Abstract
In the title compound, [Au2(C6H8N2S4)(C18H15P)4]·2CHCl3, the digold complex resides on a crystallographic inversion center and co-crystallizes with two molecules of chloroform solvent. The piperazine-1,4-dicarbodithioate linker has an almost ideal chair conformation. The geometry about the gold atoms is severely distorted tetrahedral punctuated by a very acute S—Au—S bite angle.
Related literature
For stabilization of gold salts by dithiocarbonates, see: Fernandez et al. (1998 ▶). For use of piperazine dithiocarbamates as ligands used to engineer multimetallic assemblies, see: Wilton-Ely et al. (2008 ▶); Knight et al. (2009a
▶,b
▶); Oliver et al. (2011 ▶). For the copper analgoue, see: Kumar et al. (2009 ▶). For other related gold complexes, see: Razak et al. (2000 ▶); Jian et al. (2000 ▶). A molecular geometry check was performed with Mogul, see: Bruno et al. (2002 ▶). Related compounds were found in the Cambridge Structural Database (Allen, 2002 ▶). For ring analysis, see: Cremer & Pople (1975 ▶).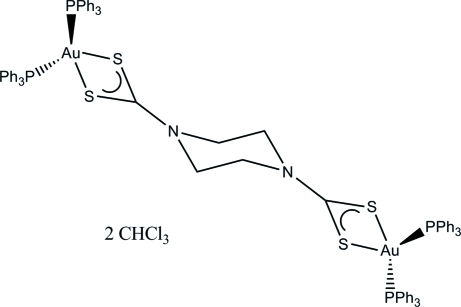
Experimental
Crystal data
[Au2(C6H8N2S4)(C18H15P)4]·2CHCl3
M r = 1918.13
Triclinic,
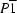
a = 12.8455 (17) Å
b = 13.2879 (10) Å
c = 13.4197 (9) Å
α = 119.572 (2)°
β = 101.544 (2)°
γ = 96.039 (2)°
V = 1895.2 (3) Å3
Z = 1
Cu Kα radiation
μ = 11.30 mm−1
T = 100 K
0.44 × 0.35 × 0.29 mm
Data collection
Bruker SMART APEXII diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2007 ▶) T min = 0.083, T max = 0.140
30338 measured reflections
7089 independent reflections
7080 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.028
wR(F 2) = 0.074
S = 1.15
7089 reflections
442 parameters
H-atom parameters constrained
Δρmax = 2.40 e Å−3
Δρmin = −1.35 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL (Sheldrick, 2008 ▶), FCF_filter (Guzei, 2007 ▶) and INSerter (Guzei, 2007 ▶); molecular graphics: SHELXTL and DIAMOND (Brandenburg, 1999 ▶); software used to prepare material for publication: SHELXTL, publCIF (Westrip, 2010 ▶) and modiCIFer (Guzei, 2007 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811044229/ng5253sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811044229/ng5253Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected geometric parameters (Å, °).
| Au1—P2 | 2.2994 (8) |
| Au1—P1 | 2.3233 (8) |
| Au1—S2 | 2.6133 (8) |
| Au1—S1 | 2.7414 (8) |
| P2—Au1—P1 | 134.65 (3) |
| P2—Au1—S2 | 116.81 (3) |
| P1—Au1—S2 | 107.10 (3) |
| P2—Au1—S1 | 107.34 (3) |
| P1—Au1—S1 | 99.39 (3) |
| S2—Au1—S1 | 67.03 (2) |
Acknowledgments
We acknowledge support from the University of Johannesburg for this work.
supplementary crystallographic information
Comment
Dithiocarbamates have long been used as ligands to stabilize gold(I) and gold(III) salts (Fernandez et al., 1998), but piperazine dithiocarbamates are currently receiving a lot more attention as ligands that can be used to engineer multimetallic assemblies including making gold nanoparticles (Wilton-Ely et al., 2008; Knight et al. 2009a; Knight et al., 2009b; Oliver et al., 2011). We recently isolated the title compound (I) via a slight modification of one of the routes described in the aforementioned literature.
The crystal structure of (I) contains the digold complex residing on a crystallographic inversion center and two molecules of solvent chloroform solvent per digold complex. The piperzine dithiocarbamate linker exhibits an almost ideal chair conformation (puckering coordinates θ=177.97 (1)° φ=0°, Cremer & Pople, 1975) similar to the analogous compounds with group ten square-planar metal centers nickel, palladium, and platinum (Knight et al., 2009a) and the tetrahedral copper analogue (Kumar et al., 2009). All bond distances and angles are typical as confirmed by a Mogul geometry check except for the S1—C1—S2, S1—C1—N1, and S2—C1—N1 angles (Bruno et al., 2002). However these angles in (I) are similar to those in the closely related compounds (N,N-diisopropyldithiocarbamato-S,S')-bis(triphenylphosphane-P)-gold(I) (Jian et al., 2000) and (piperidine-1-carbodithioato-S,S')-bis(triphenylphosphane-P)-gold(I) (Razak et al., 2000).
The geometry about the gold atom is severely distorted tetrahedral with the dihedral angle between the planes defined by atoms S1, Au1, S1 and P1, Au1, P1 measuring 88.77 (3)°. Such a distorted tetrahedral geometry and acute S—Au—S bite angle (67.03 (2)°) are typical of complexes where gold is bonded to two phosphorous atoms and two sulfur atoms of a bidentale ligand forming a four-membered metallocycle. For eight such compounds in the Cambridge Structural Database (CSD; August 2011 update; Allen, 2002) the S—Au—S bite angle has an average of 66 (3)°. These compounds also have very large P—Au—P angles with a 135 (5)° average corresponding well to the 134.65 (3)° value found in (I). The copper analogue of (I) also exhibits a similarly distorted tetrahedral geometry but with a larger S—Cu—S bite angle of 75.41 (2)° (Kumar et al., 2009). The group ten analogues exhibit distorted square planar geometries with larger S—metal—S bite angles that average 77 (3)° (Knight et al., 2009a). The Au—S distances in (I) differ by 0.1281 Å; this value agrees well with the differences in the two Au—S bonds in the eight related compounds in the CSD where such distances differ by an average of 0.18 (11) Å.
