Abstract
In the title compound, C15H14Cl2N2OS, the piperidine ring adopts a chair conformation. The dihedral angle between the thiazolidine ring and the dichlorobenzene ring is 9.30 (4)°; this near coplanar conformation is stabilized by the formation of an intramolecular C—H⋯S hydrogen bond, which generates an S(6) ring. In the crystal, molecules are linked by C—H⋯O hydrogen bonds, forming [001] chains. Weak π–π interactions [centroid–centroid separation = 3.5460 (5) Å] consolidate the structure.
Related literature
For details and properties of the 4-thiazolidinone ring system, see: Lesyk & Zimenkovsky (2004 ▶); Lesyk et al. (2007 ▶); Havrylyuk et al. (2009 ▶); Ahn et al. (2006 ▶); Park et al. (2008 ▶); Geronikaki et al. (2008 ▶); Zimenkovsky et al. (2005 ▶). For ring puckering, see: Cremer & Pople (1975 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶). For the stability of the temperature controller used in the data collection, see: Cosier & Glazer (1986 ▶).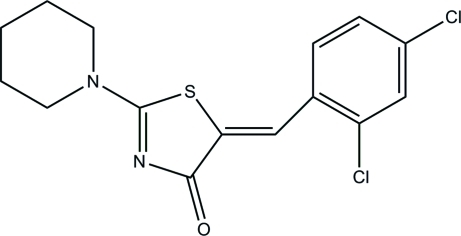
Experimental
Crystal data
C15H14Cl2N2OS
M r = 341.24
Monoclinic,

a = 28.5303 (3) Å
b = 7.4915 (1) Å
c = 15.4789 (2) Å
β = 116.407 (1)°
V = 2963.17 (6) Å3
Z = 8
Mo Kα radiation
μ = 0.58 mm−1
T = 100 K
0.44 × 0.25 × 0.13 mm
Data collection
Bruker SMART APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.783, T max = 0.928
46944 measured reflections
6673 independent reflections
5955 reflections with I > 2σ(I)
R int = 0.024
Refinement
R[F 2 > 2σ(F 2)] = 0.026
wR(F 2) = 0.074
S = 1.03
6673 reflections
190 parameters
H-atom parameters constrained
Δρmax = 0.52 e Å−3
Δρmin = −0.19 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811040785/hb6435sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811040785/hb6435Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811040785/hb6435Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C1—H1A⋯S1 | 0.95 | 2.49 | 3.2260 (8) | 134 |
| C4—H4A⋯O1i | 0.95 | 2.40 | 3.3080 (9) | 160 |
| C15—H15A⋯O1ii | 0.99 | 2.57 | 3.2778 (11) | 129 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
HKF and MH thank the Malaysian Government and Universiti Sains Malaysia for the Research University grant No. 1001/PFIZIK/811160. MH also thanks Universiti Sains Malaysia for a post-doctoral research fellowship.
supplementary crystallographic information
Comment
The 4-thiazolidinone ring system is a core structure in various synthetic compounds displaying a broad spectrum of biological activities (Lesyk & Zimenkovsky, 2004), including an anticancer effect (Lesyk et al., 2007; Havrylyuk et al., 2009). The mechanisms of antitumor activity by 4-thiazolidinones and related heterocycles may be associated with their affinities to anticancer bio-targets, such as phosphatase of a regenerating liver (PRL-3) (Ahn et al., 2006; Park et al., 2008) and nonmembrane protein tyrosine phosphatase (SHP-2)(Geronikaki et al., 2008). 5-Arylidene derivatives were previously shown as the most active group of compounds with the anticancer activity among a large pool of 4-azolidone derivatives and analogs (Zimenkovsky et al., 2005). This prompted us to synthesize the title compound, (I), (Fig. 1).
The piperidine ((N2/C11–C15) ring adopts a chair conformation [Q = 0.5462 (10) Å; θ = 5.78 (10)° and φ = 206.6 (10)°; Cremer & Pople, 1975]. The central thiazolidine (S1/N1/C8–C10) ring makes dihedral angles of 21.18 (4)° and 9.30 (4)° with the terminal piperidine (N2/C11–C15) and phenyl (C1–C6) rings. The corresponding angle between the piperidine and phenyl (N2/C11–C15)/(C1–C6) rings is 13.69 (4)°. An intramolecular C1—H1A···S1 hydrogen bond generates an S(6) (Bernstein et al., 1995) ring motif.
