Abstract
We reported recently the first polymorph of the title compound [Kuai & Cheng (2011a ▶). Acta Cryst., E67, o2787]. A second polymorph of the title compound, C15H12N2O2, was unexpectedly obtained by the hydrothermal reaction of the title compound with manganese chloride in the presence of potassium hydroxide at 413 K. The benzimidazole ring system is almost planar, with a maximum deviation from the mean plane of 0.015 (2) Å. The benzimidazole and benzene rings are inclined at a dihedral angle of 79.00 (1)°. In the crystal, adjacent molecules are connected through O—H⋯N hydrogen bonds into a one-dimensional chain along the [001] direction.
Related literature
For the synthesis of 4-[(1H-benzo[d]imidazol-1-yl)methyl]benzoic acid, see: Hua et al. (2010 ▶). For two other polymorphs of the title compound, see: Kuai & Cheng (2011a
▶,b
▶). For related structures, see Das & Bharadwaj (2009 ▶).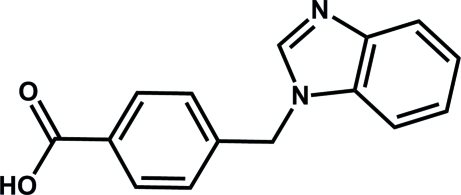
Experimental
Crystal data
C15H12N2O2
M r = 252.27
Orthorhombic,

a = 5.6969 (15) Å
b = 12.657 (3) Å
c = 17.604 (5) Å
V = 1269.4 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 293 K
0.30 × 0.18 × 0.18 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.974, T max = 0.984
7948 measured reflections
1786 independent reflections
1313 reflections with I > 2σ(I)
R int = 0.040
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.089
S = 0.99
1786 reflections
166 parameters
H-atom parameters constrained
Δρmax = 0.11 e Å−3
Δρmin = −0.15 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg, 2000 ▶); software used to prepare material for publication: SHELXTL (Sheldrick, 2008 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811042838/aa2024sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811042838/aa2024Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811042838/aa2024Isup4.cdx
Supplementary material file. DOI: 10.1107/S1600536811042838/aa2024Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H12⋯N12i | 0.82 | 1.84 | 2.649 (3) | 168 |
Symmetry code: (i)  .
.
Acknowledgments
The authors gratefully acknowledge the Natural Science Foundation of Jiangsu Province of China (grant No. BK2008195) for financial support of this work.
supplementary crystallographic information
Comment
The title compound, C15H12N2O2 (I), is usually regarded as an excellent candidate for building block in molecular self-assembly engineering due to its variable conformation and coordination modes (Das & Bharadwaj, 2009). During assembly of a coordination polymer, we accidentally obtained three polymorphs of (I), which can be proved by different unit-cell parameters and space groups. Here, we are introducing one of them. The single crystals of (I) were accidentally obtained by the hydrothermal reaction at 413 K of (I) with manganese chloride in the presence of potassium hydroxide as alkaline medium for the deprotonation. As shown in Fig. 1, the asymmetric unit of (I) consists of only one molecule. Interestingly, though crystallizing from alkaline solution, (I) remains the intact carboxylic group in the crystal structure. The flexible benzimidazolyl arm is apt to rotate. As a result, the benzimidazolyl ring and central benzene rings are inclined at a dihedral angle of 79.00 (1) °; The torsion angles N11—C11—C1—C2 and N11—C11—C1—C6 are -61.8 (2) ° and 118.0 (2) °, respectively. Adjacent molecules are connected through O—H···N hydrogen bonds into a one-dimensional chain along [001] direction (Fig. 2, Table 1).
Experimental
Reaction mixture of MnCl2 (21.5 mg, 0.1 mmol), 4-((1H-benzo[d]imidazol-1-yl)methyl)benzoic acid (25.2 mg, 0.1 mmol) and KOH (5.61 mg, 0.1 mmol) in 10 ml H2O was sealed in a 16 ml Teflon-lined stainless steel container and heated to 413 K for 3 days. After cooling to the room temperature, colorless block crystals of the title compound were obtained.
