Abstract
The title compound, C29H20O3, adopts an ‘S’ conformation with a dihedral angle of 68.5 (2)° beween the two acetone planes. The central phenyl ring forms dihedral angles of 83.8 (4) and 84.5 (4)° with the naphthalene and methoxy-substituted naphthalene mean planes, respectively. Both carbonyl-group O atoms deviate significantly from the naphthalene moiety and the methoxy-substituted naphthalene moiety [0.574 (1) and −1.053 (1) Å, respectively]. The crystal packing is stabilized by C—H⋯O intermolecular interactions, generating C(7) chain and R 2 2(10) graph-set motifs.
Related literature
For the uses and biological importance of diketones, see: Bennett et al. (1999 ▶). For related structures, see: Tsumuki et al. (2011 ▶); Jagadeesan et al. (2011 ▶); Judas et al. (1995 ▶). For graph-set notation, see: Bernstein et al. (1995 ▶).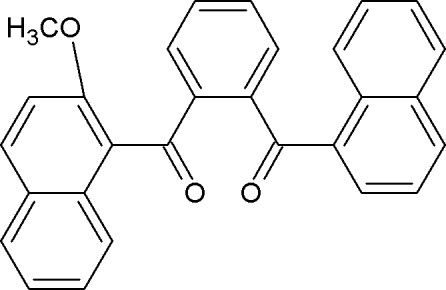
Experimental
Crystal data
C29H20O3
M r = 416.45
Monoclinic,

a = 8.3950 (3) Å
b = 8.9983 (4) Å
c = 28.5375 (11) Å
β = 97.188 (2)°
V = 2138.80 (15) Å3
Z = 4
Mo Kα radiation
μ = 0.08 mm−1
T = 293 K
0.30 × 0.25 × 0.20 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
26364 measured reflections
6299 independent reflections
3875 reflections with I > 2σ(I)
R int = 0.036
Refinement
R[F 2 > 2σ(F 2)] = 0.049
wR(F 2) = 0.137
S = 1.00
6270 reflections
290 parameters
H-atom parameters constrained
Δρmax = 0.23 e Å−3
Δρmin = −0.17 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: APEX2; data reduction: SAINT (Bruker, 2008 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811042747/bt5676sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811042747/bt5676Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811042747/bt5676Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C3—H3⋯O1i | 0.93 | 2.49 | 3.392 (2) | 164 |
| C13—H13⋯O1ii | 0.93 | 2.52 | 3.440 (2) | 170 |
| C27—H27⋯O2iii | 0.93 | 2.51 | 3.262 (3) | 138 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
GJ and KS thank Dr Babu Varghese, SAIF, IIT, Chennai, India, for the X-ray intensity data collection and Dr V. Murugan, HOD, Department of Physics, for providing facilities in the department to carry out this work.
supplementary crystallographic information
Comment
Diketones are popular in organic synthesis, for their applications in biology and medicine. They are known to exhibit antioxidant, antitumour and antibacterial activities (Bennett et al., 1999).
X-ray analysis confirms the molecular structure and atom connectivity of the title compound as illustrated in the Fig. 1. The central phenyl ring (C12–C17) of the compound forms dihedral angles of 83.8 (4)° and 84.5 (4)° with the naphthalene moiety (C1–C10) and methoxy substituted naphthalene moiety (C19–28), respectively. The central phenyl ring (C12–C17) forms dihedral angles of 69.7 (5)° and 11.1 (5)° with the mean planes of the ketone groups, (C10–C12/O1) and (C17–C19/O2), respectively. The dihedral angle between the methoxy substituted naphthalene moiety (C19–C28) and naphthlene moiety (C1–C10) is 64.2 (4)°.
The two benzene rings (C1—C4/C9/C10) and (C4–C9) are almost coplanar with a dihedral angle of 2.26 (6)° between them. The atoms C29, O2 and O3 are having deviations of 0.363 (3) Å, -1.053 (1)Å and 0.101 (1)Å from the mean plane of the methoxy substituted naphthalene ring (C19–C28), respectively. The atom O1 deviates by 0.574 (1)Å from the plane of the naphthlene ring (C1–C10). The C10–C11 and C18–C19 bond lengths of 1.49 (2)Å and 1.50 (2)Å respectively and can be considered as single C(sp2)–C(sp2) bond distances. The molecule possesses distorted S–conformation in which C19/C18/C17/C12/C11/C10/O1/O2 are in a single plane, which is determined by the dihedral angle of 68.5 (2)° beween the planes defined by C19/C18/C17/O2 and that through C10/C11/C12/O1 (Judas et al., 1995).The title compound exhibits the structural similarities with the reported related structures (Tsumuki et al., 2011 & Jagadeesan et al., 2011).
