Abstract
Perception of temperature is an important brain function for organisms to survive. Evidence suggests that temperature preference behavior (TPB) in Drosophila melanogaster, one of poikilothermal animals, is regulated by cAMP-dependent protein kinase (PKA) signaling in mushroom bodies of the brain. However, downstream targets for the PKA signaling in this behavior have not been identified. From a genome-wide search for the genes regulated by PKA activity in the mushroom bodies, we identified the cyp6a17 Cytochrome P450 gene as a new target for PKA. Our detailed analysis of mutants by genetic, molecular and behavioral assays shows that cyp6a17 is essential for temperature preference behavior. cyp6a17 expression is enriched in the mushroom bodies of the adult brain. Tissue-specific knockdown and rescue experiments demonstrate that cyp6a17 is required in the mushroom bodies for normal temperature preference behavior. This is the first study, to our knowledge, to show PKA-dependent expression of a cytochrome P450 gene in the mushroom bodies and its role as a key factor for temperature preference behavior. Taken together, this study reveals a new PKA-Cytochrome P450 pathway that regulates the temperature preference behavior.
Introduction
Behavioral responses to environmental stimuli, including light, humidity, and temperature, are important for survival of all living organisms. Especially for poikilothermal animals, extreme changes in ambient temperature or persistence for a long time at high or low temperature lead to death. These animals adapt to their body temperature changes by using molecular mechanisms to alter metabolism or by behavioral strategies to choose proper temperature conditions. Drosophila has been widely used as a genetic model for studying a variety of behaviors including learning. Recently, it has also been utilized to study the genetic basis of temperature sensation and temperature preference behavior.
In Drosophila, larvae and adult flies show strong temperature preference behavior [1]–[3]. A family of transient receptor potential (TRP) ion channels plays major roles in the sensation of temperature. For example, Painless, one of the TRP channel (TRPA1) superfamily, is required for sensing nociceptive stimuli over 38°C [4]. Another TRP channel, Pyrexia, is involved in protecting flies against noxious temperature over 40°C [5]. In contrast, Drosophila ANKTM1 TRP family channel participates in temperature selection by opening at warm temperature (24–29°C) [6], [7].
Despite extensive studies on the role of Drosophila TRP family channels in temperature sensing, it is not well understood how flies perform specific behavior to choose optimal temperature conditions. Interestingly, recent studies have shown that the mushroom bodies in the brain, which plays a critical role in learning and memory, is important for TPB [8]. Furthermore, cAMP-dependent PKA signaling in the mushroom bodies is not only essential for learning and memory but also for TPB. These studies have provided important clues to the mechanism underlying TPB, but the target genes for PKA signaling have been elusive. Hence, we carried out a genome-wide screen for the genes regulated by PKA to obtain insights into the molecular events underlying TPB. From this screen, we found cyp6a17, a cytochrome P450 superfamily gene, as a PKA downstream factor for TPB.
The cytochrome P450 (CYP) family is a diverse group of enzymes. Most CYP proteins are involved in the oxidation of a variety of organic substrates including natural products and detoxification of foreign compounds [9]–[14]. In Drosophila, there exist about 90 CYPs. Some of these CYP genes are involved in ecdysone hormones synthesis [15]–[20], male aggressive behavior [21], [22] and male mating [23]. However, no CYP genes have been implicated in specific brain functions like temperature sensing behavior.
Here, we show that cyp6a17 is regulated by PKA and is required for temperature preference behavior. We demonstrate that cyp6a17 expression in the mushroom bodies is necessary and sufficient for TPB. This study identifies cyp6a17 as an important target of PKA signaling for mediating TPB in the mushroom bodies.
Results
Identification of new genes regulated by PKA in the mushroom bodies
Temperature preference behavior in Drosophila depends on the level of PKA signaling in the mushroom bodies. To identify new components downstream to PKA, we carried out a genome-wide screen for genes regulated by PKA signaling in the mushroom bodies. Using the Gal4-UAS system, we increased or decreased PKA activity in the mushroom bodies by expressing dominant-negative (UAS-PKADN) or constitutively active PKA (UAS-PKACA), respectively. Expression of PKA transgenes was targeted to the mushroom bodies using the mushroom body-specific MB247-Gal4 driver [8]. PKA expression was induced for 12–16 hours in three-day-old adults by inactivating the temperature-sensitive Gal80 [24] at the restrictive temperature. We then analyzed gene-expression profiles to identify the genes showing altered expression levels in response to the high or low PKA activity. The Drosophila GeneChip (DrosGenome 2.0) was used to obtain gene expression profiles from fly heads of three different groups: (i) the control group with no PKA transgene expression, (ii) the low PKA activity group and (iii) the high PKA activity group. Transcripts that showed more than 2-fold changes from the control expression level were considered for further analysis (Figure 1A & Table S1).
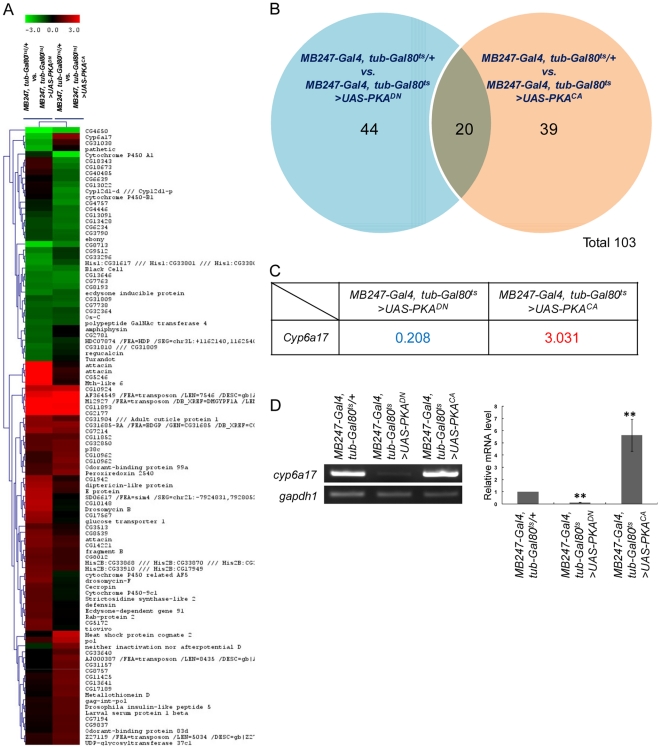
Figure 1. Genes differentially expressed by altered PKA activity in the mushroom bodies.
