Abstract
The asymmetric unit of the title co-crystal, C7H6N2·C7H4N2O6, contains two formula units of both components. The crystal structure is stabilized by intermolecular O—H⋯O, N—H⋯O, N—H⋯N and C—H⋯O hydrogen bonds, generating a two-dimensional wave-like network. π–π stacking interactions [centroid–centroid distances = 3.702 (2), 3.660 (2)and 3.671 (2) Å] stabilize the crystal packing.
Related literature
For general background to hydrogen bonding, see: Desiraju (2002 ▶); Prins et al. (2001 ▶); Steiner (2002 ▶). For background to the applications of co-crystals, see: Bhatt & Desiraju (2008 ▶); Etter & Baures (1988 ▶); Gao et al. (2004 ▶); Hori et al. (2009 ▶); Weyna et al. (2009 ▶). For the synthesis of co-crystals by complementary functional groups, see: Li et al. (2006 ▶); Roy et al. (2009 ▶); Wei (2007 ▶).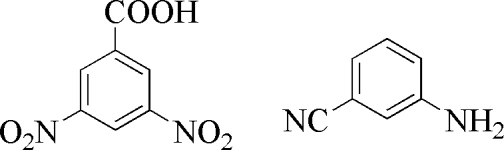
Experimental
Crystal data
C7H6N2·C7H4N2O6
M r = 330.26
Triclinic,

a = 7.4547 (15) Å
b = 14.260 (3) Å
c = 14.845 (3) Å
α = 108.01 (3)°
β = 91.90 (3)°
γ = 93.37 (3)°
V = 1496.0 (5) Å3
Z = 4
Mo Kα radiation
μ = 0.12 mm−1
T = 293 K
0.35 × 0.22 × 0.20 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.969, T max = 0.977
15550 measured reflections
6830 independent reflections
3195 reflections with I > 2σ(I)
R int = 0.056
Refinement
R[F 2 > 2σ(F 2)] = 0.062
wR(F 2) = 0.159
S = 0.99
6830 reflections
461 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.16 e Å−3
Δρmin = −0.19 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811039870/kp2355sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811039870/kp2355Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811039870/kp2355Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1A⋯O2i | 1.18 (4) | 1.41 (4) | 2.590 (2) | 173 (3) |
| N6—H6A⋯N7ii | 0.92 (3) | 2.32 (4) | 3.232 (5) | 169 (3) |
| N6—H6B⋯O12iii | 0.90 (3) | 2.57 (3) | 2.953 (4) | 107 (2) |
| N8—H8A⋯N5iv | 0.91 (3) | 2.37 (3) | 3.262 (5) | 170 (3) |
| N8—H8B⋯O5iii | 0.85 (3) | 2.48 (4) | 3.286 (4) | 157 (3) |
| O7—H9A⋯O8iv | 1.23 (5) | 1.38 (5) | 2.608 (2) | 174 (4) |
| C5—H5A⋯O4v | 0.93 | 2.44 | 3.321 (3) | 158 |
| C12—H12A⋯O11ii | 0.93 | 2.50 | 3.352 (3) | 153 |
| C18—H18A⋯O2vi | 0.96 (3) | 2.58 (3) | 3.421 (4) | 147 (2) |
| C21—H21A⋯O10vii | 0.93 | 2.60 | 3.451 (3) | 153 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  ; (vi)
; (vi)  ; (vii)
; (vii)  .
.
Acknowledgments
This work was supported by NJ210003, BJ211008 and in part by the National Basic Research Program of China (2009CB930601).
supplementary crystallographic information
Comment
The self-assembly of two or more different types of molecules to form a multi-component crystal is greatly fascinating to chemists, known as cocrystals. The considerable effort has been devoted to cocrystal formation over decades, due to its extensive applications in construction of organic solid-state materials, such as in the pharmaceutical industry (Weyna et al., 2009), in organic synthesis (Gao et al., 2004), for promoting crystal growth (Etter & Baures, 1988), as luminescent materials (Hori et al., 2009), and for absolute structure determination (Bhatt & Desiraju, 2008). One of the important ways is the utilisation of self-assembly of small molecules through intermolecular interactions to construct cocrystals, which are one-, two- or three-dimensional networks. In the study of intermolecular interactions the central role is the hydrogen bond. The simple way of preparation of cocrystals is to employ the components containing functional groups with hydrogen bonding capability (Li et al., 2006; Roy et al., 2009; Wei, 2007), such as –COOH and –NH2, which can easily result in O—H···N and N—H···O hydrogen bonds.