Refinement
All hydrogen atoms were located in geometrically idealized positions and constrained to ride on their parent atoms, with C—H = 0.93–0.97, O—H = 0.82 Å and Uiso(H) = 1.2Ueq(C, or O). Absolute structure can not be determined in this case because of no heavy atoms present. Friedel-pair data are merged with the MERG 3 instruction. The number of Friedel pairs is 1229.
Figures
Fig. 1.
: The crystal structure of (I) showing 30% probability displacement ellipsoids.
Fig. 2.
: The packing diagram of (I). Hydrogen bonds are shown as dashed lines.
Crystal data
| C15H12N2O2 | F(000) = 528 |
| Mr = 252.27 | Dx = 1.320 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 1326 reflections |
| a = 5.6969 (15) Å | θ = 2.3–19.9° |
| b = 12.657 (3) Å | µ = 0.09 mm−1 |
| c = 17.604 (5) Å | T = 293 K |
| V = 1269.4 (6) Å3 | Block, colorless |
| Z = 4 | 0.30 × 0.18 × 0.18 mm |
Data collection
| Bruker APEXII CCD diffractometer | 1786 independent reflections |
| Radiation source: fine-focus sealed tube | 1313 reflections with I > 2σ(I) |
| graphite | Rint = 0.040 |
| φ and ω scans | θmax = 28.0°, θmin = 2.0° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −7→7 |
| Tmin = 0.974, Tmax = 0.984 | k = −14→16 |
| 7948 measured reflections | l = −23→20 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.089 | H-atom parameters constrained |
| S = 0.99 | w = 1/[σ2(Fo2) + (0.0455P)2] where P = (Fo2 + 2Fc2)/3 |
| 1786 reflections | (Δ/σ)max < 0.001 |
| 166 parameters | Δρmax = 0.11 e Å−3 |
| 0 restraints | Δρmin = −0.15 e Å−3 |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. Absolute structure can not be determined in this case because of no heavy atoms present. Friedel-pair data are merged with the MERG 3 instruction. The number of Friedel pairs is 1229. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.3871 (4) | 0.37836 (13) | 1.03801 (9) | 0.0731 (6) | |
| H12 | 0.3280 | 0.3376 | 1.0688 | 0.088* | |
| O2 | 0.0975 (4) | 0.48320 (13) | 1.07621 (9) | 0.0650 (5) | |
| N11 | 0.4456 (3) | 0.78681 (13) | 0.75703 (9) | 0.0412 (4) | |
| N12 | 0.2556 (4) | 0.74457 (14) | 0.65069 (10) | 0.053 | |
| C4 | 0.3625 (4) | 0.54619 (16) | 0.98240 (11) | 0.0390 (5) | |
| C11 | 0.6074 (4) | 0.77548 (17) | 0.82087 (11) | 0.0451 (5) | |
| H6 | 0.6262 | 0.8434 | 0.8456 | 0.054* | |
| H5 | 0.7598 | 0.7537 | 0.8019 | 0.054* | |
| C13 | 0.1428 (4) | 0.83025 (16) | 0.68400 (11) | 0.0417 (5) | |