(A) CLUSTER image of 130 PKA-regulated genes. Each column represents changes in the transcript level of candidate genes. Red indicates up-regulation by PKACA, and green represents down-regulation by PKADN. Identities of all PKA-regulated genes are provided on the right side of the CLUSTER image, placed in the same order as their relative position in the CLUSTER image. (B) Van diagram for 103 candidate genes. Blue area: 44 transcripts responded to PKADN but not PKACA. Red area: 39 transcripts responded to PKACA but not PKADN. Blue and red overlap area: 20 transcripts showed changes by both PKADN and PKACA. (C) Transcript profiles of cyp6a17 by PKADN or PKACA. (D) RT-PCR confirmed the results of genome-scale microarray experiment. The relative mRNA transcript level was measured (histograms) by real-time qPCR analysis. gapdh1 was used as internal control. The number of tests: N = 4. Two asterisks, P<0.001.
A total of 103 transcripts showing more than 2-fold changes were classified into three categories based on their responses to PKA activity: (i) 44 transcripts responded to PKADN but not by PKACA. Among these, 14 showed downregulation while 30 resulted in upregulation. (ii) 39 transcripts showed altered expression by PKACA (23 increases and 16 decreases). (iii) 20 transcripts showed changes by both PKADN and PKACA conditions. Among them, 8 showed decreased and 11 showed increased transcript levels by both PKADN and PKACA conditions (Figure 1B).
Only one gene, cyp6a17, showed upregulation by PKACA and downregulation by PKADN. We were most interested in this gene since its expression was correlated with both higher and lower PKA activities (Figure 1C). To confirm these microarray data, we examined the PKA-dependent expression of cyp6a17 by RT-PCR and real-time-PCR. The expression level of cyp6a17 mRNA in the head was decreased to 12% of the wild-type level by inhibiting PKA activity with mushroom body-specific expression of PKADN. On the contrary, the transcript level was increased 5.63±1.32 times by PKACA overexpression (Figure 1D). Hence, cyp6a17 expression in the head is regulated by PKA in an activity-dependent manner.
cyp6a17 mutations affect temperature preference behavior
To test whether cyp6a17 is required for normal TPB, we analyzed the effects of cyp6a17 mutations on TPB. We generated a deletion allele (cyp6a17Δ173) by inducing imprecise excision of the P-element P{SUPor-P}KG04448 inserted at 133 bp downstream from the translation start site (Figure 2A). The cyp6a17Δ173 mutation is a deletion of 1.1 kb sequence from the P-insertion site and affects only the cyp6a17 gene (Figure 2A). Consistent with the deletion mapping, the transcript for cyp6a17 was not detected in the cyp6a17Δ173 mutant by RT-PCR analysis, suggesting that cyp6a17Δ173 is a null mutation (Figure 2B). Homozygous cyp6a17Δ173 mutant flies are viable and fertile with no visible morphological defects, suggesting that cyp6a17 is not essential for normal development and fertility.
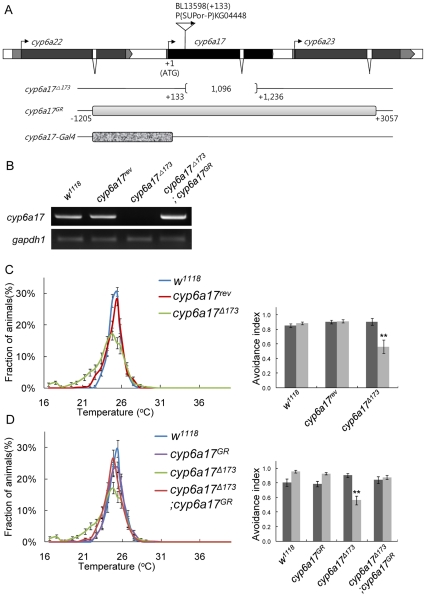
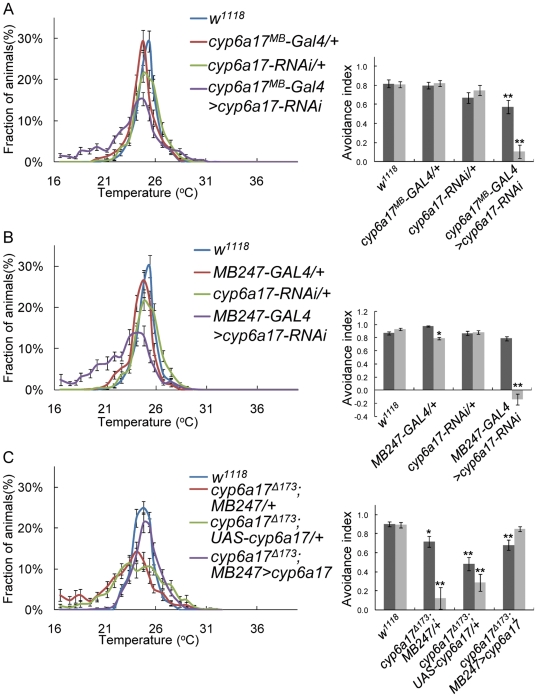
Figure 2. cyp6a17 is required for normal temperature preference behavior.
(A) Exons of cyp6a17 are indicated by boxes, and coding regions are colored black. The P{SUpor-P} line has an insertion at 133 bp downstream from the translation start site. cyp6a17Δ173 is a mutant with a deletion of 1.1 kb, as depicted by square brackets. A genomic DNA fragments used for rescue (cyp6a17GR) is indicated by gray box. The cyp6a17-Gal4 construct contains a genomic fragment of 1.2 kb (short thick stippled box). (B) Determination of the expression level of cyp6a17 by RT-PCR. gapdh1 was used as internal control: N = 4. (C) cyp6a17Δ173 showed reduced TPB. (D) The abnormal TPB of cyp6a17Δ173 was restored by expression of the cyp6a17 genomic transgene (cyp6a17GR). Black bars, AIHigh; gray bars, AILow. One asterisk, P<0.005 and two asterisks, P<0.001.
Next, we characterized the temperature preference in this deletion mutant. For TPB assays, all mutant and transgenic lines were outcrossed to the w1118 line at least four times. cyp6a17Δ173 mutant flies showed reduced AI (avoidance index) for low temperature (AILow = 0.56±0.06, p<0.001) (Figure 2C and Table S2). Immunocytochemcial analysis with anti-FasII antibody marker for the mushroom bodies did not reveal significant morphological defects in the brain of this mutant (Figure S1). Thus, the abnormal TPB shown in cyp6a17 loss-of-function mutant is probably not caused by morphological defects in the mushroom bodies.
Specificity of the cyp6a17 gene function in TPB was also confirmed by rescue of the cyp16a17 mutant phenotypes. Abnormal TPB phenotypes of cyp6a17 null mutant flies were rescued to the wild-type level by a 4.2-kb genomic DNA fragment (cyp6a17GR in Figure 2A) spanning the cyp6a17 locus (AIHigh = 0.84±0.04; AILow = 0.88±0.01) (Figure 2A, B, & D and Table S2). These data indicate that cyp6a17 is required for normal TPB.