The crystal packing is stabilized by C–H···O interactions (Table 1). The C3–H3···O1i interaction generates a C(7) chain along the a axis and the C13–H13···O1ii hydrogen bond generates R22(10) graphset motifs (Bernstein et al., 1995); the carbonyl-group O1 atom is involved in bifurcated hydrogen bonding. The Symmetry codes are: (i) x - 1, y, z; (ii) -x + 1, -y + 1, -z; (iii) -x + 1/2, y + 1/2, -z + 1/2. The packing view of the compound is shown in (Fig. 2).
Experimental
To a stirred suspension of 1-(2-methoxy-1-naphthoyl)phenyl-1-naphthyl- 2-benzofuran (1 g, 3.22 mmol) in dry THF (20 ml), lead tetraaccetate (1.52 g, 3.42 mmol) was added and refluxed at 343 K for half an hour. The reaction mixture was then poured into water (200 ml) and extracted with ethyl acetate (2x20 ml), washed with brine solution and dried (Na2SO4). The removal of solvent in vacuo afforded crude product. The crude product upon crystallization from methanol furnished the tittle compound as a colorless solid.
Refinement
Hydrogen atoms were placed in calculated positions with C–H = 0.93Å and 0.96Å and refined using a the riding model with fixed isotropic displacement parameters: Uiso(H) = 1.5 Ueq(C) for the methyl group and Uiso(H) = 1.2 Ueq(C) for other groups.
Figures
Fig. 1.
The molecular structure of the title compound with the atomic numbering scheme and displacement ellipsoids at the 30% probability level.
Fig. 2.
The packing arrangement of the title compound. Dashed lines indicates the C–H···O interactions. Symmetry code: (i) x - 1, y, z; (ii) -x + 1, -y + 1, -z; (iii) -x + 1/2, y + 1/2, -z + 1/2.
Crystal data
| C29H20O3 | F(000) = 872 |
| Mr = 416.45 | Dx = 1.293 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 3875 reflections |
| a = 8.3950 (3) Å | θ = 1.4–30.1° |
| b = 8.9983 (4) Å | µ = 0.08 mm−1 |
| c = 28.5375 (11) Å | T = 293 K |
| β = 97.188 (2)° | Block, colourless |
| V = 2138.80 (15) Å3 | 0.30 × 0.25 × 0.20 mm |
| Z = 4 |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 3875 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.036 |
| graphite | θmax = 30.1°, θmin = 1.4° |
| ω scans | h = −10→11 |
| 26364 measured reflections | k = −12→12 |
| 6299 independent reflections | l = −40→40 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.049 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.137 | H-atom parameters constrained |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.0541P)2 + 0.4431P] where P = (Fo2 + 2Fc2)/3 |
| 6270 reflections | (Δ/σ)max < 0.001 |
| 290 parameters | Δρmax = 0.23 e Å−3 |