| C14 | 0.2622 (4) | 0.85805 (15) | 0.75045 (11) | 0.0372 (5) | |
| C6 | 0.6520 (4) | 0.60602 (17) | 0.89316 (11) | 0.0458 (5) | |
| H4 | 0.7940 | 0.5954 | 0.8682 | 0.055* | |
| C5 | 0.5733 (4) | 0.53174 (17) | 0.94491 (11) | 0.0473 (6) | |
| H3 | 0.6627 | 0.4717 | 0.9545 | 0.057* | |
| C41 | 0.2678 (5) | 0.46665 (17) | 1.03690 (11) | 0.0472 (6) | |
| C15 | 0.1919 (4) | 0.94228 (17) | 0.79536 (12) | 0.0462 (6) | |
| H8 | 0.2725 | 0.9610 | 0.8393 | 0.055* | |
| C1 | 0.5221 (4) | 0.69567 (16) | 0.87823 (11) | 0.0374 (5) | |
| C17 | −0.1262 (5) | 0.9687 (2) | 0.70573 (13) | 0.0572 (7) | |
| H10 | −0.2587 | 1.0071 | 0.6920 | 0.069* | |
| C12 | 0.4308 (5) | 0.72183 (17) | 0.69610 (12) | 0.0510 (6) | |
| H7 | 0.5348 | 0.6665 | 0.6873 | 0.061* | |
| C16 | −0.0036 (5) | 0.99675 (19) | 0.77139 (13) | 0.0557 (6) | |
| H9 | −0.0557 | 1.0541 | 0.7998 | 0.067* | |
| C2 | 0.3115 (4) | 0.71014 (16) | 0.91701 (11) | 0.0437 (5) | |
| H1 | 0.2224 | 0.7704 | 0.9081 | 0.052* | |
| C3 | 0.2344 (4) | 0.63641 (15) | 0.96819 (11) | 0.0426 (5) | |
| H2 | 0.0936 | 0.6474 | 0.9937 | 0.051* | |
| C18 | −0.0555 (4) | 0.88605 (18) | 0.66126 (13) | 0.0526 (6) | |
| H11 | −0.1370 | 0.8678 | 0.6174 | 0.063* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0997 (15) | 0.0514 (10) | 0.0681 (10) | 0.0157 (11) | 0.0362 (11) | 0.0196 (8) |
| O2 | 0.0673 (13) | 0.0707 (11) | 0.0571 (9) | 0.0034 (10) | 0.0240 (10) | 0.0109 (8) |
| N11 | 0.0449 (11) | 0.0390 (9) | 0.0396 (9) | −0.0030 (9) | −0.0010 (8) | 0.0024 (8) |
| N12 | 0.072 | 0.044 | 0.042 | −0.007 | −0.005 | −0.002 |
| C4 | 0.0435 (13) | 0.0422 (11) | 0.0313 (10) | 0.0001 (10) | 0.0020 (10) | −0.0025 (8) |
| C11 | 0.0395 (13) | 0.0473 (12) | 0.0484 (12) | −0.0053 (11) | −0.0046 (11) | 0.0069 (10) |
| C13 | 0.0494 (14) | 0.0382 (11) | 0.0375 (10) | −0.0108 (10) | −0.0017 (10) | 0.0050 (9) |
| C14 | 0.0382 (12) | 0.0354 (10) | 0.0379 (10) | −0.0067 (9) | 0.0022 (10) | 0.0054 (9) |
| C6 | 0.0354 (13) | 0.0556 (13) | 0.0462 (12) | 0.0063 (11) | 0.0079 (11) | 0.0022 (11) |
| C5 | 0.0509 (15) | 0.0467 (12) | 0.0442 (12) | 0.0138 (11) | 0.0048 (11) | 0.0069 (10) |
| C41 | 0.0569 (15) | 0.0505 (14) | 0.0341 (10) | 0.0001 (12) | 0.0032 (12) | 0.0005 (10) |
| C15 | 0.0542 (16) | 0.0442 (12) | 0.0404 (11) | −0.0009 (11) | −0.0001 (11) | 0.0002 (10) |
| C1 | 0.0360 (12) | 0.0400 (11) | 0.0363 (10) | −0.0036 (9) | −0.0045 (9) | −0.0012 (9) |