Expression of cyp6a17 is enriched in the mushroom bodies
To visualize the expression pattern of cyp6a17, we generated a cyp6a17MB-Gal4 transgenic reporter strain in which Gal4 is induced by a cyp6a17 genomic region containing the promoter. The translation start site for cyp6a17 is only 612 bp apart from the adjacent upstream gene cyp6a22. cyp6a17MB-Gal4 was created by cloning an 1,201 bp fragment upstream of the cyp6a17 translation start site into pCaSpeR vector. This fragment contains the entire 5′ region of cyp6a17 including a 3′ part of the adjacent cyp6a22 gene. Therefore, the 5′ region of cyp6a17 includes regulatory sequences necessary for the cyp6a17 function in TPB (Figure 2A).
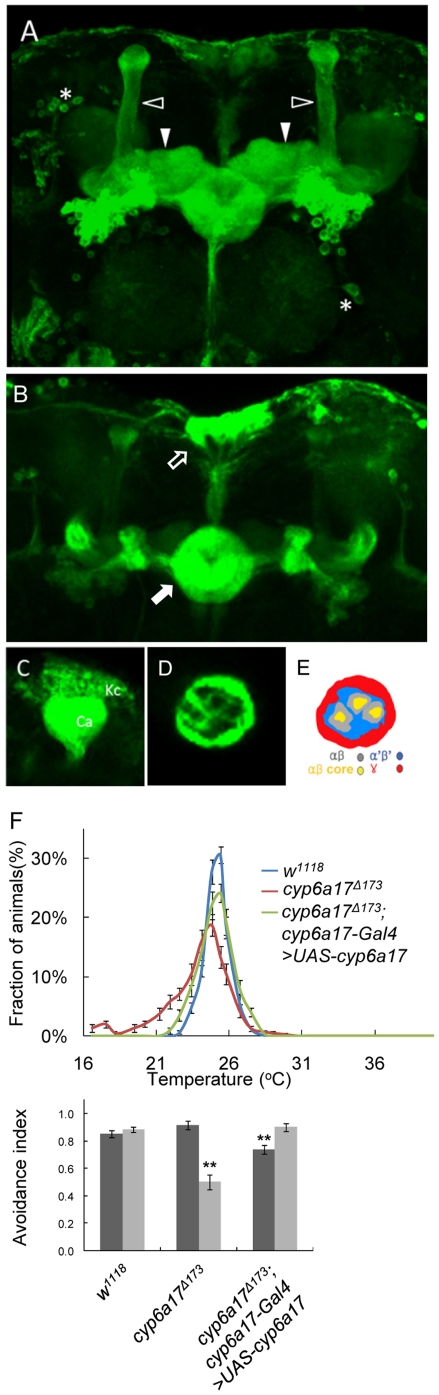
To detect cyp6a17 expression, we crossed cyp6a17MB-Gal4 transgenic flies to a reporter line UAS-mCD8.GFP. Confocal analysis of GFP expression in the adult brain revealed a complex pattern of expression in the mushroom bodies (Figure 3A). GAL4 is expressed in the mushroom body neurons that project to α-, β- and γ-lobes (Figure 3D & E), although it is not clear whether the cyp6a17MB-Gal4 is expressed in all or a subset of mushroom body neurons. In addition to the mushroom bodies, GFP was also expressed in the ellipsoid bodies (EB), pars intercerebralis (PI) and several unknown lateral neurons (Figure 3B). Abnormal TPB phenotypes in the cyp6a17Δ173 mutant was rescued by expressing wild-type cyp6a17 using the cyp6a17MB-Gal4 driver (AIHigh = 0.74±0.03, p<0.001; AILow = 0.90±0.03) (Figure 3F and Table S2). These data suggest that the cyp6a17MB-Gal4 reporter represents the pattern of endogenous cyp6a17 expression.
Figure 3. cyp6a17 is expressed in the mushroom bodies.
(A–E) cyp6a17 reporter expression by cyp6a17MB-Gal4>UAS-mCD8:GFP (green). (A) cyp6a17 was strongly expressed in the αβ lobe (white and open triangle) and γ lobe (white triangle) of the mushroom bodies. (B) The GFP expression was also detected in the ellipsoid bodies (EB, white arrowhead) and pars intercerebralis (PI, white and open arrowhead). (C) cyp6a17MB-Gal4 derived GFP was expressed in the Kenyon cells (Kc) and Calyx (Ca) of the mushroom bodies. (D) A cross section of the peduncle shows GFP staining along the outer rim that may correspond to the γ lobe (red in Fig. 3E). It also shows weak circular staining in three ring structures that appear to be αβ lobes. Note that there is no staining inside the rings that may correspond to the αβ core. (E) Schematics of Figure (D). Red may correspond to axon fibers for γ lobe; blue for α′β′; gray for αβ; yellow for αβ core based on their characteristic position in the peduncle. (F) Rescue of cyp6a17 mutant phenotypes by cyp6a17MB-Gal4>UAS-cyp6a17. Temperature preference profile and avoidance index (AI) of the mutant line expressing the cyp6a17 transgene (cyp6a17Δ173; cyp6a17MB-Gal4>UAS-cyp6a17) are shown. Black bars, AIHigh; gray bars, AILow. Two asterisks, P<0.001.
cyp6a17 is required in the mushroom bodies for temperature preference behavior
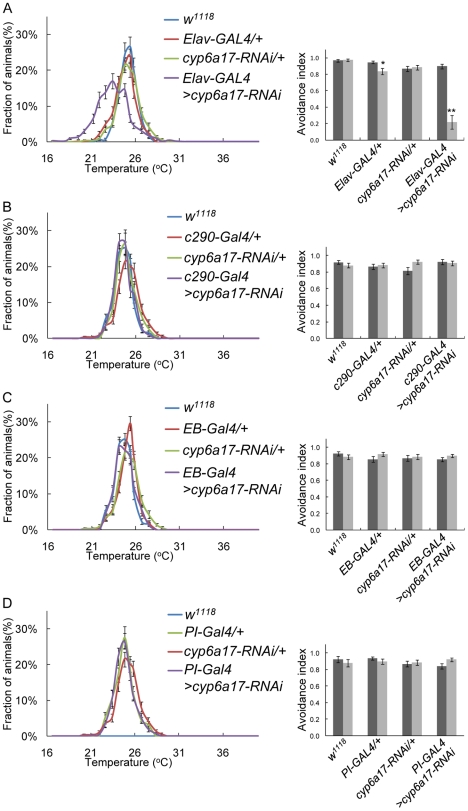
To test whether cyp6a17 is required for TPB in specific tissues, we examined the effects of reduced cyp6a17 expression by targeted knockdown using cyp6a17-specific UAS-RNAi transgenic flies. To minimize potential off-target effects on other Drosophila CYP genes, we selected a relatively small unconserved region (222 base pairs) of cyp6a17 as an RNAi target. Expression of cyp6a17-RNAi in the neuronal cells using a pan-neural Elav-Gal4 driver resulted in a significant reduction of TPB (AIHigh = 0.90±0.02; AILow = 0.22±0.14, p<0.001) (Figure 4A and Table S2). In contrast, cyp6a17 RNAi expression in the whole body except the head by using c290-Gal4 showed no effect on TPB (Figure 4B). These data suggest that cyp6a17 function for normal TPB is required in neurons of the brain.