In this report we have established unambiguously the structure of the cocrystal 3,5-dinitrobenzoic acid with 3-aminobenzonitrile in the solid state by X-ray diffraction analysis. An asymmetric unit of the title compound contains two 3,5-dinitrobenzoic acid and two 3-aminobenzonitrile (Fig. 1). Intermolecular O—H···O, N—H···O, N—H···N and C—H···O hydrogen bonds are observed (Fig. 2, Table 1). The hydrogen bonds O—H···O between carboxyl groups results in the dimerization of 3,5-dinitrobenzoic acid. Extensive hydrogen-bonding interactions generate a two-dimensional wave-like network (Fig. 3).
Additionally, the crystal packing is stabilised by aromatic π–π stacking interactions involving the rings of the asymmetric unit with separation distances between their centroids: Cg1(C2→ C7)···Cg3 (C16→ C21) of 3.702 (2) Å, Cg2(C9→ C14)··· Cg3(C16→ C21) of 3.660 (2)Å, and Cg2(C9→ C14)···Cg4(C23→ C28) of 3.671 (2) Å.
Experimental
The cocrystals were prepared from stoichiometric amounts of components in water mixed with metanol (in volume ratio 1:1) and left to evaporate slowly at room temperature.
Refinement
Aromatic H atoms were placed in calculated positions with C—H = 0.93 Å, and refined in riding mode with Uiso(H) = 1.2Ueq(C). H atoms involved in hydrogen bonds were located from differential Fourier maps and refined isotropically.
Figures
Fig. 1.
The molecular structure of the title cocrystal with the numbering scheme and 30% probability displacement ellipsoids.
Fig. 2.
The packing diagram of the molecules viewed along the b axis. Hydrogen bonds are drawn as dashed lines.
Fig. 3.
Packing diagram of the molecules. Hydrogen bonds are drawn as dashed lines.
Crystal data
| C7H6N2·C7H4N2O6 | Z = 4 |
| Mr = 330.26 | F(000) = 680 |
| Triclinic, P1 | Dx = 1.466 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.4547 (15) Å | Cell parameters from 6830 reflections |
| b = 14.260 (3) Å | θ = 3.0–27.5° |
| c = 14.845 (3) Å | µ = 0.12 mm−1 |
| α = 108.01 (3)° | T = 293 K |
| β = 91.90 (3)° | Block, colorless |
| γ = 93.37 (3)° | 0.35 × 0.22 × 0.20 mm |
| V = 1496.0 (5) Å3 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 6830 independent reflections |
| Radiation source: fine-focus sealed tube | 3195 reflections with I > 2σ(I) |
| graphite | Rint = 0.056 |
| Detector resolution: 8.366 pixels mm-1 | θmax = 27.5°, θmin = 3.0° |
| φ and ω scans | h = −9→9 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | k = −18→18 |
| Tmin = 0.969, Tmax = 0.977 | l = −19→19 |