| 0 restraints | Δρmin = −0.17 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.15231 (17) | 0.63183 (17) | 0.06709 (6) | 0.0491 (4) | |
| H1 | 0.2003 | 0.7251 | 0.0696 | 0.059* | |
| C2 | −0.01600 (18) | 0.6217 (2) | 0.06134 (7) | 0.0585 (4) | |
| H2 | −0.0780 | 0.7075 | 0.0606 | 0.070* | |
| C3 | −0.08746 (17) | 0.4876 (2) | 0.05692 (6) | 0.0523 (4) | |
| H3 | −0.1989 | 0.4816 | 0.0521 | 0.063* | |
| C4 | 0.00426 (16) | 0.35663 (17) | 0.05949 (5) | 0.0437 (3) | |
| C5 | −0.0706 (2) | 0.2162 (2) | 0.05524 (7) | 0.0606 (5) | |
| H5 | −0.1820 | 0.2108 | 0.0499 | 0.073* | |
| C6 | 0.0158 (2) | 0.0898 (2) | 0.05872 (8) | 0.0763 (6) | |
| H6 | −0.0359 | −0.0016 | 0.0553 | 0.092* | |
| C7 | 0.1836 (2) | 0.0961 (2) | 0.06743 (8) | 0.0708 (5) | |
| H7 | 0.2426 | 0.0084 | 0.0707 | 0.085* | |
| C8 | 0.26131 (18) | 0.22922 (17) | 0.07110 (6) | 0.0507 (4) | |
| H8 | 0.3728 | 0.2313 | 0.0768 | 0.061* | |
| C9 | 0.17494 (15) | 0.36407 (15) | 0.06640 (5) | 0.0367 (3) | |
| C10 | 0.24751 (14) | 0.50808 (15) | 0.06903 (5) | 0.0356 (3) | |
| C11 | 0.42381 (15) | 0.52955 (15) | 0.07111 (5) | 0.0360 (3) | |
| C12 | 0.49178 (14) | 0.68277 (14) | 0.08141 (5) | 0.0335 (3) | |
| C13 | 0.54639 (16) | 0.75872 (17) | 0.04437 (5) | 0.0430 (3) | |
| H13 | 0.5386 | 0.7151 | 0.0146 | 0.052* | |
| C14 | 0.61245 (17) | 0.89905 (18) | 0.05125 (5) | 0.0476 (4) | |
| H14 | 0.6483 | 0.9495 | 0.0261 | 0.057* | |
| C15 | 0.62522 (18) | 0.96406 (17) | 0.09500 (6) | 0.0509 (4) | |
| H15 | 0.6696 | 1.0585 | 0.0995 | 0.061* | |
| C16 | 0.57230 (17) | 0.88951 (16) | 0.13223 (5) | 0.0458 (3) | |
| H16 | 0.5815 | 0.9340 | 0.1619 | 0.055* | |
| C17 | 0.50522 (14) | 0.74850 (14) | 0.12605 (5) | 0.0351 (3) | |
| C18 | 0.45546 (15) | 0.66581 (15) | 0.16664 (5) | 0.0383 (3) | |
| C19 | 0.44871 (19) | 0.74572 (17) | 0.21243 (5) | 0.0473 (4) | |
| C20 | 0.5827 (2) | 0.74335 (17) | 0.24785 (5) | 0.0514 (4) | |
| C21 | 0.7276 (2) | 0.67257 (19) | 0.24152 (6) | 0.0592 (4) | |
| H21 | 0.7352 | 0.6202 | 0.2138 | 0.071* | |
| C22 | 0.8574 (3) | 0.6795 (2) | 0.27548 (7) | 0.0767 (6) | |
| H22 | 0.9527 | 0.6329 | 0.2706 | 0.092* | |
| C23 | 0.8476 (4) | 0.7560 (3) | 0.31743 (8) | 0.0913 (8) | |
| H23 | 0.9369 | 0.7610 | 0.3402 | 0.110* | |
| C24 | 0.7105 (4) | 0.8225 (2) | 0.32528 (7) | 0.0904 (8) | |
| H24 | 0.7059 | 0.8718 | 0.3537 | 0.108* | |
| C25 | 0.5721 (3) | 0.8193 (2) | 0.29106 (6) | 0.0696 (5) | |
| C26 | 0.4270 (4) | 0.8903 (3) | 0.29698 (8) | 0.0916 (8) | |
| H26 | 0.4179 | 0.9379 | 0.3255 | 0.110* | |
| C27 | 0.3004 (3) | 0.8919 (3) | 0.26284 (9) | 0.0871 (7) | |
| H27 | 0.2061 | 0.9403 | 0.2679 | 0.104* | |