Experimental
To a solution of potassium piperazine-1,4-bis(dithiocarbamate) (0.17 g, 0.57 mmol) in water (10 mL) was added a solution of [AuCl(PPh3)] (0.40 g, 0.81 mmol) in dichloromethane (10 mL). The biphasic reaction mixture was stirred for 30 minutes. The organic layer was separated and layered with chloroform and hexane to yield a yellow solid. Yield: 0.32 g (69%). 1H NMR (CDCl3): δ 7.53 (m, 12H), 7.42 (m, 18H), 4.30 (s, 8H). 13C{1H} NMR (CDCl3): δ 208.2 (2 C, C=S), 134.1, 130.7, 128.9, 50.1. 31 P{1H} NMR (CDCl3): δ 28.9. ESI-MS (m/z): 1155 ([M], 5%), 721 [Au(PPh3)2]+, 100%). IR (ATR, cm-1): υ(C—N) = 1451, υ(C=S) = 1026, υ(C—S) = 997. Anal. Calc. for C78H68Au2N2P2S4.CHCl3: C 50.09, H 3.68, N 1.46. Found: C 49.73, H 3.76, N 1.60%.
Refinement
All H-atoms attached to carbon atoms were placed in idealized locations and refined as riding with appropriate thermal displacement coefficients Uiso(H) = 1.2 times Ueq(bearing atom). Default effective X—H distances for T = -173.0°C C(sp 3)–1H=1.00, C(sp 3)–2H=0.99, C(sp 2)–H=0.95. The final difference map had a peak and a hole in the vicinities of Au.
Figures
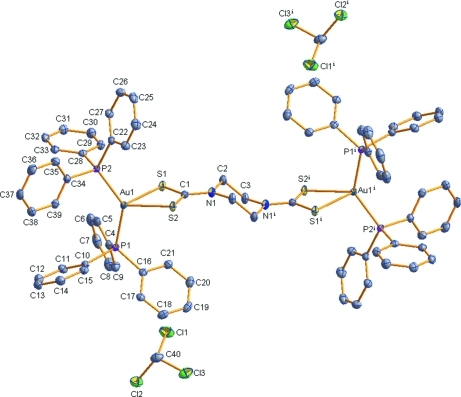
Fig. 1.
Molecular structure of (I) (Brandenburg, 1999). The thermal ellipsoids are shown at 50% probability level. All hydrogen atoms were omitted. Symmetry code: (i) -x,2-y, 2-z.
Crystal data
| [Au2(C6H8N2S4)(C18H15P)4]·2CHCl3 | Z = 1 |
| Mr = 1918.13 | F(000) = 948 |
| Triclinic, P1 | Dx = 1.681 Mg m−3 |
| Hall symbol: -P 1 | Cu Kα radiation, λ = 1.54178 Å |
| a = 12.8455 (17) Å | Cell parameters from 9936 reflections |
| b = 13.2879 (10) Å | θ = 3.6–71.7° |
| c = 13.4197 (9) Å | µ = 11.30 mm−1 |
| α = 119.572 (2)° | T = 100 K |
| β = 101.544 (2)° | Block, colourless |
| γ = 96.039 (2)° | 0.44 × 0.35 × 0.29 mm |
| V = 1895.2 (3) Å3 |
Data collection
| Bruker SMART APEXII diffractometer | 7089 independent reflections |
| Radiation source: fine-focus sealed tube | 7080 reflections with I > 2σ(I) |
| graphite | Rint = 0.031 |
| 0.50° ω and 0.5 ° φ scans | θmax = 72.3°, θmin = 3.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 2007) | h = −15→14 |
| Tmin = 0.083, Tmax = 0.140 | k = −16→16 |
| 30338 measured reflections | l = −16→16 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.028 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.074 | H-atom parameters constrained |
| S = 1.15 | w = 1/[σ2(Fo2) + (0.0416P)2 + 3.5159P] where P = (Fo2 + 2Fc2)/3 |
| 7089 reflections | (Δ/σ)max = 0.001 |
| 442 parameters | Δρmax = 2.40 e Å−3 |
| 0 restraints | Δρmin = −1.35 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Au1 | 0.234436 (10) | 0.776500 (10) | 0.653197 (10) | 0.01426 (6) | |
| S1 | 0.25387 (6) | 0.94442 (7) | 0.88645 (7) | 0.01770 (16) | |
| S2 | 0.04873 (6) | 0.82819 (7) | 0.68645 (7) | 0.01689 (16) | |
| P1 | 0.24564 (7) | 0.61270 (7) | 0.67150 (7) | 0.01428 (16) | |
| P2 | 0.32654 (7) | 0.86933 (7) | 0.57904 (7) | 0.01350 (16) | |
| N1 | 0.0585 (2) | 0.9761 (2) | 0.9150 (2) | 0.0168 (5) | |
| C1 | 0.1155 (3) | 0.9217 (3) | 0.8365 (3) | 0.0156 (6) | |
| C2 | 0.1115 (3) | 1.0645 (3) | 1.0418 (3) | 0.0195 (7) | |
| H2AB | 0.1051 | 1.1446 | 1.0584 | 0.023* | |
| H2AA | 0.1903 | 1.0655 | 1.0614 | 0.023* | |
| C3 | 0.0590 (3) | 1.0359 (3) | 1.1199 (3) | 0.0192 (7) | |
| H3AA | 0.0737 | 0.9608 | 1.1113 | 0.023* | |