| 15550 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.062 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.159 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.99 | w = 1/[σ2(Fo2) + (0.0591P)2] where P = (Fo2 + 2Fc2)/3 |
| 6830 reflections | (Δ/σ)max < 0.001 |
| 461 parameters | Δρmax = 0.16 e Å−3 |
| 0 restraints | Δρmin = −0.19 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.4080 (3) | 0.01643 (18) | 0.11993 (16) | 0.0553 (6) | |
| C2 | 0.3392 (3) | 0.02905 (17) | 0.21487 (15) | 0.0490 (6) | |
| C3 | 0.1637 (3) | −0.00114 (17) | 0.22396 (15) | 0.0490 (6) | |
| H3A | 0.0862 | −0.0291 | 0.1707 | 0.059* | |
| C4 | 0.1058 (3) | 0.01088 (17) | 0.31324 (16) | 0.0506 (6) | |
| C5 | 0.2159 (3) | 0.05067 (18) | 0.39350 (16) | 0.0578 (6) | |
| H5A | 0.1751 | 0.0569 | 0.4534 | 0.069* | |
| C6 | 0.3888 (3) | 0.08091 (18) | 0.38155 (16) | 0.0544 (6) | |
| C7 | 0.4516 (3) | 0.07230 (17) | 0.29418 (16) | 0.0555 (6) | |
| H7A | 0.5686 | 0.0953 | 0.2884 | 0.067* | |
| O1 | 0.3036 (2) | −0.03099 (14) | 0.04900 (12) | 0.0726 (6) | |
| H1A | 0.363 (5) | −0.035 (3) | −0.025 (3) | 0.160 (14)* | |
| O2 | 0.5615 (2) | 0.05235 (14) | 0.11410 (11) | 0.0736 (5) | |
| N1 | −0.0818 (3) | −0.01956 (18) | 0.32279 (18) | 0.0721 (6) | |
| O3 | −0.1815 (2) | −0.04676 (16) | 0.25263 (16) | 0.0915 (7) | |
| O4 | −0.1288 (3) | −0.0171 (2) | 0.40020 (16) | 0.1302 (10) | |
| N2 | 0.5102 (3) | 0.12590 (18) | 0.46559 (15) | 0.0768 (7) | |
| O5 | 0.4669 (3) | 0.11480 (19) | 0.53961 (14) | 0.1112 (8) | |
| O6 | 0.6490 (3) | 0.16961 (19) | 0.45609 (15) | 0.1188 (9) | |
| C8 | 0.3501 (3) | 0.50842 (18) | 0.10190 (17) | 0.0544 (6) | |
| C9 | 0.2362 (3) | 0.50677 (16) | 0.18160 (15) | 0.0488 (6) | |
| C10 | 0.0604 (3) | 0.46720 (17) | 0.16271 (16) | 0.0536 (6) | |
| H10A | 0.0117 | 0.4431 | 0.1007 | 0.064* | |
| C11 | −0.0412 (3) | 0.46423 (18) | 0.23756 (17) | 0.0527 (6) | |
| C12 | 0.0242 (3) | 0.49980 (17) | 0.33016 (16) | 0.0544 (6) | |
| H12A | −0.0470 | 0.4979 | 0.3800 | 0.065* | |
| C13 | 0.1991 (3) | 0.53816 (17) | 0.34579 (15) | 0.0527 (6) | |
| C14 | 0.3074 (3) | 0.54240 (17) | 0.27369 (16) | 0.0517 (6) | |
| H14A | 0.4260 | 0.5687 | 0.2868 | 0.062* | |
| N3 | −0.2260 (3) | 0.41929 (18) | 0.21744 (18) | 0.0714 (6) | |
| O7 | 0.2803 (2) | 0.47150 (15) | 0.01875 (13) | 0.0800 (6) | |
| H9A | 0.379 (6) | 0.469 (3) | −0.047 (3) | 0.203 (18)* | |
| O8 | 0.5087 (2) | 0.54478 (14) | 0.12114 (12) | 0.0714 (5) | |
| O9 | −0.2761 (2) | 0.37659 (17) | 0.13605 (16) | 0.0941 (7) | |