| C28 | 0.3114 (2) | 0.8202 (2) | 0.21948 (7) | 0.0639 (5) | |
| C29 | 0.0565 (3) | 0.9120 (4) | 0.18389 (11) | 0.1213 (11) | |
| H29A | 0.0035 | 0.8871 | 0.2108 | 0.182* | |
| H29B | −0.0166 | 0.8989 | 0.1555 | 0.182* | |
| H29C | 0.0913 | 1.0137 | 0.1863 | 0.182* | |
| O1 | 0.51456 (11) | 0.43299 (12) | 0.06066 (4) | 0.0542 (3) | |
| O2 | 0.42381 (13) | 0.53442 (11) | 0.16317 (4) | 0.0503 (3) | |
| O3 | 0.19135 (16) | 0.81810 (18) | 0.18259 (6) | 0.0838 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0369 (7) | 0.0401 (8) | 0.0693 (11) | −0.0038 (6) | 0.0026 (7) | −0.0014 (7) |
| C2 | 0.0351 (7) | 0.0559 (10) | 0.0836 (13) | 0.0070 (7) | 0.0041 (7) | 0.0041 (9) |
| C3 | 0.0290 (6) | 0.0676 (11) | 0.0598 (10) | −0.0062 (7) | 0.0033 (6) | 0.0063 (8) |
| C4 | 0.0375 (7) | 0.0522 (9) | 0.0420 (8) | −0.0141 (6) | 0.0069 (6) | 0.0021 (7) |
| C5 | 0.0463 (8) | 0.0634 (11) | 0.0731 (12) | −0.0245 (8) | 0.0110 (8) | 0.0027 (9) |
| C6 | 0.0722 (12) | 0.0518 (11) | 0.1073 (17) | −0.0308 (10) | 0.0205 (11) | −0.0035 (11) |
| C7 | 0.0673 (11) | 0.0412 (9) | 0.1065 (16) | −0.0086 (8) | 0.0215 (10) | 0.0029 (10) |
| C8 | 0.0452 (8) | 0.0419 (8) | 0.0661 (11) | −0.0069 (6) | 0.0115 (7) | 0.0009 (7) |
| C9 | 0.0355 (6) | 0.0399 (7) | 0.0349 (7) | −0.0074 (5) | 0.0058 (5) | −0.0014 (6) |
| C10 | 0.0304 (6) | 0.0382 (7) | 0.0378 (7) | −0.0053 (5) | 0.0025 (5) | −0.0038 (6) |
| C11 | 0.0329 (6) | 0.0389 (7) | 0.0368 (7) | −0.0056 (5) | 0.0064 (5) | −0.0080 (6) |
| C12 | 0.0274 (5) | 0.0355 (7) | 0.0384 (7) | −0.0051 (5) | 0.0066 (5) | −0.0052 (6) |
| C13 | 0.0413 (7) | 0.0510 (9) | 0.0375 (7) | −0.0073 (6) | 0.0087 (6) | −0.0048 (6) |
| C14 | 0.0480 (8) | 0.0503 (9) | 0.0466 (9) | −0.0121 (7) | 0.0137 (6) | 0.0055 (7) |
| C15 | 0.0554 (9) | 0.0406 (8) | 0.0590 (10) | −0.0179 (7) | 0.0154 (7) | −0.0045 (7) |
| C16 | 0.0531 (8) | 0.0402 (8) | 0.0456 (8) | −0.0131 (6) | 0.0121 (6) | −0.0104 (6) |
| C17 | 0.0345 (6) | 0.0339 (7) | 0.0379 (7) | −0.0054 (5) | 0.0089 (5) | −0.0053 (5) |
| C18 | 0.0382 (7) | 0.0380 (7) | 0.0399 (7) | −0.0026 (5) | 0.0098 (6) | −0.0026 (6) |
| C19 | 0.0647 (9) | 0.0411 (8) | 0.0397 (8) | −0.0047 (7) | 0.0207 (7) | −0.0036 (6) |
| C20 | 0.0812 (11) | 0.0394 (8) | 0.0356 (8) | −0.0111 (8) | 0.0146 (7) | −0.0007 (6) |
| C21 | 0.0761 (11) | 0.0516 (10) | 0.0475 (10) | −0.0066 (9) | −0.0009 (8) | 0.0005 (8) |
| C22 | 0.0938 (14) | 0.0626 (12) | 0.0669 (13) | −0.0111 (11) | −0.0165 (11) | 0.0125 (10) |
| C23 | 0.142 (2) | 0.0629 (13) | 0.0579 (13) | −0.0311 (14) | −0.0316 (14) | 0.0148 (11) |
| C24 | 0.173 (3) | 0.0596 (13) | 0.0351 (10) | −0.0294 (15) | −0.0001 (13) | −0.0008 (9) |