| H3AB | 0.0913 | 1.1005 | 1.2050 | 0.023* | |
| C4 | 0.3628 (3) | 0.6456 (3) | 0.7941 (3) | 0.0183 (7) | |
| C5 | 0.4568 (3) | 0.7301 (3) | 0.8254 (3) | 0.0215 (7) | |
| H5AA | 0.4581 | 0.7723 | 0.7854 | 0.026* | |
| C6 | 0.5488 (3) | 0.7527 (4) | 0.9152 (3) | 0.0285 (8) | |
| H6AA | 0.6130 | 0.8098 | 0.9357 | 0.034* | |
| C7 | 0.5471 (3) | 0.6927 (4) | 0.9744 (3) | 0.0311 (9) | |
| H7AA | 0.6101 | 0.7085 | 1.0357 | 0.037* | |
| C8 | 0.4537 (3) | 0.6093 (4) | 0.9447 (3) | 0.0301 (8) | |
| H8AA | 0.4526 | 0.5687 | 0.9863 | 0.036* | |
| C9 | 0.3620 (3) | 0.5849 (3) | 0.8550 (3) | 0.0238 (7) | |
| H9AA | 0.2985 | 0.5270 | 0.8346 | 0.029* | |
| C10 | 0.2675 (3) | 0.4871 (3) | 0.5424 (3) | 0.0159 (6) | |
| C11 | 0.3632 (3) | 0.4487 (3) | 0.5474 (3) | 0.0203 (7) | |
| H11A | 0.4171 | 0.4825 | 0.6223 | 0.024* | |
| C12 | 0.3813 (3) | 0.3601 (3) | 0.4429 (3) | 0.0245 (7) | |
| H12A | 0.4475 | 0.3346 | 0.4470 | 0.029* | |
| C13 | 0.3025 (3) | 0.3101 (3) | 0.3339 (3) | 0.0229 (7) | |
| H13A | 0.3145 | 0.2498 | 0.2631 | 0.027* | |
| C14 | 0.2067 (3) | 0.3477 (3) | 0.3280 (3) | 0.0215 (7) | |
| H14A | 0.1527 | 0.3128 | 0.2530 | 0.026* | |
| C15 | 0.1887 (3) | 0.4362 (3) | 0.4311 (3) | 0.0190 (7) | |
| H15A | 0.1228 | 0.4624 | 0.4262 | 0.023* | |
| C16 | 0.1298 (3) | 0.5531 (3) | 0.7022 (3) | 0.0168 (6) | |
| C17 | 0.0658 (3) | 0.4376 (3) | 0.6282 (3) | 0.0187 (7) | |
| H17A | 0.0823 | 0.3836 | 0.5574 | 0.022* | |
| C18 | −0.0229 (3) | 0.4005 (3) | 0.6578 (3) | 0.0236 (7) | |
| H18A | −0.0670 | 0.3215 | 0.6065 | 0.028* | |
| C19 | −0.0467 (3) | 0.4788 (3) | 0.7617 (3) | 0.0243 (7) | |
| H19A | −0.1067 | 0.4534 | 0.7820 | 0.029* | |
| C20 | 0.0174 (3) | 0.5941 (3) | 0.8355 (3) | 0.0248 (7) | |
| H20A | 0.0017 | 0.6474 | 0.9072 | 0.030* | |
| C21 | 0.1042 (3) | 0.6322 (3) | 0.8060 (3) | 0.0204 (7) | |
| H21A | 0.1464 | 0.7121 | 0.8561 | 0.025* | |
| C22 | 0.2798 (3) | 0.9894 (3) | 0.5672 (3) | 0.0169 (6) | |
| C23 | 0.1719 (3) | 0.9678 (3) | 0.5049 (3) | 0.0218 (7) | |
| H23A | 0.1233 | 0.8929 | 0.4726 | 0.026* | |
| C24 | 0.1338 (3) | 1.0550 (3) | 0.4894 (3) | 0.0269 (8) | |
| H24A | 0.0604 | 1.0382 | 0.4438 | 0.032* | |
| C25 | 0.2025 (3) | 1.1658 (3) | 0.5399 (3) | 0.0262 (8) | |
| H25A | 0.1764 | 1.2252 | 0.5293 | 0.031* | |
| C26 | 0.3099 (3) | 1.1904 (3) | 0.6063 (3) | 0.0242 (7) | |
| H26A | 0.3567 | 1.2672 | 0.6431 | 0.029* | |
| C27 | 0.3491 (3) | 1.1018 (3) | 0.6189 (3) | 0.0207 (7) | |
| H27A | 0.4230 | 1.1181 | 0.6628 | 0.025* | |
| C28 | 0.4678 (3) | 0.9384 (3) | 0.6736 (3) | 0.0157 (6) | |
| C29 | 0.4856 (3) | 1.0177 (3) | 0.7967 (3) | 0.0187 (7) | |
| H29A | 0.4249 | 1.0341 | 0.8276 | 0.022* | |
| C30 | 0.5910 (3) | 1.0720 (3) | 0.8732 (3) | 0.0214 (7) | |
| H30A | 0.6027 | 1.1262 | 0.9563 | 0.026* | |
| C31 | 0.6797 (3) | 1.0473 (3) | 0.8284 (3) | 0.0208 (7) | |
| H31A | 0.7520 | 1.0844 | 0.8812 | 0.025* | |
| C32 | 0.6637 (3) | 0.9691 (3) | 0.7077 (3) | 0.0206 (7) | |
| H32A | 0.7247 | 0.9524 | 0.6776 | 0.025* | |
| C33 | 0.5574 (3) | 0.9145 (3) | 0.6298 (3) | 0.0177 (6) | |
| H33A | 0.5464 | 0.8610 | 0.5467 | 0.021* | |
| C34 | 0.3386 (3) | 0.7666 (3) | 0.4312 (3) | 0.0154 (6) | |
| C35 | 0.3435 (3) | 0.7995 (3) | 0.3476 (3) | 0.0205 (7) | |
| H35A | 0.3372 | 0.8769 | 0.3657 | 0.025* | |
| C36 | 0.3575 (3) | 0.7189 (3) | 0.2381 (3) | 0.0247 (7) | |