| O10 | −0.3199 (2) | 0.42753 (18) | 0.28450 (15) | 0.1015 (7) | |
| O11 | 0.1742 (3) | 0.58417 (17) | 0.50753 (13) | 0.1032 (7) | |
| O12 | 0.4324 (3) | 0.6007 (2) | 0.45542 (14) | 0.1205 (9) | |
| N4 | 0.2750 (3) | 0.57692 (17) | 0.44339 (15) | 0.0744 (6) | |
| N5 | 0.3740 (4) | 0.2057 (2) | −0.0240 (2) | 0.1150 (10) | |
| C15 | 0.2997 (4) | 0.2144 (2) | 0.0436 (2) | 0.0796 (9) | |
| C16 | 0.2076 (3) | 0.22522 (19) | 0.12900 (18) | 0.0599 (7) | |
| C17 | 0.0298 (4) | 0.1905 (2) | 0.1228 (2) | 0.0735 (8) | |
| H17A | −0.0302 | 0.1603 | 0.0641 | 0.088* | |
| C18 | −0.0566 (4) | 0.2014 (2) | 0.2050 (2) | 0.0765 (8) | |
| H18A | −0.180 (4) | 0.1780 (19) | 0.2036 (17) | 0.086 (9)* | |
| C19 | 0.0298 (4) | 0.24544 (19) | 0.2909 (2) | 0.0685 (7) | |
| H19A | −0.0323 | 0.2521 | 0.3456 | 0.082* | |
| C20 | 0.2079 (3) | 0.28052 (17) | 0.29893 (18) | 0.0596 (7) | |
| C21 | 0.2961 (3) | 0.27005 (18) | 0.21579 (18) | 0.0610 (7) | |
| H21A | 0.4159 | 0.2936 | 0.2189 | 0.073* | |
| N6 | 0.2934 (5) | 0.3273 (2) | 0.3859 (2) | 0.0891 (8) | |
| H6A | 0.240 (5) | 0.317 (3) | 0.437 (3) | 0.127 (14)* | |
| H6B | 0.410 (4) | 0.341 (2) | 0.378 (2) | 0.109 (13)* | |
| N7 | −0.1478 (4) | 0.7302 (2) | 0.4340 (2) | 0.1190 (11) | |
| C22 | −0.0799 (4) | 0.7346 (2) | 0.3675 (2) | 0.0822 (9) | |
| C23 | 0.0139 (3) | 0.73855 (18) | 0.28564 (19) | 0.0606 (7) | |
| C24 | −0.0753 (4) | 0.7084 (2) | 0.1977 (2) | 0.0732 (8) | |
| H24A | −0.1966 | 0.6869 | 0.1906 | 0.088* | |
| C25 | 0.0187 (4) | 0.7110 (2) | 0.1210 (2) | 0.0771 (8) | |
| H25A | −0.0403 | 0.6921 | 0.0613 | 0.093* | |
| C26 | 0.1980 (4) | 0.74096 (19) | 0.13031 (18) | 0.0664 (7) | |
| H26A | 0.2596 | 0.7403 | 0.0767 | 0.080* | |
| C27 | 0.2891 (3) | 0.77223 (18) | 0.21843 (18) | 0.0584 (6) | |
| C28 | 0.1939 (3) | 0.76999 (17) | 0.29630 (17) | 0.0588 (6) | |
| H28A | 0.2519 | 0.7899 | 0.3563 | 0.071* | |
| N8 | 0.4678 (4) | 0.8027 (2) | 0.2284 (3) | 0.0883 (8) | |
| H8A | 0.524 (4) | 0.806 (2) | 0.176 (2) | 0.103 (12)* | |
| H8B | 0.517 (5) | 0.825 (3) | 0.284 (3) | 0.126 (15)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0514 (14) | 0.0662 (17) | 0.0432 (14) | −0.0022 (12) | 0.0060 (12) | 0.0104 (13) |
| C2 | 0.0527 (14) | 0.0535 (15) | 0.0398 (13) | 0.0060 (11) | 0.0078 (11) | 0.0119 (11) |
| C3 | 0.0552 (14) | 0.0509 (14) | 0.0424 (13) | 0.0036 (11) | 0.0047 (10) | 0.0165 (11) |
| C4 | 0.0527 (13) | 0.0536 (15) | 0.0512 (15) | 0.0053 (11) | 0.0119 (11) | 0.0233 (12) |
| C5 | 0.0707 (17) | 0.0647 (17) | 0.0426 (14) | 0.0115 (13) | 0.0127 (12) | 0.0215 (13) |