Figure 4. cyp6a17 knockdown in the neurons results in abnormal temperature preference behavior.
(A–D) Effects of tissue-specific knockdown of cyp6a17 on TPB. cyp6a17 was knockdowned by RNAi in pan-neuronal cell by Elav-Gal4 (A), whole body except head c290-Gal4 (B), ellipsoid bodies by c232-Gal4 (EB-Gal4) (C) and pars intercerebralis by dilp2-Gal4 (PI-Gal4) (D). Temperature preference profile and avoidance index (AI) are shown. Black bars, AIHigh; gray bars, AILow. One asterisks, P<0.005 and two asterisks, P<0.001.
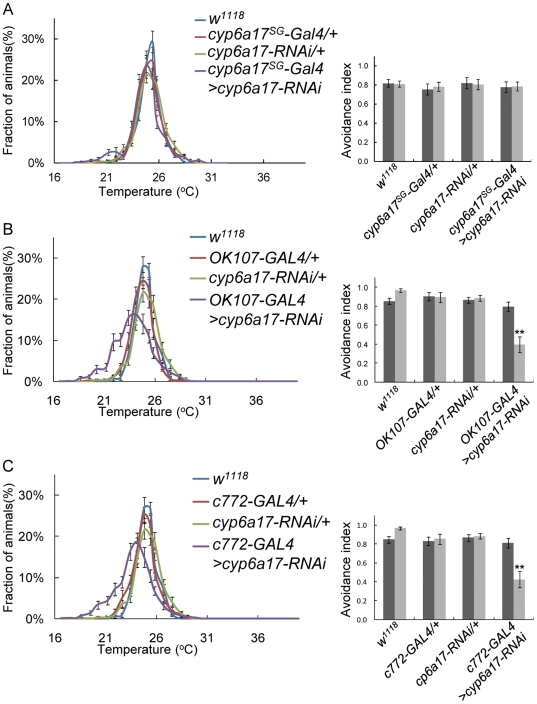
In the adult brain, cyp6a17MB-Gal4 expression is enriched in the mushroom bodies as well as EB and PI (Figure 3A). cyp6a17-RNAi knockdown in subsets of EB and PI neurons labeled by c232-Gal4 and dilp2-Gal4, showed little effect on TPB (Figure 4C & D and Table S2). This result is consistent with the previous report that EB and PI are not essential for TPB [8]. To exclude the possibility that cyp6a17 RNAi affects locomotor activity, we examined the motor activity of the flies by climbing behavior test. In these control assays, locomotor activity was normal at all temperature conditions tested.
Next, we induced cyp6a17-RNAi in the cyp6a17-expressing cells by using cyp6a17MB-Gal4. These flies showed a reduced AI index, as seen in cyp6a17 mutants (AIHigh = 0.57±0.07, p<0.001; AILow = 0.11±0.07, p<0.001) (Figure 5A and Table S2). Thus, cyp6a17 function is indeed required for TPB in the cells identified by the cyp6a17 reporter. To determine whether cyp6a17 is required in the mushroom bodies, we induced cyp6a17-RNAi preferentially in the mushroom bodies using MB247-Gal4. These flies with knockdown of cyp6a17 in the mushroom bodies showed a reduced avoidance index in the TPB assay (AIHigh = 0.89±0.03; AILow = 0.14±0.09, p<0.001) (Figure 5B and Table S2). In the heads of these flies, cyp6a17 mRNA expression level was reduced compared with wild-type (0.56±0.06, p<0.001) (Figure S2A). cyp6a17MB-Gal4 and MB247 show additional expression in surface glia [25]. To check whether cyp6a17 in surface glia is required for TPB, we induced cyp6a17-RNAi in surface glia by cyp6a17SG-Gal4 which drives strong Gal4 expression in surface glia but very weakly in the mushroom bodies (Figure S3). These flies showed normal TPB (Figure 6A and Table S2). In addition, cyp6a17 RNAi knockdown by other mushroom body Gal4 lines like OK107- and c772-Gal4 also resulted in slightly weaker but significant TPB defects: OK107-Gal4 (AIHigh = 0.80±0.02; AILow = 0.40±0.20, p<0.001) (Figure 6B and Table S2) and c772-Gal4 (AIHigh = 0.81±0.05; AILow = 0.43±0.13, p<0.001) (Figure 6C & Table S2). Furthermore, cyp6a17 mutant phenotype was rescued by expressing the wild-type cyp6a17 cDNA using the MB247-Gal4 mushroom body driver (Figure 5C and Table S2). Taken together, these data suggest that cyp6a17 expression in the mushroom bodies is necessary for normal low temperature TPB.
Figure 5. cyp6a17 is required in the mushroom bodies for temperature preference behavior.
(A–C) Effects of mushroom bodies-specific knockdown of cyp6a17 on TPB. cyp6a17 was knockdowned by RNAi in the cyp6a17-specific cells by cyp6a17MB-Gal4 (A) or in the mushroom bodies by MB247-Gal4 (B). (C) Rescue of cyp6a17 mutant phenotype. Expression of cyp6a17 transgene in the mushroom bodies by MB247-Gal4 restored the cyp6a17Δ173 phenotype. Black bars, AIHigh; gray bars, AILow. One asterisks, P<0.005 and two asterisks, P<0.001.
Figure 6. Effects of cyp6a17 knockdown in surface glia cells and the mushroom bodies.
(A) cyp6a17 was knockdowned by RNAi in surface glia by cyp6a17SG-Gal4. (B–C) cyp6a17 RNAi knockdown by two mushroom body-specific Gal4 lines, OK107-Gal4(B) and c772-Gal4 (C), causes significant TPB defects in the distribution profiles and AI. Black bars, AIHigh; grey bars, AILow. Two asterisks, P<0.001.