| H36A | 0.3597 | 0.7410 | 0.1810 | 0.030* | |
| C37 | 0.3683 (3) | 0.6067 (3) | 0.2119 (3) | 0.0220 (7) | |
| H37A | 0.3790 | 0.5524 | 0.1374 | 0.026* | |
| C38 | 0.3635 (3) | 0.5729 (3) | 0.2941 (3) | 0.0220 (7) | |
| H38A | 0.3715 | 0.4960 | 0.2763 | 0.026* | |
| C39 | 0.3470 (3) | 0.6526 (3) | 0.4026 (3) | 0.0184 (7) | |
| H39A | 0.3413 | 0.6288 | 0.4578 | 0.022* | |
| Cl1 | 0.15739 (10) | 0.41053 (9) | 0.95175 (11) | 0.0451 (3) | |
| Cl2 | 0.14677 (11) | 0.15846 (10) | 0.81701 (11) | 0.0455 (3) | |
| Cl3 | −0.05265 (9) | 0.24127 (10) | 0.82114 (9) | 0.0383 (2) | |
| C40 | 0.0848 (3) | 0.2721 (4) | 0.8232 (4) | 0.0305 (8) | |
| H40A | 0.0863 | 0.2762 | 0.7511 | 0.037* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Au1 | 0.01735 (9) | 0.01435 (8) | 0.01405 (8) | 0.00500 (5) | 0.00798 (6) | 0.00817 (6) |
| S1 | 0.0143 (4) | 0.0222 (4) | 0.0133 (3) | 0.0047 (3) | 0.0051 (3) | 0.0065 (3) |
| S2 | 0.0149 (4) | 0.0209 (4) | 0.0123 (3) | 0.0050 (3) | 0.0044 (3) | 0.0066 (3) |
| P1 | 0.0157 (4) | 0.0147 (4) | 0.0139 (4) | 0.0043 (3) | 0.0058 (3) | 0.0080 (3) |
| P2 | 0.0156 (4) | 0.0141 (4) | 0.0136 (4) | 0.0045 (3) | 0.0063 (3) | 0.0084 (3) |
| N1 | 0.0135 (14) | 0.0212 (14) | 0.0138 (13) | 0.0050 (11) | 0.0049 (10) | 0.0073 (11) |
| C1 | 0.0168 (16) | 0.0153 (15) | 0.0155 (15) | 0.0025 (12) | 0.0057 (12) | 0.0085 (13) |
| C2 | 0.0175 (17) | 0.0210 (16) | 0.0151 (16) | 0.0036 (13) | 0.0056 (13) | 0.0058 (14) |
| C3 | 0.0156 (17) | 0.0260 (17) | 0.0147 (15) | 0.0079 (13) | 0.0062 (12) | 0.0086 (14) |
| C4 | 0.0197 (17) | 0.0199 (16) | 0.0145 (15) | 0.0068 (13) | 0.0062 (13) | 0.0077 (13) |
| C5 | 0.0224 (18) | 0.0199 (16) | 0.0170 (16) | 0.0042 (13) | 0.0077 (13) | 0.0054 (14) |
| C6 | 0.0201 (19) | 0.0315 (19) | 0.0225 (18) | 0.0079 (15) | 0.0067 (14) | 0.0057 (16) |
| C7 | 0.028 (2) | 0.041 (2) | 0.0168 (17) | 0.0185 (17) | 0.0045 (15) | 0.0091 (16) |
| C8 | 0.035 (2) | 0.043 (2) | 0.0243 (19) | 0.0203 (18) | 0.0115 (16) | 0.0225 (18) |
| C9 | 0.027 (2) | 0.0289 (18) | 0.0237 (18) | 0.0109 (15) | 0.0102 (15) | 0.0175 (16) |
| C10 | 0.0218 (18) | 0.0128 (14) | 0.0159 (15) | 0.0054 (12) | 0.0090 (13) | 0.0080 (12) |
| C11 | 0.0209 (18) | 0.0190 (16) | 0.0211 (17) | 0.0069 (13) | 0.0052 (14) | 0.0105 (14) |
| C12 | 0.0243 (19) | 0.0223 (17) | 0.0285 (19) | 0.0087 (14) | 0.0117 (15) | 0.0124 (15) |
| C13 | 0.030 (2) | 0.0157 (15) | 0.0219 (17) | 0.0062 (14) | 0.0130 (15) | 0.0071 (14) |
| C14 | 0.0243 (19) | 0.0198 (16) | 0.0166 (16) | 0.0027 (14) | 0.0037 (13) | 0.0084 (14) |
| C15 | 0.0199 (18) | 0.0184 (16) | 0.0200 (16) | 0.0053 (13) | 0.0060 (13) | 0.0108 (14) |
| C16 | 0.0155 (17) | 0.0200 (16) | 0.0204 (16) | 0.0062 (13) | 0.0053 (12) | 0.0143 (14) |
| C17 | 0.0193 (18) | 0.0195 (16) | 0.0206 (16) | 0.0062 (13) | 0.0061 (13) | 0.0125 (14) |
| C18 | 0.0196 (18) | 0.0260 (18) | 0.0286 (19) | 0.0013 (14) | 0.0043 (14) | 0.0185 (16) |
| C19 | 0.0173 (18) | 0.037 (2) | 0.0321 (19) | 0.0067 (15) | 0.0089 (14) | 0.0270 (17) |
| C20 | 0.026 (2) | 0.0334 (19) | 0.0237 (18) | 0.0115 (15) | 0.0123 (15) | 0.0189 (16) |
| C21 | 0.0233 (18) | 0.0206 (16) | 0.0202 (16) | 0.0052 (13) | 0.0079 (13) | 0.0122 (14) |
| C22 | 0.0216 (18) | 0.0187 (15) | 0.0169 (15) | 0.0095 (13) | 0.0105 (13) | 0.0113 (13) |