| C6 | 0.0657 (15) | 0.0574 (16) | 0.0389 (13) | 0.0058 (12) | 0.0027 (11) | 0.0129 (12) |
| C7 | 0.0528 (14) | 0.0614 (16) | 0.0509 (15) | 0.0007 (12) | 0.0048 (12) | 0.0158 (13) |
| O1 | 0.0636 (10) | 0.1044 (15) | 0.0372 (10) | −0.0161 (10) | 0.0058 (8) | 0.0072 (10) |
| O2 | 0.0595 (11) | 0.1067 (15) | 0.0433 (10) | −0.0163 (10) | 0.0104 (8) | 0.0101 (10) |
| N1 | 0.0651 (15) | 0.0872 (17) | 0.0704 (17) | −0.0002 (12) | 0.0185 (13) | 0.0335 (14) |
| O3 | 0.0583 (11) | 0.1231 (18) | 0.0878 (15) | −0.0115 (11) | 0.0058 (11) | 0.0283 (14) |
| O4 | 0.0954 (16) | 0.225 (3) | 0.0855 (16) | −0.0204 (16) | 0.0314 (13) | 0.0746 (18) |
| N2 | 0.0839 (17) | 0.0953 (19) | 0.0439 (14) | −0.0009 (14) | −0.0024 (12) | 0.0128 (13) |
| O5 | 0.1111 (17) | 0.174 (2) | 0.0451 (12) | −0.0154 (15) | −0.0068 (11) | 0.0355 (14) |
| O6 | 0.1040 (17) | 0.166 (2) | 0.0704 (14) | −0.0510 (17) | −0.0168 (12) | 0.0260 (15) |
| C8 | 0.0540 (15) | 0.0601 (16) | 0.0460 (15) | 0.0010 (12) | 0.0094 (12) | 0.0121 (12) |
| C9 | 0.0539 (14) | 0.0493 (14) | 0.0433 (13) | 0.0032 (11) | 0.0108 (11) | 0.0140 (11) |
| C10 | 0.0548 (14) | 0.0584 (16) | 0.0475 (14) | 0.0034 (12) | 0.0030 (11) | 0.0163 (12) |
| C11 | 0.0449 (13) | 0.0606 (16) | 0.0584 (16) | 0.0046 (11) | 0.0109 (11) | 0.0260 (13) |
| C12 | 0.0607 (15) | 0.0575 (16) | 0.0506 (15) | 0.0067 (12) | 0.0184 (12) | 0.0231 (13) |
| C13 | 0.0616 (15) | 0.0559 (15) | 0.0408 (14) | −0.0003 (12) | 0.0070 (11) | 0.0157 (12) |
| C14 | 0.0508 (13) | 0.0543 (15) | 0.0487 (14) | −0.0002 (11) | 0.0077 (11) | 0.0143 (12) |
| N3 | 0.0505 (13) | 0.0916 (18) | 0.0756 (17) | 0.0002 (12) | 0.0066 (13) | 0.0320 (15) |
| O7 | 0.0728 (12) | 0.1110 (16) | 0.0450 (11) | −0.0172 (10) | 0.0127 (9) | 0.0115 (11) |
| O8 | 0.0577 (11) | 0.0959 (14) | 0.0535 (11) | −0.0099 (10) | 0.0156 (8) | 0.0143 (10) |
| O9 | 0.0639 (12) | 0.1283 (19) | 0.0836 (15) | −0.0143 (11) | −0.0095 (11) | 0.0288 (14) |
| O10 | 0.0590 (12) | 0.158 (2) | 0.0921 (16) | −0.0105 (12) | 0.0214 (11) | 0.0475 (15) |
| O11 | 0.1285 (17) | 0.132 (2) | 0.0450 (11) | −0.0182 (14) | 0.0226 (12) | 0.0245 (12) |
| O12 | 0.0902 (15) | 0.187 (3) | 0.0614 (13) | −0.0423 (16) | −0.0114 (11) | 0.0159 (14) |
| N4 | 0.0881 (17) | 0.0858 (18) | 0.0440 (13) | −0.0099 (14) | 0.0041 (13) | 0.0154 (12) |
| N5 | 0.121 (2) | 0.157 (3) | 0.080 (2) | 0.017 (2) | 0.0235 (18) | 0.053 (2) |
| C15 | 0.085 (2) | 0.091 (2) | 0.070 (2) | 0.0088 (17) | 0.0048 (17) | 0.0367 (19) |
