google Scholar]
- Abe H. In: The Wild Mammals of Japan. Ohdachi SD, Ishibashi Y, Iwasa MA, Saitoh T, editor. Kyoto, Japan: Shoukadoh; 2009. Chimarrogale platycephala (Temminck, 1842) pp. 16–17. [Google Scholar]
- Douady CJ, Douzery EJP. Molecular estimation of eulipotyphlan divergence times and the evolution of. Mol Phylogenet Evol. 2003;28:285–296. doi: 10.1016/S1055-7903(03)00119-2. [DOI] [PubMed] [Google Scholar]
- Douady CJ, Douzery EJP. In: The Timetree of Life. Hedges SB, Kumar S, editor. Vol. 157. New York, NY: Oxford University Press; 2009. Hedgehogs, shrews, moles, and solenodons (Eulipotyphla) pp. 1011–1018. [Google Scholar]
- MacPhee RDE, Novacek MJ. In: Mammal Phylogeny: Placentals. Szalay FS, Novacek MJ, McKenna MC, editor. New York: Springer-Verlag; 1993. Definition and relationships of Lipotyphla; pp. 13–31. [Google Scholar]
- Sánchez-Villagra MR. Comparative patterns of postcranial ontogeny in therian mammals: an analysis of relative timing of ossification events. J Exp Zool B Mol Dev Evol. 2002;294:264–273. doi: 10.1002/jez.10147. [DOI] [PubMed] [Google Scholar]
- Davies M. Ontogeny of bone and the role of heterochrony in the myobatrachine genera Uperoleia, Crinia, and Pseudophryne (Anura: Leptodactylidae: Myobatrachinae) J Morphol. 1989;200:269–300. doi: 10.1002/jmor.1052000305. [DOI] [PubMed] [Google Scholar]
- Mabee PM, Olmstead KL, Cubbage CC. An experimental study of intraspecific variation, developmental timing, and heterochrony in fishes. Evolution. 2000;54:2091–2106. doi: 10.1111/j.0014-3820.2000.tb01252.x. [DOI] [PubMed] [Google Scholar]
- Rieppel O. Studies on skeleton formation in reptiles, v. Patterns of ossification in the skeleton of Alligator mississippiensis Daudin (Reptilia, Crocodylia) Zool J Linn Soc. 1993;109:301–325. doi: 10.1111/j.1096-3642.1993.tb02537.x. [DOI] [Google Scholar]
- Goswami A. Cranial modularity shifts during mammalian evolution. Am Nat. 2006;168:270–280. doi: 10.1086/505758. [DOI] [PubMed] [Google Scholar]
- Smith KK. Integration of craniofacial structures during development in mammals. Am Zool. 1996;36:70–79. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frequency distribution of specimens for each stage. Ossification events are skewed toward earlier stages of sequences, suggesting a greater concentration of ossification events and/or less resolution early in the sequence.
Heterochronies reported by the ACCTRAN and DELTRAN. All the heterochronic events detected by the ACCTRAN and DELTRAN are summarized.
Phenogram obtained from parsimony analysis of event-pair characters. The consensus tree is obtained from parsimony analysis of event-paring scores. Although all six mole species were clustered together, the phenogram was mostly incongruent with the commonly accepted phylogenetic relationships.