| C23 | 0.0222 (18) | 0.0218 (17) | 0.0239 (17) | 0.0069 (14) | 0.0073 (14) | 0.0132 (15) |
| C24 | 0.027 (2) | 0.032 (2) | 0.0281 (19) | 0.0129 (16) | 0.0090 (15) | 0.0191 (17) |
| C25 | 0.032 (2) | 0.0299 (19) | 0.0308 (19) | 0.0190 (16) | 0.0162 (16) | 0.0216 (17) |
| C26 | 0.030 (2) | 0.0188 (16) | 0.0276 (18) | 0.0067 (14) | 0.0125 (15) | 0.0134 (15) |
| C27 | 0.0203 (18) | 0.0218 (17) | 0.0244 (17) | 0.0077 (14) | 0.0080 (14) | 0.0142 (15) |
| C28 | 0.0176 (17) | 0.0152 (14) | 0.0173 (15) | 0.0045 (12) | 0.0047 (12) | 0.0108 (13) |
| C29 | 0.0180 (17) | 0.0222 (16) | 0.0182 (16) | 0.0062 (13) | 0.0068 (13) | 0.0114 (14) |
| C30 | 0.0228 (18) | 0.0239 (17) | 0.0165 (16) | 0.0060 (14) | 0.0044 (13) | 0.0104 (14) |
| C31 | 0.0158 (17) | 0.0238 (17) | 0.0240 (17) | 0.0056 (13) | 0.0027 (13) | 0.0143 (15) |
| C32 | 0.0203 (18) | 0.0236 (17) | 0.0252 (18) | 0.0114 (14) | 0.0110 (14) | 0.0154 (15) |
| C33 | 0.0199 (17) | 0.0192 (15) | 0.0199 (16) | 0.0070 (13) | 0.0080 (13) | 0.0132 (14) |
| C34 | 0.0129 (16) | 0.0172 (15) | 0.0152 (15) | 0.0041 (12) | 0.0048 (12) | 0.0075 (13) |
| C35 | 0.0275 (19) | 0.0193 (16) | 0.0176 (16) | 0.0062 (13) | 0.0086 (14) | 0.0109 (14) |
| C36 | 0.031 (2) | 0.0309 (19) | 0.0186 (17) | 0.0078 (15) | 0.0111 (14) | 0.0162 (15) |
| C37 | 0.0231 (18) | 0.0237 (17) | 0.0152 (16) | 0.0053 (14) | 0.0097 (13) | 0.0059 (14) |
| C38 | 0.0239 (19) | 0.0192 (16) | 0.0224 (17) | 0.0064 (13) | 0.0102 (14) | 0.0093 (14) |
| C39 | 0.0196 (17) | 0.0208 (16) | 0.0169 (15) | 0.0051 (13) | 0.0068 (13) | 0.0109 (14) |
| Cl1 | 0.0397 (6) | 0.0299 (5) | 0.0451 (6) | −0.0012 (4) | 0.0001 (5) | 0.0112 (5) |
| Cl2 | 0.0608 (7) | 0.0342 (5) | 0.0547 (7) | 0.0180 (5) | 0.0278 (6) | 0.0276 (5) |
| Cl3 | 0.0355 (5) | 0.0420 (5) | 0.0346 (5) | −0.0020 (4) | 0.0028 (4) | 0.0230 (4) |
| C40 | 0.035 (2) | 0.030 (2) | 0.0264 (19) | 0.0009 (16) | 0.0048 (16) | 0.0183 (17) |
Geometric parameters (Å, °)
| Au1—P2 | 2.2994 (8) | C17—H17A | 0.9500 |
| Au1—P1 | 2.3233 (8) | C18—C19 | 1.388 (6) |
| Au1—S2 | 2.6133 (8) | C18—H18A | 0.9500 |
| Au1—S1 | 2.7414 (8) | C19—C20 | 1.384 (5) |
| S1—C1 | 1.706 (3) | C19—H19A | 0.9500 |
| S2—C1 | 1.718 (3) | C20—C21 | 1.383 (5) |
| P1—C10 | 1.818 (3) | C20—H20A | 0.9500 |
| P1—C4 | 1.823 (4) | C21—H21A | 0.9500 |
| P1—C16 | 1.825 (3) | C22—C23 | 1.386 (5) |
| P2—C34 | 1.822 (3) | C22—C27 | 1.398 (5) |
| P2—C28 | 1.823 (3) | C23—C24 | 1.393 (5) |
| P2—C22 | 1.829 (3) | C23—H23A | 0.9500 |
| N1—C1 | 1.354 (4) | C24—C25 | 1.382 (6) |
| N1—C3i | 1.454 (4) | C24—H24A | 0.9500 |
| N1—C2 | 1.460 (4) | C25—C26 | 1.389 (6) |
| C2—C3 | 1.524 (5) | C25—H25A | 0.9500 |
| C2—H2AB | 0.9900 | C26—C27 | 1.397 (5) |
| C2—H2AA | 0.9900 | C26—H26A | 0.9500 |
| C3—N1i | 1.454 (4) | C27—H27A | 0.9500 |
| C3—H3AA | 0.9900 | C28—C33 | 1.392 (5) |
| C3—H3AB | 0.9900 | C28—C29 | 1.403 (5) |
| C4—C5 | 1.393 (5) | C29—C30 | 1.385 (5) |
| C4—C9 | 1.405 (5) | C29—H29A | 0.9500 |
| C5—C6 | 1.392 (5) | C30—C31 | 1.387 (5) |
| C5—H5AA | 0.9500 | C30—H30A | 0.9500 |
| C6—C7 | 1.378 (6) | C31—C32 | 1.381 (5) |
| C6—H6AA | 0.9500 | C31—H31A | 0.9500 |
| C7—C8 | 1.386 (6) | C32—C33 | 1.397 (5) |
| C7—H7AA | 0.9500 | C32—H32A | 0.9500 |
| C8—C9 | 1.380 (5) | C33—H33A | 0.9500 |
| C8—H8AA | 0.9500 | C34—C39 | 1.390 (5) |
| C9—H9AA | 0.9500 | C34—C35 | 1.400 (5) |
| C10—C11 | 1.381 (5) | C35—C36 | 1.390 (5) |