| C16 | 0.0650 (16) | 0.0612 (17) | 0.0546 (16) | 0.0017 (13) | 0.0008 (13) | 0.0205 (14) |
| C17 | 0.0744 (18) | 0.0696 (19) | 0.0684 (19) | −0.0075 (15) | −0.0126 (15) | 0.0137 (15) |
| C18 | 0.0634 (18) | 0.075 (2) | 0.087 (2) | −0.0125 (15) | 0.0020 (17) | 0.0222 (18) |
| C19 | 0.0726 (18) | 0.0608 (18) | 0.0721 (19) | 0.0004 (14) | 0.0124 (15) | 0.0207 (15) |
| C20 | 0.0746 (17) | 0.0462 (15) | 0.0561 (17) | 0.0016 (13) | −0.0079 (13) | 0.0148 (13) |
| C21 | 0.0554 (14) | 0.0606 (17) | 0.0694 (18) | −0.0029 (12) | −0.0033 (13) | 0.0258 (14) |
| N6 | 0.107 (2) | 0.093 (2) | 0.0613 (18) | −0.0088 (18) | −0.0128 (17) | 0.0202 (15) |
| N7 | 0.146 (3) | 0.112 (3) | 0.107 (2) | 0.0211 (19) | 0.064 (2) | 0.039 (2) |
| C22 | 0.091 (2) | 0.071 (2) | 0.087 (2) | 0.0170 (16) | 0.0310 (18) | 0.0229 (18) |
| C23 | 0.0673 (16) | 0.0535 (16) | 0.0616 (17) | 0.0104 (13) | 0.0117 (14) | 0.0168 (13) |
| C24 | 0.0606 (16) | 0.0712 (19) | 0.083 (2) | 0.0050 (14) | −0.0036 (15) | 0.0177 (17) |
| C25 | 0.090 (2) | 0.076 (2) | 0.0602 (18) | 0.0097 (16) | −0.0166 (16) | 0.0141 (16) |
| C26 | 0.0819 (19) | 0.0663 (18) | 0.0503 (16) | 0.0071 (15) | 0.0052 (14) | 0.0169 (14) |
| C27 | 0.0653 (16) | 0.0498 (15) | 0.0593 (17) | 0.0047 (12) | 0.0038 (13) | 0.0156 (13) |
| C28 | 0.0711 (17) | 0.0541 (16) | 0.0477 (15) | 0.0046 (13) | −0.0056 (12) | 0.0117 (13) |
| N8 | 0.0720 (18) | 0.102 (2) | 0.084 (2) | −0.0103 (15) | 0.0072 (17) | 0.0221 (19) |
Geometric parameters (Å, °)
| C1—O2 | 1.243 (3) | N3—O10 | 1.216 (3) |
| C1—O1 | 1.271 (3) | O7—H9A | 1.23 (5) |
| C1—C2 | 1.478 (3) | O11—N4 | 1.216 (3) |
| C2—C7 | 1.376 (3) | O12—N4 | 1.194 (3) |
| C2—C3 | 1.377 (3) | N5—C15 | 1.139 (3) |
| C3—C4 | 1.371 (3) | C15—C16 | 1.433 (4) |
| C3—H3A | 0.9300 | C16—C21 | 1.374 (3) |
| C4—C5 | 1.370 (3) | C16—C17 | 1.377 (3) |
| C4—N1 | 1.464 (3) | C17—C18 | 1.370 (4) |
| C5—C6 | 1.369 (3) | C17—H17A | 0.9300 |
| C5—H5A | 0.9300 | C18—C19 | 1.356 (4) |
| C6—C7 | 1.366 (3) | C18—H18A | 0.96 (3) |
| C6—N2 | 1.466 (3) | C19—C20 | 1.379 (3) |
| C7—H7A | 0.9300 | C19—H19A | 0.9300 |
| O1—H1A | 1.18 (4) | C20—N6 | 1.371 (3) |
| N1—O4 | 1.203 (3) | C20—C21 | 1.389 (3) |
| N1—O3 | 1.206 (3) | C21—H21A | 0.9300 |
| N2—O5 | 1.209 (3) | N6—H6A | 0.92 (3) |
| N2—O6 | 1.211 (3) | N6—H6B | 0.90 (3) |
| C8—O8 | 1.250 (3) | N7—C22 | 1.140 (3) |
| C8—O7 | 1.263 (3) | C22—C23 | 1.437 (4) |
| C8—C9 | 1.484 (3) | C23—C24 | 1.376 (4) |
| C9—C14 | 1.378 (3) | C23—C28 | 1.377 (3) |
| C9—C10 | 1.380 (3) | C24—C25 | 1.365 (4) |
| C10—C11 | 1.375 (3) | C24—H24A | 0.9300 |