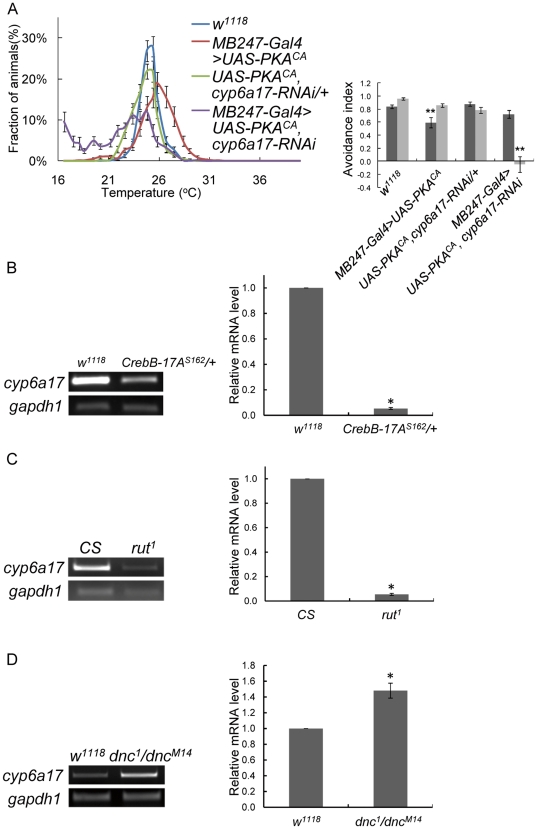
Previous studies proposed that the PKA signaling cascade is important for TPB [8]. According to our microarray data, cyp6a17 expression is regulated by PKA in an activity-dependent manner. Thus we tested epistatic relationship between PKA and cyp6a17. It is noteworthy that TPB phenotypes induced by PKACA overexpression were strongly suppressed by cyp6a17-RNAi (AIHigh = 0.72±0.06, p = 0.08; AILow = −0.05±0.12, p<0.001) (Figure 7A & Table S2). Furthermore, the transcript of cyp6a17 was significantly decreased to 0.044±0.026 (p<0.001, t test) in dCrebB-17As162 mutant (Figure 7B). In rut1, which is a loss-of-function mutant for rutabaga, encoding calcium-calmodulin responsive adenylate cyclase, cyp6a17 expression level was reduced compared with the wild-type level (0.054±0.008, p<0.001, t test) (Figure 7C). On the contrary, in the dnc1/dncM14 transheterozygote, which can increase cAMP level due to a defect in Dunce phosphodiesterase, cyp6a17 expression level was slightly increased (1.48±0.05, p<0.001, t test) (Figure 7D). These results suggest that cyp6a17 transcription is regulated by the cAMP-PKA signaling pathway and cyp6a17 functions downstream to PKA for TPB.
Figure 7. cyp6a17 expression is regulated by PKA signaling pathway.
(A) Temperature preference behavior of MB247>UAS-PKACA, cyp6a17-RNAi. Temperature preference profile and avoidance index (AI) are shown. Black bars, AIHigh; gray bars, AILow. Two asterisks, P<0.001. (B–D) Determination of expression level of cyp6a17 in the PKA signaling pathway mutant background. CrebB-17AS162 (B); rut1 (C); and dncM14/dnc 1(D). Relative transcript level was measured by real-time PCR (histogram). gapdh1 was used as internal control. The number of tests: N = 3∼4. Asterisks, P<0.001 by t test. Error bars indicates S.E.M.
Discussion
Temperature preference is an important behavioral strategy for optimizing the body temperature in poikilothermal animals like Drosophila. TPB involves temperature sensing and processing of the sensory input at higher levels in the brain. Recent studies have implicated that PKA signaling in the mushroom bodies is required for TPB. However, it is largely unknown how PKA signaling pathway contributes to the behavioral response. In this study, we identified cyp6a17 as a new gene regulated by PKA signaling. Our analysis provides several pieces of evidence that cyp6a17 is required in the mushroom bodies for TPB. First, mutations in the cyp6a17 gene showed reduced TPB. Second, abnormal TPB phenotypes in cyp6a17 mutants were rescued with a wild-type cyp6a17 transgene. Third, tissue-specific RNAi knockdown of cyp6a17 expression in the mushroom bodies resulted in TPB defects.
TPB assays are measured by two indices, AILow and AIHigh, for avoiding low and high temperatures, respectively. It is interesting to note that loss of cyp6a17 results in a preferential reduction of the AILow value compared with the AIHigh score. This asymmetric effect of cyp6a17 mutations suggests that cyp6a17 might be selectively required for avoidance of low temperatures. We noted that the transgenic expression by MB247>cyp6a17 from the cyp6a17 mutant significantly lowers the AIHigh index. However, one of the control strains for this test, cyp6a17Δ173; UAS-cyp6a17/+, shows a very low AI-High value in the absence of any Gal4 driver. Thus, the reduction of AIHigh in this strain is unlikely to be due to cyp6a17 overexpression. cyp6a17 mutant and this transgene were outcrossed at least four times, but the effect of UAS-cyp6a17/+ in reducing the AIHigh still remained. It is unknown why this strain shows non-specific low AIHigh. Importantly, however, transgenic overexpression of Cyp6a17 in the presence of Gal4 restores the low AILow defect of the cyp6a17Δ173 mutant (Fig. 5C). Previous studies have shown that increased levels of cAMP or PKA signaling cause stronger avoidance from low temperature whereas lower cAMP-PKA signaling levels result in the opposite trend with reduced avoidance. This is consistent with our data that cyp6a17 mutants show lower AILow. cyp6a17 is necessary for TPB but may not be sufficient to induce AIHigh. Since cyp6a17 is the only one of multiple downstream factors for PKACA, it may need additional PKA downstream factors to induce AIHigh.
It has been reported that many cytochrome P450 proteins are expressed in the midgut, malpighian tubules and fat body whereas only one gene cyp4g15 is detected in the brain of developing larvae [26]. We found that cyp6a17 mRNA is expressed in the adult body including hindgut, but about 30% more expression is detected in the head (Figure S2B). Using a reporter construct regulated by a 5′ region of the cyp6a17 gene, we showed that cyp6a17 expression is highly enriched in the mushroom bodies, especially αβ and γ lobes. Three observations strongly support that this reporter reflects the pattern of cyp6a17 gene expression. Firstly, cyp6a17 RNAi driven by mushroom body-specific Gal4 lines causes TPB defects. Secondly, cyp6a17 RNAi using cyp6a17MB-Gal4 also results in similar TPB phenotypes. Thirdly, cyp6a17 expression induced by cyp6a17MB-Gal4 rescues cyp6a17 null mutant phenotypes. These data indicate that the mushroom body regions labeled by cyp6a17 reporter are the sites where cyp6a17 function is required for TPB. To our knowledge, cyp6a17 is the first gene of this family shown to be expressed in the mushroom bodies with specific function in TPB. However, it is possible that other cell types in addition to the mushroom bodies might also be involved in the TPB phenotype.
Cytochrome P450 proteins are known to participate in the synthesis of steroid hormone, thereby regulating homeostasis and thermal responses [27], [28]. Because cyp6a17 is also expressed in PI and EB regions that have been implicated in neuroendocrine regulation [29], cyp6a17 may play a role in the neuroendocrine system. However, it has been shown that the PI and EB are not required for TPB [8]. Furthermore, our data show that the cyp6a17 mutant deficits in TPB are rescued by cyp6a17 expression in the mushroom bodies but not in subsets of PI and EB neurons. Thus, it appears that the cyp6a17 function for TPB is required mainly in the mushroom bodies rather than the PI and EB cells.