| C25 | 0.1247 (17) | 0.0506 (10) | 0.0361 (9) | −0.0165 (11) | 0.0207 (10) | −0.0034 (7) |
| C26 | 0.156 (2) | 0.0736 (14) | 0.0544 (12) | −0.0021 (15) | 0.0489 (15) | −0.0182 (11) |
| C27 | 0.1146 (18) | 0.0784 (15) | 0.0797 (16) | 0.0114 (13) | 0.0576 (14) | −0.0141 (12) |
| C28 | 0.0750 (11) | 0.0614 (11) | 0.0618 (11) | 0.0034 (9) | 0.0339 (10) | −0.0052 (9) |
| C29 | 0.0917 (17) | 0.142 (3) | 0.139 (3) | 0.0545 (17) | 0.0487 (16) | 0.023 (2) |
| O1 | 0.0376 (5) | 0.0475 (6) | 0.0803 (8) | −0.0058 (4) | 0.0181 (5) | −0.0230 (6) |
| O2 | 0.0664 (7) | 0.0387 (6) | 0.0471 (6) | −0.0084 (5) | 0.0125 (5) | 0.0002 (5) |
| O3 | 0.0645 (8) | 0.0969 (11) | 0.0935 (11) | 0.0205 (8) | 0.0227 (8) | −0.0104 (9) |
Geometric parameters (Å, °)
| C1—C10 | 1.3678 (19) | C15—H15 | 0.9300 |
| C1—C2 | 1.405 (2) | C16—C17 | 1.3904 (18) |
| C1—H1 | 0.9300 | C16—H16 | 0.9300 |
| C2—C3 | 1.347 (2) | C17—C18 | 1.4804 (19) |
| C2—H2 | 0.9300 | C18—O2 | 1.2131 (16) |
| C3—C4 | 1.405 (2) | C18—C19 | 1.499 (2) |
| C3—H3 | 0.9300 | C19—C28 | 1.370 (2) |
| C4—C5 | 1.410 (2) | C19—C20 | 1.415 (2) |
| C4—C9 | 1.4233 (18) | C20—C21 | 1.404 (2) |
| C5—C6 | 1.345 (3) | C20—C25 | 1.422 (2) |
| C5—H5 | 0.9300 | C21—C22 | 1.366 (3) |
| C6—C7 | 1.401 (3) | C21—H21 | 0.9300 |
| C6—H6 | 0.9300 | C22—C23 | 1.392 (3) |
| C7—C8 | 1.362 (2) | C22—H22 | 0.9300 |
| C7—H7 | 0.9300 | C23—C24 | 1.340 (4) |
| C8—C9 | 1.411 (2) | C23—H23 | 0.9300 |
| C8—H8 | 0.9300 | C24—C25 | 1.421 (3) |
| C9—C10 | 1.4298 (18) | C24—H24 | 0.9300 |
| C10—C11 | 1.4864 (17) | C25—C26 | 1.404 (3) |
| C11—O1 | 1.2170 (16) | C26—C27 | 1.349 (4) |
| C11—C12 | 1.5069 (18) | C26—H26 | 0.9300 |
| C12—C13 | 1.3842 (19) | C27—C28 | 1.409 (3) |
| C12—C17 | 1.3961 (18) | C27—H27 | 0.9300 |
| C13—C14 | 1.383 (2) | C28—O3 | 1.364 (2) |
| C13—H13 | 0.9300 | C29—O3 | 1.417 (3) |
| C14—C15 | 1.371 (2) | C29—H29A | 0.9600 |
| C14—H14 | 0.9300 | C29—H29B | 0.9600 |
| C15—C16 | 1.376 (2) | C29—H29C | 0.9600 |
| C10—C1—C2 | 121.74 (14) | C15—C16—H16 | 119.6 |
| C10—C1—H1 | 119.1 | C17—C16—H16 | 119.6 |
| C2—C1—H1 | 119.1 | C16—C17—C12 | 119.18 (12) |
| C3—C2—C1 | 119.91 (15) | C16—C17—C18 | 120.59 (12) |
| C3—C2—H2 | 120.0 | C12—C17—C18 | 120.18 (11) |
| C1—C2—H2 | 120.0 | O2—C18—C17 | 120.42 (12) |
| C2—C3—C4 | 120.81 (13) | O2—C18—C19 | 120.46 (13) |
| C2—C3—H3 | 119.6 | C17—C18—C19 | 119.10 (12) |
| C4—C3—H3 | 119.6 | C28—C19—C20 | 120.71 (15) |
| C3—C4—C5 | 120.80 (13) | C28—C19—C18 | 119.15 (15) |
| C3—C4—C9 | 120.24 (13) | C20—C19—C18 | 120.14 (13) |
| C5—C4—C9 | 118.96 (14) | C21—C20—C19 | 122.59 (14) |