| C10—H10A | 0.9300 | C25—C26 | 1.369 (4) |
| C11—C12 | 1.371 (3) | C25—H25A | 0.9300 |
| C11—N3 | 1.466 (3) | C26—C27 | 1.384 (3) |
| C12—C13 | 1.368 (3) | C26—H26A | 0.9300 |
| C12—H12A | 0.9300 | C27—N8 | 1.365 (3) |
| C13—C14 | 1.376 (3) | C27—C28 | 1.384 (3) |
| C13—N4 | 1.462 (3) | C28—H28A | 0.9300 |
| C14—H14A | 0.9300 | N8—H8A | 0.91 (3) |
| N3—O9 | 1.208 (3) | N8—H8B | 0.85 (3) |
| O2—C1—O1 | 124.3 (2) | O9—N3—C11 | 118.6 (2) |
| O2—C1—C2 | 118.9 (2) | O10—N3—C11 | 117.5 (2) |
| O1—C1—C2 | 116.8 (2) | C8—O7—H9A | 116.9 (18) |
| C7—C2—C3 | 120.3 (2) | O12—N4—O11 | 123.8 (2) |
| C7—C2—C1 | 119.4 (2) | O12—N4—C13 | 118.0 (2) |
| C3—C2—C1 | 120.3 (2) | O11—N4—C13 | 118.3 (2) |
| C4—C3—C2 | 118.5 (2) | N5—C15—C16 | 179.5 (3) |
| C4—C3—H3A | 120.8 | C21—C16—C17 | 120.7 (2) |
| C2—C3—H3A | 120.8 | C21—C16—C15 | 120.2 (2) |
| C5—C4—C3 | 122.7 (2) | C17—C16—C15 | 119.1 (3) |
| C5—C4—N1 | 119.0 (2) | C18—C17—C16 | 118.5 (3) |
| C3—C4—N1 | 118.4 (2) | C18—C17—H17A | 120.7 |
| C6—C5—C4 | 117.1 (2) | C16—C17—H17A | 120.7 |
| C6—C5—H5A | 121.5 | C19—C18—C17 | 121.1 (3) |
| C4—C5—H5A | 121.5 | C19—C18—H18A | 118.0 (15) |
| C7—C6—C5 | 122.4 (2) | C17—C18—H18A | 120.9 (15) |
| C7—C6—N2 | 118.6 (2) | C18—C19—C20 | 121.4 (3) |
| C5—C6—N2 | 118.9 (2) | C18—C19—H19A | 119.3 |
| C6—C7—C2 | 119.0 (2) | C20—C19—H19A | 119.3 |
| C6—C7—H7A | 120.5 | N6—C20—C19 | 121.1 (3) |
| C2—C7—H7A | 120.5 | N6—C20—C21 | 121.1 (3) |
| C1—O1—H1A | 113.5 (16) | C19—C20—C21 | 117.7 (2) |
| O4—N1—O3 | 123.0 (2) | C16—C21—C20 | 120.5 (2) |
| O4—N1—C4 | 118.5 (3) | C16—C21—H21A | 119.7 |
| O3—N1—C4 | 118.5 (2) | C20—C21—H21A | 119.7 |
| O5—N2—O6 | 124.3 (2) | C20—N6—H6A | 116 (2) |
| O5—N2—C6 | 117.6 (2) | C20—N6—H6B | 109 (2) |
| O6—N2—C6 | 118.1 (2) | H6A—N6—H6B | 129 (3) |
| O8—C8—O7 | 124.3 (2) | N7—C22—C23 | 177.1 (4) |
| O8—C8—C9 | 118.2 (2) | C24—C23—C28 | 121.0 (2) |
| O7—C8—C9 | 117.5 (2) | C24—C23—C22 | 119.8 (3) |
| C14—C9—C10 | 120.4 (2) | C28—C23—C22 | 119.2 (3) |
| C14—C9—C8 | 120.0 (2) | C25—C24—C23 | 118.4 (3) |
| C10—C9—C8 | 119.6 (2) | C25—C24—H24A | 120.8 |
| C11—C10—C9 | 118.6 (2) | C23—C24—H24A | 120.8 |
| C11—C10—H10A | 120.7 | C24—C25—C26 | 121.3 (3) |
| C9—C10—H10A | 120.7 | C24—C25—H25A | 119.3 |
| C12—C11—C10 | 122.8 (2) | C26—C25—H25A | 119.3 |
| C12—C11—N3 | 118.6 (2) | C25—C26—C27 | 120.9 (3) |
| C10—C11—N3 | 118.6 (2) | C25—C26—H26A | 119.5 |
| C13—C12—C11 | 116.8 (2) | C27—C26—H26A | 119.5 |
| C13—C12—H12A | 121.6 | N8—C27—C26 | 121.3 (3) |