There are 83 CYP family genes in Drosophila, excluding 7 pseudogenes. Of these, only several CYP genes have been characterized for their expression patterns and functions. Some CYP proteins function together in a protein complex system for catalyzing oxidative reactions [30]. However, many CYP genes show tissue-specific expression profiles, suggesting that CYP genes may play distinct functions in diverse tissues. This possibility is supported by our finding that cyp6a17 has a specific function in the mushroom bodies as a necessary component for controlling the temperature preference behavior. In addition to cyp6a17, other CYP genes might also play important roles in TPB.
cAMP-PKA signaling in the mushroom bodies is important for learning and memory in Drosophila [31], but downstream events are not well understood. Our study identified a cytochrome P450 gene as a downstream factor for PKA signaling in TPB. Given the potential relationship between temperature and stress related genes [32]–[34], this work could provide clues to understanding the relationship between stress response and temperature preference behavior. Although we focused on characterizing the function of cyp6a17 in this study, our genome-wide screen identified many other genes that showed strong response to either PKACA or PKADN (Table S1). Further analysis of these genes might identify additional genetic factors involved in PKA-dependent TPB and their relationship with cyp6a17.
Until now, there is no report to our knowledge that Drosophila CYP is involved in temperature preference behavior. It is an intriguing question how PKA-dependent regulation of cyp6a17 is functionally related to TPB. Since steroid hormone can modulate several kinds of behavior, neuronal homeostasis could be an important factor for TPB. Consistent with this idea, it has been shown that Drosophila dystroglycan plays a role in modulating energy homeostasis and the thermal responses [35]. Cyp6a17 is known to have monooxygenase activity [13]. Hence, it is plausible that cyp6a17 might be involved in the synthesis of steroid hormones, thereby modulating temperature preference behavior. cyp6a17 may also participate in the regulation of ATP level to modulate energy homeostasis in neurons. Detailed analysis of these possibilities in the future will help provide mechanistic insights into the function of cyp6a17 for TPB.
Materials and Methods
Drosophila strains and transgenic lines
Fly stocks were raised on standard cornmeal food at 25°C and 40–50% relative humidity. w1118 and Canton-S were used as wild-type strains. UAS-PKADN and PKACA were provided by D. Kalderon; other lines were provided as follows: MB247, c772 (T. Zars), c309 (L. C. Griffith), MB247; tub-GAL80ts (G. Roman), P{SUpor-P}KG04448, c232, c290 and dilp2-Gal4 (Bloomington stock center). All mutant and transgenic alleles used in this study were outcrossed to the w1118 line at least four times.
Microarray analysis
Total RNA was extracted from adult heads. After extracting total RNA, microarrays were generated by Digital Genomics Inc. (Seoul, Korea). Modified cDNA from total RNA was hybridized to GeneChip® Drosophila Genome 2.0 Array. Image acquisition and data extraction from microarray experiments were analyzed by Affymetrix GeneChip® Scanner 3000 7G and Affymetrix GCOS1.4 software, respectively. Data were normalized by MAS5 and RMA (Robust Multi-array Average), and differentially expressed genes were selected by comparing control and transgenic lines. The functional category of selected genes was classified using the DAVID2007 (http://david.abcc.ncifcrf.gov/home.jsp) program.
Temperature preference behavior assay and climbing assay
TPB assays were performed as described previously [8], [36]. 1–3 day old adult files of mixed gender were collected without anesthetization in vials containing fresh food during morning hours. After 3 days under a 12 hr light/dark incubation cycle, the collected flies were placed in the test device. A temperature gradient from 15 to 45°C with a slope of 0.75°C/cm was produced in an aluminum block (42 length×24 width×7 cm heights). Electronic thermal sensors were embedded in the block every 3 cm, and the gradient was established using cold and hot circulating water chambers at each end. A glass plate with five separate lanes was placed 2.5 mm above the block, creating suitable space for flies to walk. The glass plate was coated with quinine sulfate powder, an aversive stimulus for Drosophila, to prevent flies from escaping the temperature gradient by resting on the walls or roof of the lane. 1% yeast paste was applied on the block for flies to stay on the block. Adult flies were placed between the aluminum block and the glass plate, allowed to move for 25 min in the dark, and photographed with a digital camera with 2–5 msec exposure at the end of each assay. Avoidance indices against low temperature (AILow) and high temperature (AIHigh) were calculated from the following formulae: AILow = NInt−NLow/NInt+NLow and AIHigh = NInt−NHigh/NInt+NHigh. NLow, NHigh and NInt are the numbers of flies in the region below 23.4°C, above 26.7°C, and between 23.4 and 26.7°C, respectively. A population of mixed sexes was tested for TPB. The number of flies was recorded by using digital photographs and processed with Microsoft Excel, and one-way ANOVA followed by Dunnett's post test was performed. All data points in the figures are represented as the mean and the standard error of the mean (s.e.m.), unless mentioned otherwise.
For climbing test to check the locomotor activity, groups of ten flies were transferred into vials and incubated for 1 hr at 25°C for environmental acclimatization. Then flies were transferred to 18°C and 32°C incubator for 15 min. After tapping the flies down to the bottom, we counted the number of flies that climb more than 10 cm from the bottom in 1 min. Three trials were performed for each genotype and repeated with 3 different batches. All mutant strains used in our TPB assays showed similar climbing rates as the wild-type. A horizontal locomotor activity test was done as described previously [37]. It showed that cyp6a17Δ173 mutant flies are normal in the horizontal locomotor activity.
It has been reported that the mushroom bodies are involved in suppressing hyperactivity of flies [38]. We found that test flies of the same genotype placed at high and low temperatures show different locomotor activity at the beginning of the trial but results in the same distribution profiles at the end of the assay. This indicates that the difference in the initial locomotor activity does not affect the temperature preference behavior in the standard assay conditions that allow time for test flies to choose and settle at their preferred temperature.
Construction of transgenic flies
cyp6a17-RNAi construct was generated as described [39]. cyp6a17-RNAi flies contain two copies of a 222 bp fragment of cyp6a17 cDNA. PCR primers used to create the target region for cyp6a17 knockdown were 5′- TGCTCTAGACAGCTTGTACGATCCAAA-3′ and 5′- TGCTCTAGAAATCATTTCGCTTTTCCT-3′. A PCR fragment was cloned into pWIZ vector. To construct UAS-cyp6a17, 1.5 kb fragment of a cyp6a17 cDNA obtained from the Drosophila Genome Research Center was cloned into pUAST. A cyp6a17-Gal4 construct was generated using pCaSper vector and 1,201 bp sequence upstream from the transcription start site of the cyp6a17 gene. cyp6a17SG-Gal4 was inserted at 78A1. Unlike cyp6a17MB, Gal4 expression in the mushroom bodies was strongly repressed, thus showing a preferential expression in the surface glia. To make a cyp6a17 genomic rescue construct, a 4.2 kb SacI-digested genomic fragment containing about 1.2 kb upstream from the transcription start site and about 1.5 kb downstream from the transcription stop site, was cloned into pCaSper vector. Transgenic lines were generated in the w1118 background using standard germline transformation techniques.