| C10—C15 | 1.403 (5) | C35—H35A | 0.9500 |
| C11—C12 | 1.402 (5) | C36—C37 | 1.383 (5) |
| C11—H11A | 0.9500 | C36—H36A | 0.9500 |
| C12—C13 | 1.382 (5) | C37—C38 | 1.389 (5) |
| C12—H12A | 0.9500 | C37—H37A | 0.9500 |
| C13—C14 | 1.378 (5) | C38—C39 | 1.392 (5) |
| C13—H13A | 0.9500 | C38—H38A | 0.9500 |
| C14—C15 | 1.390 (5) | C39—H39A | 0.9500 |
| C14—H14A | 0.9500 | Cl1—C40 | 1.758 (4) |
| C15—H15A | 0.9500 | Cl2—C40 | 1.754 (4) |
| C16—C17 | 1.386 (5) | Cl3—C40 | 1.762 (4) |
| C16—C21 | 1.402 (5) | C40—H40A | 1.0000 |
| C17—C18 | 1.398 (5) | ||
| P2—Au1—P1 | 134.65 (3) | C21—C16—P1 | 116.5 (3) |
| P2—Au1—S2 | 116.81 (3) | C16—C17—C18 | 120.1 (3) |
| P1—Au1—S2 | 107.10 (3) | C16—C17—H17A | 120.0 |
| P2—Au1—S1 | 107.34 (3) | C18—C17—H17A | 120.0 |
| P1—Au1—S1 | 99.39 (3) | C19—C18—C17 | 120.2 (3) |
| S2—Au1—S1 | 67.03 (2) | C19—C18—H18A | 119.9 |
| C1—S1—Au1 | 84.64 (11) | C17—C18—H18A | 119.9 |
| C1—S2—Au1 | 88.55 (12) | C20—C19—C18 | 119.6 (3) |
| C10—P1—C4 | 103.04 (16) | C20—C19—H19A | 120.2 |
| C10—P1—C16 | 106.19 (15) | C18—C19—H19A | 120.2 |
| C4—P1—C16 | 103.85 (15) | C21—C20—C19 | 120.6 (3) |
| C10—P1—Au1 | 113.94 (10) | C21—C20—H20A | 119.7 |
| C4—P1—Au1 | 112.18 (11) | C19—C20—H20A | 119.7 |
| C16—P1—Au1 | 116.33 (11) | C20—C21—C16 | 120.1 (3) |
| C34—P2—C28 | 104.10 (15) | C20—C21—H21A | 120.0 |
| C34—P2—C22 | 104.72 (15) | C16—C21—H21A | 120.0 |
| C28—P2—C22 | 103.69 (15) | C23—C22—C27 | 119.1 (3) |
| C34—P2—Au1 | 113.39 (11) | C23—C22—P2 | 118.6 (3) |
| C28—P2—Au1 | 109.59 (10) | C27—C22—P2 | 122.3 (3) |
| C22—P2—Au1 | 119.85 (11) | C22—C23—C24 | 120.6 (3) |
| C1—N1—C3i | 123.8 (3) | C22—C23—H23A | 119.7 |
| C1—N1—C2 | 122.7 (3) | C24—C23—H23A | 119.7 |
| C3i—N1—C2 | 113.0 (3) | C25—C24—C23 | 120.2 (4) |
| N1—C1—S1 | 120.2 (2) | C25—C24—H24A | 119.9 |
| N1—C1—S2 | 120.3 (3) | C23—C24—H24A | 119.9 |
| S1—C1—S2 | 119.56 (19) | C24—C25—C26 | 119.9 (3) |
| N1—C2—C3 | 110.7 (3) | C24—C25—H25A | 120.1 |
| N1—C2—H2AB | 109.5 | C26—C25—H25A | 120.1 |
| C3—C2—H2AB | 109.5 | C25—C26—C27 | 119.9 (3) |
| N1—C2—H2AA | 109.5 | C25—C26—H26A | 120.0 |
| C3—C2—H2AA | 109.5 | C27—C26—H26A | 120.0 |
| H2AB—C2—H2AA | 108.1 | C26—C27—C22 | 120.2 (3) |
| N1i—C3—C2 | 110.2 (3) | C26—C27—H27A | 119.9 |
| N1i—C3—H3AA | 109.6 | C22—C27—H27A | 119.9 |
| C2—C3—H3AA | 109.6 | C33—C28—C29 | 119.1 (3) |
| N1i—C3—H3AB | 109.6 | C33—C28—P2 | 123.2 (3) |
| C2—C3—H3AB | 109.6 | C29—C28—P2 | 117.7 (3) |
| H3AA—C3—H3AB | 108.1 | C30—C29—C28 | 120.4 (3) |
| C5—C4—C9 | 119.1 (3) | C30—C29—H29A | 119.8 |
| C5—C4—P1 | 119.3 (3) | C28—C29—H29A | 119.8 |
| C9—C4—P1 | 121.6 (3) | C29—C30—C31 | 119.9 (3) |
| C6—C5—C4 | 120.2 (3) | C29—C30—H30A | 120.0 |
| C6—C5—H5AA | 119.9 | C31—C30—H30A | 120.0 |
| C4—C5—H5AA | 119.9 | C32—C31—C30 | 120.5 (3) |
| C7—C6—C5 | 120.2 (4) | C32—C31—H31A | 119.8 |
| C7—C6—H6AA | 119.9 | C30—C31—H31A | 119.8 |
| C5—C6—H6AA | 119.9 | C31—C32—C33 | 119.9 (3) |
| C6—C7—C8 | 120.0 (4) | C31—C32—H32A | 120.0 |
| C6—C7—H7AA | 120.0 | C33—C32—H32A | 120.0 |
| C8—C7—H7AA | 120.0 | C28—C33—C32 | 120.2 (3) |
| C9—C8—C7 | 120.5 (4) | C28—C33—H33A | 119.9 |
| C9—C8—H8AA | 119.8 | C32—C33—H33A | 119.9 |
| C7—C8—H8AA | 119.8 | C39—C34—C35 | 119.2 (3) |
| C8—C9—C4 | 119.9 (4) | C39—C34—P2 | 118.2 (2) |
| C8—C9—H9AA | 120.0 | C35—C34—P2 | 122.6 (2) |