| C6—C5—C4 | 121.39 (15) | C21—C20—C25 | 118.58 (17) |
| C6—C5—H5 | 119.3 | C19—C20—C25 | 118.81 (17) |
| C4—C5—H5 | 119.3 | C22—C21—C20 | 120.98 (18) |
| C5—C6—C7 | 119.99 (16) | C22—C21—H21 | 119.5 |
| C5—C6—H6 | 120.0 | C20—C21—H21 | 119.5 |
| C7—C6—H6 | 120.0 | C21—C22—C23 | 120.3 (2) |
| C8—C7—C6 | 120.71 (17) | C21—C22—H22 | 119.8 |
| C8—C7—H7 | 119.6 | C23—C22—H22 | 119.8 |
| C6—C7—H7 | 119.6 | C24—C23—C22 | 120.6 (2) |
| C7—C8—C9 | 120.89 (15) | C24—C23—H23 | 119.7 |
| C7—C8—H8 | 119.6 | C22—C23—H23 | 119.7 |
| C9—C8—H8 | 119.6 | C23—C24—C25 | 121.4 (2) |
| C8—C9—C4 | 117.97 (12) | C23—C24—H24 | 119.3 |
| C8—C9—C10 | 124.32 (12) | C25—C24—H24 | 119.3 |
| C4—C9—C10 | 117.70 (12) | C26—C25—C20 | 118.2 (2) |
| C1—C10—C9 | 119.52 (11) | C26—C25—C24 | 123.7 (2) |
| C1—C10—C11 | 118.01 (12) | C20—C25—C24 | 118.1 (2) |
| C9—C10—C11 | 122.38 (12) | C27—C26—C25 | 122.34 (18) |
| O1—C11—C10 | 123.24 (12) | C27—C26—H26 | 118.8 |
| O1—C11—C12 | 117.88 (11) | C25—C26—H26 | 118.8 |
| C10—C11—C12 | 118.53 (11) | C26—C27—C28 | 119.8 (2) |
| C13—C12—C17 | 119.41 (12) | C26—C27—H27 | 120.1 |
| C13—C12—C11 | 117.14 (12) | C28—C27—H27 | 120.1 |
| C17—C12—C11 | 123.43 (11) | O3—C28—C19 | 115.73 (15) |
| C14—C13—C12 | 120.49 (13) | O3—C28—C27 | 124.13 (18) |
| C14—C13—H13 | 119.8 | C19—C28—C27 | 120.1 (2) |
| C12—C13—H13 | 119.8 | O3—C29—H29A | 109.5 |
| C15—C14—C13 | 120.21 (14) | O3—C29—H29B | 109.5 |
| C15—C14—H14 | 119.9 | H29A—C29—H29B | 109.5 |
| C13—C14—H14 | 119.9 | O3—C29—H29C | 109.5 |
| C14—C15—C16 | 119.94 (14) | H29A—C29—H29C | 109.5 |
| C14—C15—H15 | 120.0 | H29B—C29—H29C | 109.5 |
| C16—C15—H15 | 120.0 | C28—O3—C29 | 119.16 (19) |
| C15—C16—C17 | 120.77 (14) | ||
| C10—C1—C2—C3 | −0.9 (3) | C11—C12—C17—C16 | 178.86 (12) |
| C1—C2—C3—C4 | 2.1 (3) | C13—C12—C17—C18 | −176.75 (12) |
| C2—C3—C4—C5 | 179.49 (17) | C11—C12—C17—C18 | 1.67 (19) |
| C2—C3—C4—C9 | −0.6 (2) | C16—C17—C18—O2 | −166.95 (14) |
| C3—C4—C5—C6 | −178.45 (18) | C12—C17—C18—O2 | 10.20 (19) |
| C9—C4—C5—C6 | 1.6 (3) | C16—C17—C18—C19 | 11.53 (19) |
| C4—C5—C6—C7 | 0.9 (3) | C12—C17—C18—C19 | −171.32 (12) |
| C5—C6—C7—C8 | −1.9 (3) | O2—C18—C19—C28 | −94.73 (19) |
| C6—C7—C8—C9 | 0.2 (3) | C17—C18—C19—C28 | 86.79 (18) |
| C7—C8—C9—C4 | 2.3 (2) | O2—C18—C19—C20 | 85.03 (18) |
| C7—C8—C9—C10 | −178.96 (16) | C17—C18—C19—C20 | −93.45 (17) |
| C3—C4—C9—C8 | 176.89 (14) | C28—C19—C20—C21 | −177.85 (16) |
| C5—C4—C9—C8 | −3.2 (2) | C18—C19—C20—C21 | 2.4 (2) |
| C3—C4—C9—C10 | −1.9 (2) | C28—C19—C20—C25 | 0.2 (2) |
| C5—C4—C9—C10 | 177.98 (14) | C18—C19—C20—C25 | −179.52 (14) |