Relative quantitative RT-PCR
4–6 day old adult flies were collected and frozen immediately in liquid nitrogen. Heads were removed from bodies by vortex-mixing and sorting through a chilled sieve. Total RNA was extracted from heads with a Trizol (Invitrogen) and reverse transcribed using the AMV reverse transcriptase system (Promega). RT-PCR was performed with the following primers: for cyp6a17, 5′-GTTGTTACTGGCGCTAATCGT-3′ and 5′-TTCCATCACAACCTGCTCCA-3′; for cyp6a22, 5′-TTCGGGAAACTGTGAAGCAG-3′ and 5′-GATAAAAACATTTGATTGTGT-3′; for cyp6a23, 5′-TCGCTGTTACTAACGTTGAT-3′ and 5′-TTTCCATGACGACCTGCTCCA-3′; for gapdh1, 5′-ACCGTCGACGGTCCCTCT-3′ and 5′-GTGTAGCCCAGGATTCCCT-3′. Real-time PCR was performed with iCycler iQ (Bio-Rad) using the KAPA SYBR FAST Bio-Rad iCycler qPCR kit (Kapa Biosystems). The level of gapdh1 mRNA was measured as an internal control for the RNA amount in each sample. Data processing was performed with Microsoft Excel. All quantitative analysis were done more than three times.
Immunohistochemistry
Adult brains expressing transgenic cyp6a17-Gal4>UAS-mCD8.GFP were removed from head capsules and fixed in 4% paraformaldehyde in PBS for 30 min, and rinsed in PBS containing 0.5% Triton X-100. For staining with anti-FasII (Hybridoma Bank), adult brains were fixed for 30 min in 4% paraformaldehyde in PBS containing 0.5% Triton X-100. Fixed samples were washed for 10 min three times and incubated with 5% normal goat serum. Antibody dilution was 1∶50 for mouse anti-FasII and 1∶250 for goat anti-mouse Alexa Fluor 568 (Molecular Probe). Confocal analysis was performed on a Zeiss LSM710 microscope. Confocal stacks were processed with ZEN 2009 Light Edition and Adobe Photoshop.
Supporting Information
Effects on cyp6a17 mutation on mushroom body morphology. Mushroom body morphology in cyp6a17Δ173 mutant. Mushroom body axons are visualized using FASII antisera. Compared with wild type (A), no significant defects were found in mutant (B). Doral is up.
(PDF)
Effects of cyp6a17 RNAi and tissue distribution of cyp6a17 expression. (A) Reduction of the expression level by MB247>cyp6a17-RNAi. gapdh1 was used as internal control. The number of tests: N = 4. Two asterisks, P<0.001. (B) Transcriptional profiles of cyp6a17 in the head and body assayed by real-time PCR. The histogram shows that cyp6a17 mRNA is slightly more abundant in the head compared with the body. gapdh1 was used as internal control. The number of tests: N = 4. Two asterisks, P<0.001. All data are means, and the error bars indicate s.e.m.
(PDF)
The expression pattern of cyp6a17SG - Gal4 . cyp6a17 reporter expression by cyp6a17SG-Gal4>UAS-mCD8:GFP (green). GFP expression was detected strongly in surface glial cells (filled triangle) and weakly in Kenyon cells (open triangle).
(PDF)
Total list of the 103 probes.
(PDF)
Statistical analysis of the results shown in Figures.
(PDF)
Acknowledgments
We would like to thank S.-T. Hong for advice and comments, Y.K. Oh and J. Choe for help with locomotor test, and D. Kalderon, T. Zars, L.C. Griffith, G. Roman and the Bloomington stock center for providing fly stocks.
Footnotes
Competing Interests: Jaeseob Kim is employed by a commercial company (Aprogen Inc.). This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and material.
Funding: This research was supported by grants from the World Class University Program through the National Research Foundation of Korea funded by the Ministry of Education, Science & Technology, contract grant number R31-2008-000-10071-0. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sayeed O, Benzer S. Behavioral genetics of thermosensation and hygrosensation in Drosophila. Proc Natl Acad Sci USA. 1996;93:6079–6084. doi: 10.1073/pnas.93.12.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zars T. Two thermosensors in Drosophila have different behavioral functions. J Comp Phyiol A. 2001;187:235–242. doi: 10.1007/s003590100194. [DOI] [PubMed] [Google Scholar]
- 3.Liu L, Yeromlaieva O, Johnson WA, Abboud FM, Welsh MJ. Identification and function of thermosensory neurons in Drosophila larvae. Nat Neurosci. 2003;6:267–273. doi: 10.1038/nn1009. [DOI] [PubMed] [Google Scholar]
- 4.Tracey WD, Jr, Wilson RI, Laurent G, Benzer S. The painless, a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/s0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y, Lee Y, Lee J, Bang S, Hyun S, et al. Pyrexia is a new thermal transient receptor potential channel endowing tolerance to high temperatures in Drosophila melanogaster. Nature Genetics. 2005;37(3):305–310. doi: 10.1038/ng1513. [DOI] [PubMed] [Google Scholar]
- 6.Viswanath V, Story GM, Peier AM, Petrus MJ, Lee VM, et al. Opposite thermosensor in fruitfly and mouse. Nature. 2003;423(2942):822–3. doi: 10.1038/423822a. [DOI] [PubMed] [Google Scholar]
- 7.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454(7201):217–20. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong ST, Bang S, Hyun S, Kang J, Jeong K, et al. cAMP signaling in mushroom bodies modulates temperature preference behavior in Drosophila. Nature. 2008;454:771–775. doi: 10.1038/nature07090. [DOI] [PubMed] [Google Scholar]
- 9.Mansuy D. The great diversity of reactions catalyzed by cytochromes P450. Comp biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;121:5–14. doi: 10.1016/s0742-8413(98)10026-9. [DOI] [PubMed] [Google Scholar]
- 10.Hannemann F, Bichet A, Ewen KM, Bernhardt R. Cytochrome P450 systems-biological variations of electron transport chains. Biochim biophys Acta. 2007;1770:330–344. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 11.Isin EM, Guengerich FP. Complex reactions catalyzed by cytochrome P450 enzymes. Biochim biophys Acta. 2007;1770:314–329. doi: 10.1016/j.bbagen.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Nebert DW. Proposed role of drug-metabolizing enzymes: regulation of steady state levels of the ligands that effect growth, homeostasis, differentiation, and neuroendocrine functions. Mol Endocrinol. 1991;5:1203–1214. doi: 10.1210/mend-5-9-1203. [DOI] [PubMed] [Google Scholar]
- 13.Feyereisens R. Insect P450 enzymes. Annu Rev entomol. 1999;44:507–533. doi: 10.1146/annurev.ento.44.1.507. [DOI] [PubMed] [Google Scholar]
- 14.Tijet N, helvig C, Feyereisen R. The cytochrome P450 gene superfamily in Drosophila melanogaster: annotation, intron-exon organization and phylogeny. Gene. 2001;262:189–198. doi: 10.1016/s0378-1119(00)00533-3. [DOI] [PubMed] [Google Scholar]