| C4—C9—H9AA | 120.0 | C36—C35—C34 | 120.0 (3) |
| C11—C10—C15 | 119.0 (3) | C36—C35—H35A | 120.0 |
| C11—C10—P1 | 122.6 (3) | C34—C35—H35A | 120.0 |
| C15—C10—P1 | 118.0 (3) | C37—C36—C35 | 120.2 (3) |
| C10—C11—C12 | 120.6 (3) | C37—C36—H36A | 119.9 |
| C10—C11—H11A | 119.7 | C35—C36—H36A | 119.9 |
| C12—C11—H11A | 119.7 | C36—C37—C38 | 120.3 (3) |
| C13—C12—C11 | 119.8 (3) | C36—C37—H37A | 119.8 |
| C13—C12—H12A | 120.1 | C38—C37—H37A | 119.8 |
| C11—C12—H12A | 120.1 | C37—C38—C39 | 119.5 (3) |
| C14—C13—C12 | 120.1 (3) | C37—C38—H38A | 120.2 |
| C14—C13—H13A | 120.0 | C39—C38—H38A | 120.2 |
| C12—C13—H13A | 120.0 | C34—C39—C38 | 120.7 (3) |
| C13—C14—C15 | 120.4 (3) | C34—C39—H39A | 119.6 |
| C13—C14—H14A | 119.8 | C38—C39—H39A | 119.6 |
| C15—C14—H14A | 119.8 | Cl2—C40—Cl1 | 110.6 (2) |
| C14—C15—C10 | 120.1 (3) | Cl2—C40—Cl3 | 110.7 (2) |
| C14—C15—H15A | 120.0 | Cl1—C40—Cl3 | 110.3 (2) |
| C10—C15—H15A | 120.0 | Cl2—C40—H40A | 108.4 |
| C17—C16—C21 | 119.4 (3) | Cl1—C40—H40A | 108.4 |
| C17—C16—P1 | 124.1 (3) | Cl3—C40—H40A | 108.4 |
| P2—Au1—S1—C1 | 115.18 (11) | C13—C14—C15—C10 | −0.8 (5) |
| P1—Au1—S1—C1 | −101.89 (11) | C11—C10—C15—C14 | 0.7 (5) |
| S2—Au1—S1—C1 | 2.79 (11) | P1—C10—C15—C14 | 173.9 (3) |
| P2—Au1—S2—C1 | −101.30 (11) | C10—P1—C16—C17 | −7.7 (3) |
| P1—Au1—S2—C1 | 90.39 (11) | C4—P1—C16—C17 | −116.0 (3) |
| S1—Au1—S2—C1 | −2.76 (11) | Au1—P1—C16—C17 | 120.2 (3) |
| P2—Au1—P1—C10 | −40.63 (13) | C10—P1—C16—C21 | 174.0 (3) |
| S2—Au1—P1—C10 | 124.64 (13) | C4—P1—C16—C21 | 65.7 (3) |
| S1—Au1—P1—C10 | −166.64 (13) | Au1—P1—C16—C21 | −58.0 (3) |
| P2—Au1—P1—C4 | 75.96 (12) | C21—C16—C17—C18 | −0.4 (5) |
| S2—Au1—P1—C4 | −118.77 (12) | P1—C16—C17—C18 | −178.6 (3) |
| S1—Au1—P1—C4 | −50.05 (12) | C16—C17—C18—C19 | −0.6 (5) |
| P2—Au1—P1—C16 | −164.70 (12) | C17—C18—C19—C20 | 0.4 (5) |
| S2—Au1—P1—C16 | 0.57 (13) | C18—C19—C20—C21 | 0.7 (5) |
| S1—Au1—P1—C16 | 69.29 (13) | C19—C20—C21—C16 | −1.7 (5) |
| P1—Au1—P2—C34 | 48.64 (13) | C17—C16—C21—C20 | 1.6 (5) |
| S2—Au1—P2—C34 | −115.56 (12) | P1—C16—C21—C20 | 179.9 (3) |
| S1—Au1—P2—C34 | 171.92 (12) | C34—P2—C22—C23 | 74.3 (3) |
| P1—Au1—P2—C28 | −67.20 (12) | C28—P2—C22—C23 | −176.8 (3) |
| S2—Au1—P2—C28 | 128.61 (11) | Au1—P2—C22—C23 | −54.3 (3) |
| S1—Au1—P2—C28 | 56.08 (11) | C34—P2—C22—C27 | −105.8 (3) |
| P1—Au1—P2—C22 | 173.21 (13) | C28—P2—C22—C27 | 3.1 (3) |
| S2—Au1—P2—C22 | 9.01 (13) | Au1—P2—C22—C27 | 125.6 (3) |
| S1—Au1—P2—C22 | −63.52 (13) | C27—C22—C23—C24 | 2.8 (5) |
| C3i—N1—C1—S1 | 178.5 (2) | P2—C22—C23—C24 | −177.3 (3) |
| C2—N1—C1—S1 | 6.3 (4) | C22—C23—C24—C25 | −2.4 (6) |
| C3i—N1—C1—S2 | −2.2 (4) | C23—C24—C25—C26 | 0.1 (6) |
| C2—N1—C1—S2 | −174.4 (2) | C24—C25—C26—C27 | 1.8 (5) |
| Au1—S1—C1—N1 | 174.8 (3) | C25—C26—C27—C22 | −1.5 (5) |
| Au1—S1—C1—S2 | −4.50 (17) | C23—C22—C27—C26 | −0.8 (5) |
| Au1—S2—C1—N1 | −174.6 (3) | P2—C22—C27—C26 | 179.3 (3) |
| Au1—S2—C1—S1 | 4.70 (18) | C34—P2—C28—C33 | 2.9 (3) |
| C1—N1—C2—C3 | −131.2 (3) | C22—P2—C28—C33 | −106.4 (3) |
| C3i—N1—C2—C3 | 55.8 (4) | Au1—P2—C28—C33 | 124.5 (2) |
| N1—C2—C3—N1i | −54.2 (4) | C34—P2—C28—C29 | −175.9 (2) |
| C10—P1—C4—C5 | 92.7 (3) | C22—P2—C28—C29 | 74.8 (3) |
| C16—P1—C4—C5 | −156.7 (3) | Au1—P2—C28—C29 | −54.3 (3) |
| Au1—P1—C4—C5 | −30.2 (3) | C33—C28—C29—C30 | 0.5 (5) |