| C2—C1—C10—C9 | −1.7 (2) | C19—C20—C21—C22 | 176.34 (16) |
| C2—C1—C10—C11 | 174.95 (15) | C25—C20—C21—C22 | −1.7 (2) |
| C8—C9—C10—C1 | −175.73 (15) | C20—C21—C22—C23 | 0.7 (3) |
| C4—C9—C10—C1 | 3.0 (2) | C21—C22—C23—C24 | 0.7 (3) |
| C8—C9—C10—C11 | 7.8 (2) | C22—C23—C24—C25 | −1.0 (3) |
| C4—C9—C10—C11 | −173.44 (12) | C21—C20—C25—C26 | 179.73 (17) |
| C1—C10—C11—O1 | −160.14 (15) | C19—C20—C25—C26 | 1.6 (2) |
| C9—C10—C11—O1 | 16.4 (2) | C21—C20—C25—C24 | 1.4 (2) |
| C1—C10—C11—C12 | 12.87 (19) | C19—C20—C25—C24 | −176.73 (16) |
| C9—C10—C11—C12 | −170.62 (12) | C23—C24—C25—C26 | −178.3 (2) |
| O1—C11—C12—C13 | 65.58 (17) | C23—C24—C25—C20 | −0.1 (3) |
| C10—C11—C12—C13 | −107.81 (14) | C20—C25—C26—C27 | −1.8 (3) |
| O1—C11—C12—C17 | −112.87 (15) | C24—C25—C26—C27 | 176.4 (2) |
| C10—C11—C12—C17 | 73.74 (17) | C25—C26—C27—C28 | 0.2 (4) |
| C17—C12—C13—C14 | −0.6 (2) | C20—C19—C28—O3 | 178.49 (15) |
| C11—C12—C13—C14 | −179.09 (12) | C18—C19—C28—O3 | −1.7 (2) |
| C12—C13—C14—C15 | 0.3 (2) | C20—C19—C28—C27 | −1.9 (3) |
| C13—C14—C15—C16 | 0.1 (2) | C18—C19—C28—C27 | 177.88 (16) |
| C14—C15—C16—C17 | −0.2 (2) | C26—C27—C28—O3 | −178.7 (2) |
| C15—C16—C17—C12 | −0.1 (2) | C26—C27—C28—C19 | 1.7 (3) |
| C15—C16—C17—C18 | 177.11 (14) | C19—C28—O3—C29 | −169.66 (19) |
| C13—C12—C17—C16 | 0.44 (19) | C27—C28—O3—C29 | 10.7 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C3—H3···O1i | 0.93 | 2.49 | 3.392 (2) | 164 |
| C13—H13···O1ii | 0.93 | 2.52 | 3.440 (2) | 170 |
| C27—H27···O2iii | 0.93 | 2.51 | 3.262 (3) | 138 |
Symmetry codes: (i) x−1, y, z; (ii) −x+1, −y+1, −z; (iii) −x+1/2, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5676).
References
- Bennett, I., Broom, N. J. P., Cassels, R., Elder, J. S., Masson, N. D. & O’Hanlon, P. J. (1999). Bioorg. Med. Chem. Lett. 9, 1847–1852. [DOI] [PubMed]
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2008). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Jagadeesan, G., Sethusankar, K., Sivasakthikumaran, R. & Mohanakrishnan, A. K. (2011). Acta Cryst. E67, o2737. [DOI] [PMC free article] [PubMed]
- Judas, N., Kaitner, B. & Mestrovic, E. (1995). Acta Cryst. C51, 2123–2125.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Tsumuki, T., Hijikata, D., Okamoto, A., Oike, H. & Yonezawa, N. (2011). Acta Cryst. E67, o2095. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811042747/bt5676sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811042747/bt5676Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811042747/bt5676Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