- 15.Chavez VM, Marques G, Delbecque JP, Kobayashi K, Hollingsworth M, et al. The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development. 2000;127:4115–4126. doi: 10.1242/dev.127.19.4115. [DOI] [PubMed] [Google Scholar]
- 16.Warren JT, Petryk A, Marques G, Jarcho M, Parvy JP, et al. Molecular and biochemical characterization of to P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:11043–11048. doi: 10.1073/pnas.162375799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petryk A, Warren JT, Marques G, Jarcho MP, Gilbert LI, et al. Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect moltinghormone 20-hydroxyecdysone. Proc Natl Acad Sci USA. 2003;100:13773–13778. doi: 10.1073/pnas.2336088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert LI. Halloween genes encode P450 enzymes that mediate steroid hormone biosynthesis in Drosophila melanogaster. Mol Cell Endocrinol. 2004;215:1–10. doi: 10.1016/j.mce.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Warren JT, Petryk A, Marques G, Parvy JP, Shinoda T, et al. phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: a P450 enzyme critical in ecdysone biosynthesis. Insect biochem Mol boil. 2004;34:991–1010. doi: 10.1016/j.ibmb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Ono H, Rewitz KF, Shinoda T, Itoyama K, Petryk A, et al. Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in dipteral. Dev Biol. 2006;298:555–570. doi: 10.1016/j.ydbio.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 21.Derick HA, Greenspan RJ. Molecular analysis of flies selected for aggressive behavior. Nat Genet. 2006;38:1023–1031. doi: 10.1038/ng1864. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Dankert H, Perona P, Anderson DJ. A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proc Natl Acad Sci USA. 2008;105:5657–5663. doi: 10.1073/pnas.0801327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujii S, Toyama A, Amrein H. A male-specific fatty acid {omega}-hydroxylase, SXE1, is necessary for efficient male mating in Drosophila melanogaster. Genetics. 2008;180:179–190. doi: 10.1534/genetics.108.089177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGuire SE, Le PT, Osbron AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 25.Thum AS, Jenett A, Ito K, Heisenberg M, Tanimoto H. Multiple memory traces for olfactory reward learning in Drosophila. Journal of Neuroscience. 2007;27:11132–11138. doi: 10.1523/JNEUROSCI.2712-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung H, Sztal T, Pasricha S, Sridhar M, Batterham P, et al. Characterization of Drosophila melanogaster cytochrome P450 genes. Proc Natl Acad Sci USA. 2009;106:5731–5736. doi: 10.1073/pnas.0812141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snyder MJ, Girvetz E, Mulder EP. Induction of marine mollusk stress proteins by chemical or physical stress. Arch Environ Contam Toxical. 2001;41(1):22–29. doi: 10.1007/s002440010217. [DOI] [PubMed] [Google Scholar]
- 28.Ando M, Katagiri K, Yamamoto S, Wakamatsu K, Kawahara I, et al. Age-related effects of heat stress on protective enzymes for peroxides and microsomal monooxygenase in rat liver. Environ Health Perspect. 1997;105(7):726–733. doi: 10.1289/ehp.97105726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartenstein V. The neuroendocrine system of invertebrates: a developmental and evolutionary perspective. J Endocrinol. 2006;190(3):555–570. doi: 10.1677/joe.1.06964. [DOI] [PubMed] [Google Scholar]
- 30.Degtyarenko KN, Kulikova TA. Evolution of bioinorganic motifs in P450-containing systems. Biochem Soc Trans. 2001;29:139–47. doi: 10.1042/0300-5127:0290139. [DOI] [PubMed] [Google Scholar]
- 31.Davis RL. Olfactory memory formation in Drosophila: From molecular to systems Neuroscience. Annual Review of Neuroscience. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- 32.Clark AG. Stress and metabolic regulation in Drosophila. EXS. 1997;83:117–132. doi: 10.1007/978-3-0348-8882-0_7. [DOI] [PubMed] [Google Scholar]
- 33.Sinclair BJ, Gibbs AG, Roberts SP. Gene transcription during exposure to, and recovery from, cold and desiccation stress in Drosophila melanogaster. Insect Mol Biol. 2007;16(4):435–443. doi: 10.1111/j.1365-2583.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- 34.Pedersen KS, Kristensen TN, Loeschcke V, Petersen BO, Duus JO, et al. Metabolomic signatures of inbreeding at benign and stressful temperatures in Drosophila melanogaster. Genetics. 2008;180(2):1233–1243. doi: 10.1534/genetics.108.089144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeuchi K, Nakano Y, Kato U, Kaneda M, Aizu M, et al. Changes in temperature preferences and energy homeostasis in dystroglycan mutants. Science. 2009;323:1740–1743. doi: 10.1126/science.1165712. [DOI] [PubMed] [Google Scholar]
- 36.Hong ST, Bang S, Paik D, Kang J, Hwang S, et al. Histamine and its receptors modulate temperature preference behavior in Drosophila. J Neurosci. 2006;26(27):7245–7256. doi: 10.1523/JNEUROSCI.5426-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin JR, Ernst R, Heisenberg M. Mushroom bodies suppress locomotor activity in Drosophila melanogaster. Learning and Memory. 1998;5:179–191. [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30(4):322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects on cyp6a17 mutation on mushroom body morphology. Mushroom body morphology in cyp6a17Δ173 mutant. Mushroom body axons are visualized using FASII antisera. Compared with wild type (A), no significant defects were found in mutant (B). Doral is up.
(PDF)
Effects of cyp6a17 RNAi and tissue distribution of cyp6a17 expression. (A) Reduction of the expression level by MB247>cyp6a17-RNAi. gapdh1 was used as internal control. The number of tests: N = 4. Two asterisks, P<0.001. (B) Transcriptional profiles of cyp6a17 in the head and body assayed by real-time PCR. The histogram shows that cyp6a17 mRNA is slightly more abundant in the head compared with the body. gapdh1 was used as internal control. The number of tests: N = 4. Two asterisks, P<0.001. All data are means, and the error bars indicate s.e.m.
(PDF)
The expression pattern of cyp6a17SG - Gal4 . cyp6a17 reporter expression by cyp6a17SG-Gal4>UAS-mCD8:GFP (green). GFP expression was detected strongly in surface glial cells (filled triangle) and weakly in Kenyon cells (open triangle).
(PDF)
Total list of the 103 probes.
(PDF)
Statistical analysis of the results shown in Figures.
(PDF)