| C10—P1—C4—C9 | −85.0 (3) | P2—C28—C29—C30 | 179.4 (3) |
| C16—P1—C4—C9 | 25.6 (3) | C28—C29—C30—C31 | −0.6 (5) |
| Au1—P1—C4—C9 | 152.0 (3) | C29—C30—C31—C32 | 0.3 (5) |
| C9—C4—C5—C6 | 0.6 (5) | C30—C31—C32—C33 | 0.1 (5) |
| P1—C4—C5—C6 | −177.2 (3) | C29—C28—C33—C32 | −0.1 (5) |
| C4—C5—C6—C7 | −0.7 (5) | P2—C28—C33—C32 | −178.9 (2) |
| C5—C6—C7—C8 | 0.1 (6) | C31—C32—C33—C28 | −0.2 (5) |
| C6—C7—C8—C9 | 0.6 (6) | C28—P2—C34—C39 | 86.8 (3) |
| C7—C8—C9—C4 | −0.6 (6) | C22—P2—C34—C39 | −164.7 (3) |
| C5—C4—C9—C8 | 0.0 (5) | Au1—P2—C34—C39 | −32.3 (3) |
| P1—C4—C9—C8 | 177.8 (3) | C28—P2—C34—C35 | −91.1 (3) |
| C4—P1—C10—C11 | −8.8 (3) | C22—P2—C34—C35 | 17.5 (3) |
| C16—P1—C10—C11 | −117.7 (3) | Au1—P2—C34—C35 | 149.9 (3) |
| Au1—P1—C10—C11 | 113.0 (3) | C39—C34—C35—C36 | −0.6 (5) |
| C4—P1—C10—C15 | 178.2 (3) | P2—C34—C35—C36 | 177.2 (3) |
| C16—P1—C10—C15 | 69.4 (3) | C34—C35—C36—C37 | −0.9 (6) |
| Au1—P1—C10—C15 | −60.0 (3) | C35—C36—C37—C38 | 1.0 (6) |
| C15—C10—C11—C12 | 0.0 (5) | C36—C37—C38—C39 | 0.5 (6) |
| P1—C10—C11—C12 | −172.9 (3) | C35—C34—C39—C38 | 2.1 (5) |
| C10—C11—C12—C13 | −0.5 (5) | P2—C34—C39—C38 | −175.8 (3) |
| C11—C12—C13—C14 | 0.3 (5) | C37—C38—C39—C34 | −2.0 (5) |
| C12—C13—C14—C15 | 0.3 (5) |
Symmetry codes: (i) −x, −y+2, −z+2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NG5253).
References
- Allen, F. H. (2002). Acta Cryst. B58, 380–388. [DOI] [PubMed]
- Brandenburg, K. (1999). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2007). APEX2, SADABS and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruno, I. J., Cole, J. C., Edgington, P. R., Kessler, M., Macrae, C. F., McCabe, P., Pearson, J. & Taylor, R. (2002). Acta Cryst. B58, 389–397. [DOI] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1358–1367.
- Fernandez, E. J., Lopez-de-Luzuriaga, J. M., Monge, M., Olmos, E., Gimeno, M. C., Laguna, A. & Jones, P. G. (1998). Inorg. Chem. 37, 5532–5536. [DOI] [PubMed]
- Guzei, I. A. (2007). In-house Crystallographic Programs: FCF_filter, INSerter and modiCIFer Molecular Structure Laboratory, University of Wisconsin-Madison, Madison, Wisconsin, USA.
- Jian, F., Lu, L., Wang, X., Shanmuga Sundara Raj, S., Razak, I. A. & Fun, H.-K. (2000). Acta Cryst. C56, 939–940. [DOI] [PubMed]
- Knight, E. R., Leung, N. H., Lin, Y. H., Cowley, A. R., Watkins, D. J., Thompson, A. L., Hogarth, G. & Wilton-Ely, J. D. E. T. (2009a). Dalton Trans. pp. 3688–3697. [DOI] [PubMed]
- Knight, E. R., Leung, N. H., Thompson, A. L., Hogarth, G. & Wilton-Ely, J. D. E. T. (2009b). Inorg. Chem. 48, 3866–3874. [DOI] [PubMed]
- Kumar, A., Mayer-Figge, H., Sheldrick, W. S. & Singh, N. (2009). Eur. J. Inorg. Chem. pp. 2720–2725.
- Oliver, K., White, A. J. P., Hogarth, G. & Wilton-Ely, J. D. E. T. (2011). Dalton Trans. 40, 5852–5864. [DOI] [PubMed]
- Razak, I. A., Shanmuga Sundara Raj, S., Fun, H.-K., Jian, F., Bei, F., Yang, X., Lu, L. & Wang, X. (2000). Acta Cryst. C56, 666–667.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Wilton-Ely, J. D. E. T., Solanki, D., Knight, E. R., Holt, K. B., Thompson, A. L. & Hogarth, G. (2008). Inorg. Chem. 47, 9642–9653. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811044229/ng5253sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811044229/ng5253Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report