| C17 | 0.0488 (15) | 0.0614 (15) | 0.0613 (15) | 0.0054 (13) | −0.0005 (13) | 0.0225 (13) |
| C12 | 0.0672 (17) | 0.0371 (12) | 0.0487 (12) | −0.0035 (12) | 0.0066 (12) | −0.0026 (10) |
| C16 | 0.0621 (16) | 0.0493 (13) | 0.0557 (14) | 0.0082 (12) | 0.0102 (14) | 0.0079 (11) |
| C2 | 0.0441 (14) | 0.0394 (12) | 0.0476 (11) | 0.0064 (10) | 0.0007 (11) | 0.0004 (10) |
| C3 | 0.0395 (12) | 0.0478 (12) | 0.0403 (11) | 0.0048 (10) | 0.0054 (11) | −0.0039 (10) |
| C18 | 0.0535 (16) | 0.0557 (14) | 0.0487 (13) | −0.0134 (13) | −0.0128 (12) | 0.0152 (12) |
Geometric parameters (Å, °)
| O1—C41 | 1.308 (3) | C6—C1 | 1.380 (3) |
| O1—H12 | 0.8200 | C6—C5 | 1.384 (3) |
| O2—C41 | 1.210 (3) | C6—H4 | 0.9300 |
| N11—C12 | 1.354 (3) | C5—H3 | 0.9300 |
| N11—C14 | 1.385 (3) | C15—C16 | 1.376 (3) |
| N11—C11 | 1.461 (3) | C15—H8 | 0.9300 |
| N12—C12 | 1.311 (3) | C1—C2 | 1.392 (3) |
| N12—C13 | 1.390 (3) | C17—C18 | 1.367 (3) |
| C4—C3 | 1.378 (3) | C17—C16 | 1.396 (3) |
| C4—C5 | 1.382 (3) | C17—H10 | 0.9300 |
| C4—C41 | 1.492 (3) | C12—H7 | 0.9300 |
| C11—C1 | 1.509 (3) | C16—H9 | 0.9300 |
| C11—H6 | 0.9700 | C2—C3 | 1.369 (3) |
| C11—H5 | 0.9700 | C2—H1 | 0.9300 |
| C13—C18 | 1.391 (3) | C3—H2 | 0.9300 |
| C13—C14 | 1.398 (3) | C18—H11 | 0.9300 |
| C14—C15 | 1.386 (3) | ||
| C41—O1—H12 | 109.5 | O2—C41—C4 | 122.8 (2) |
| C12—N11—C14 | 106.40 (18) | O1—C41—C4 | 113.5 (2) |
| C12—N11—C11 | 126.09 (19) | C16—C15—C14 | 116.4 (2) |
| C14—N11—C11 | 127.18 (16) | C16—C15—H8 | 121.8 |
| C12—N12—C13 | 105.42 (18) | C14—C15—H8 | 121.8 |
| C3—C4—C5 | 118.88 (19) | C6—C1—C2 | 118.50 (19) |
| C3—C4—C41 | 119.0 (2) | C6—C1—C11 | 120.3 (2) |
| C5—C4—C41 | 122.1 (2) | C2—C1—C11 | 121.2 (2) |
| N11—C11—C1 | 112.17 (17) | C18—C17—C16 | 121.4 (2) |
| N11—C11—H6 | 109.2 | C18—C17—H10 | 119.3 |
| C1—C11—H6 | 109.2 | C16—C17—H10 | 119.3 |
| N11—C11—H5 | 109.2 | N12—C12—N11 | 113.4 (2) |
| C1—C11—H5 | 109.2 | N12—C12—H7 | 123.3 |
| H6—C11—H5 | 107.9 | N11—C12—H7 | 123.3 |
| N12—C13—C18 | 130.5 (2) | C15—C16—C17 | 122.1 (2) |
| N12—C13—C14 | 108.94 (19) | C15—C16—H9 | 119.0 |
| C18—C13—C14 | 120.5 (2) | C17—C16—H9 | 119.0 |
| N11—C14—C15 | 132.13 (19) | C3—C2—C1 | 120.6 (2) |
| N11—C14—C13 | 105.84 (17) | C3—C2—H1 | 119.7 |
| C15—C14—C13 | 122.0 (2) | C1—C2—H1 | 119.7 |
| C1—C6—C5 | 120.7 (2) | C2—C3—C4 | 121.0 (2) |
| C1—C6—H4 | 119.7 | C2—C3—H2 | 119.5 |