| C11—C12—H12A | 121.6 | N8—C27—C28 | 120.8 (3) |
| C12—C13—C14 | 122.9 (2) | C26—C27—C28 | 117.8 (2) |
| C12—C13—N4 | 118.8 (2) | C23—C28—C27 | 120.6 (2) |
| C14—C13—N4 | 118.2 (2) | C23—C28—H28A | 119.7 |
| C13—C14—C9 | 118.5 (2) | C27—C28—H28A | 119.7 |
| C13—C14—H14A | 120.7 | C27—N8—H8A | 118 (2) |
| C9—C14—H14A | 120.7 | C27—N8—H8B | 118 (2) |
| O9—N3—O10 | 123.9 (2) | H8A—N8—H8B | 123 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···O2i | 1.18 (4) | 1.41 (4) | 2.590 (2) | 173 (3) |
| N6—H6A···N7ii | 0.92 (3) | 2.32 (4) | 3.232 (5) | 169 (3) |
| N6—H6B···O12iii | 0.90 (3) | 2.57 (3) | 2.953 (4) | 107 (2) |
| N8—H8A···N5iv | 0.91 (3) | 2.37 (3) | 3.262 (5) | 170 (3) |
| N8—H8B···O5iii | 0.85 (3) | 2.48 (4) | 3.286 (4) | 157 (3) |
| O7—H9A···O8iv | 1.23 (5) | 1.38 (5) | 2.608 (2) | 174 (4) |
| C5—H5A···O4v | 0.93 | 2.44 | 3.321 (3) | 158. |
| C12—H12A···O11ii | 0.93 | 2.50 | 3.352 (3) | 153. |
| C18—H18A···O2vi | 0.96 (3) | 2.58 (3) | 3.421 (4) | 147 (2) |
| C21—H21A···O10vii | 0.93 | 2.60 | 3.451 (3) | 153. |
Symmetry codes: (i) −x+1, −y, −z; (ii) −x, −y+1, −z+1; (iii) −x+1, −y+1, −z+1; (iv) −x+1, −y+1, −z; (v) −x, −y, −z+1; (vi) x−1, y, z; (vii) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: KP2355).
References
- Bhatt, P. M. & Desiraju, G. R. (2008). CrystEngComm, 10, 1747–1749.
- Bruker (2007). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Desiraju, G. R. (2002). Acc. Chem. Res. 35, 565–573. [DOI] [PubMed]
- Etter, M. C. & Baures, P. W. (1988). J. Am. Chem. Soc. 110, 639–640.
- Gao, X. C., Friscic, T. & Macgillivray, L. R. (2004). Angew. Chem. Int. Ed. 43, 232–236. [DOI] [PubMed]
- Hori, A., Takatani, S., Miyamoto, T. K. & Hasegawa, M. (2009). CrystEngComm, 11, 567–569.
- Li, C., Robinson, P. D. & Dyer, D. J. (2006). Acta Cryst. C62, o336–o338. [DOI] [PubMed]
- Prins, L. J., Reinhoudt, D. N. & Timmerman, P. (2001). Angew. Chem. Int. Ed. 40, 2382–2426. [DOI] [PubMed]
- Roy, S., Mahata, G. & Biradha, K. (2009). Cryst. Growth Des. 9, 5006–5008.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Steiner, T. (2002). Angew. Chem. Int. Ed. 41, 48–76.
- Wei, L.-H. (2007). Acta Cryst. E63, o4174.
- Weyna, D. R., Shattock, T., Vishweshwar, P. & Zaworotko, M. J. (2009). Cryst. Growth Des. 9, 1106–1123.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811039870/kp2355sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811039870/kp2355Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811039870/kp2355Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