| C5—C6—H4 | 119.7 | C4—C3—H2 | 119.5 |
| C4—C5—C6 | 120.4 (2) | C17—C18—C13 | 117.6 (2) |
| C4—C5—H3 | 119.8 | C17—C18—H11 | 121.2 |
| C6—C5—H3 | 119.8 | C13—C18—H11 | 121.2 |
| O2—C41—O1 | 123.8 (2) | ||
| C12—N11—C11—C1 | −80.9 (3) | N11—C14—C15—C16 | −179.6 (2) |
| C14—N11—C11—C1 | 91.6 (2) | C13—C14—C15—C16 | 0.6 (3) |
| C12—N12—C13—C18 | −178.9 (2) | C5—C6—C1—C2 | 0.7 (3) |
| C12—N12—C13—C14 | 1.0 (2) | C5—C6—C1—C11 | −179.09 (19) |
| C12—N11—C14—C15 | −179.4 (2) | N11—C11—C1—C6 | 118.0 (2) |
| C11—N11—C14—C15 | 7.0 (3) | N11—C11—C1—C2 | −61.8 (2) |
| C12—N11—C14—C13 | 0.4 (2) | C13—N12—C12—N11 | −0.7 (2) |
| C11—N11—C14—C13 | −173.21 (18) | C14—N11—C12—N12 | 0.2 (2) |
| N12—C13—C14—N11 | −0.9 (2) | C11—N11—C12—N12 | 173.93 (19) |
| C18—C13—C14—N11 | 179.04 (18) | C14—C15—C16—C17 | 0.4 (3) |
| N12—C13—C14—C15 | 179.0 (2) | C18—C17—C16—C15 | −0.9 (4) |
| C18—C13—C14—C15 | −1.1 (3) | C6—C1—C2—C3 | −0.7 (3) |
| C3—C4—C5—C6 | −1.0 (3) | C11—C1—C2—C3 | 179.11 (19) |
| C41—C4—C5—C6 | 178.2 (2) | C1—C2—C3—C4 | −0.2 (3) |
| C1—C6—C5—C4 | 0.1 (3) | C5—C4—C3—C2 | 1.0 (3) |
| C3—C4—C41—O2 | −9.2 (3) | C41—C4—C3—C2 | −178.2 (2) |
| C5—C4—C41—O2 | 171.7 (2) | C16—C17—C18—C13 | 0.3 (3) |
| C3—C4—C41—O1 | 171.1 (2) | N12—C13—C18—C17 | −179.5 (2) |
| C5—C4—C41—O1 | −8.0 (3) | C14—C13—C18—C17 | 0.6 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H12···N12i | 0.82 | 1.84 | 2.649 (3) | 168. |
Symmetry codes: (i) −x+1/2, −y+1, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: AA2024).
References
- Brandenburg, K. (2000). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2008). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Das, M. C. & Bharadwaj, P. K. (2009). J. Am. Chem. Soc. 131, 10942–10943. [DOI] [PubMed]
- Hua, Q., Zhao, Y., Xu, G.-C., Chen, M.-S., Su, Z., Cai, K. & Sun, W.-Y. (2010). Cryst. Growth Des. 10, 2553–2562.
- Kuai, H.-W. & Cheng, X.-C. (2011a). Acta Cryst. E67, o2787. [DOI] [PMC free article] [PubMed]
- Kuai, H.-W. & Cheng, X.-C. (2011b). Acta Cryst. E67 In the press.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811042838/aa2024sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811042838/aa2024Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811042838/aa2024Isup4.cdx
Supplementary material file. DOI: 10.1107/S1600536811042838/aa2024Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