In the crystal structure, (Fig. 2), the molecules are connected via intermolecular C—H···O (Table 1) hydrogen bonds forming one-dimensional supramolecular chains along the c-axis. The crystal structure is further stabilized by weak π–π interactions between the thiazolidine (Cg1; S1/N1/C8–C10) and phenyl (Cg3; C1–C6) rings [Cg1···Cg3 = 3.5460 (5) Å; 1/2-x, 3/2-y, 1-z].
Experimental
An equimolar mixture of 2-(4-methylsulfanylphenyl)acetohydrazide and 4-chlorobenzaldehyde was refluxed for four hours in the presence of few drops of acid catalyst and ethanol as solvent. The compound obtained was filtered, washed, dried and recrystalised from ethanol to yield brown blocks of (I).
Refinement
All hydrogen atoms were positioned geometrically [ C–H = 0.95 or 0.99 Å] and were refined using a riding model, with Uiso(H) = 1.2 Ueq(C).
Figures
Fig. 1.
The asymmetric unit of the title compound, showing 50% probability displacement ellipsoids. An intramolecular hydrogen bond is shown by a dashed line.
Fig. 2.
The crystal packing of the title compound (I).
Crystal data
| C15H14Cl2N2OS | F(000) = 1408 |
| Mr = 341.24 | Dx = 1.530 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2yc | Cell parameters from 9844 reflections |
| a = 28.5303 (3) Å | θ = 2.7–35.2° |
| b = 7.4915 (1) Å | µ = 0.58 mm−1 |
| c = 15.4789 (2) Å | T = 100 K |
| β = 116.407 (1)° | Block, brown |
| V = 2963.17 (6) Å3 | 0.44 × 0.25 × 0.13 mm |
| Z = 8 |
Data collection
| Bruker SMART APEXII CCD diffractometer | 6673 independent reflections |
| Radiation source: fine-focus sealed tube | 5955 reflections with I > 2σ(I) |
| graphite | Rint = 0.024 |
| φ and ω scans | θmax = 35.4°, θmin = 2.7° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −45→46 |
| Tmin = 0.783, Tmax = 0.928 | k = −12→12 |
| 46944 measured reflections | l = −25→25 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.026 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.074 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0364P)2 + 1.7424P] where P = (Fo2 + 2Fc2)/3 |
| 6673 reflections | (Δ/σ)max = 0.002 |
| 190 parameters | Δρmax = 0.52 e Å−3 |
| 0 restraints | Δρmin = −0.19 e Å−3 |
Special details
| Experimental. The crystal was placed in the cold stream of an Oxford Cryosystems Cobra open-flow nitrogen cryostat (Cosier & Glazer, 1986) operating at 100.0 (1) K. |
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.430128 (8) | 0.28966 (3) | 0.729616 (15) | 0.02400 (5) | |
| Cl2 | 0.269296 (8) | 0.51920 (3) | 0.790208 (13) | 0.02382 (5) | |
| S1 | 0.176998 (7) | 0.57324 (3) | 0.374212 (12) | 0.01596 (4) | |
| O1 | 0.11470 (2) | 0.76041 (9) | 0.52929 (4) | 0.02178 (12) | |
| N1 | 0.09082 (3) | 0.72148 (9) | 0.36763 (5) | 0.01686 (11) | |
| N2 | 0.08873 (3) | 0.66216 (10) | 0.21783 (4) | 0.01825 (12) | |
| C1 | 0.28809 (3) | 0.46653 (11) | 0.54975 (5) | 0.01796 (13) | |
| H1A | 0.2688 | 0.4850 | 0.4822 | 0.022* | |
| C2 | 0.33866 (3) | 0.40104 (11) | 0.58547 (6) | 0.01884 (13) | |
| H2A | 0.3540 | 0.3774 | 0.5433 | 0.023* | |
| C3 | 0.36659 (3) | 0.37051 (10) | 0.68400 (6) | 0.01726 (12) | |
| C4 | 0.34481 (3) | 0.40355 (11) | 0.74672 (5) | 0.01740 (12) | |
| H4A | 0.3639 | 0.3797 | 0.8138 | 0.021* | |
| C5 | 0.29438 (3) | 0.47245 (10) | 0.70876 (5) | 0.01601 (12) | |
| C6 | 0.26411 (3) | 0.50678 (10) | 0.60961 (5) | 0.01517 (12) | |
| C7 | 0.21245 (3) | 0.58738 (10) | 0.57455 (5) | 0.01636 (12) | |
| H7A | 0.2028 | 0.6221 | 0.6235 | 0.020* | |
| C8 | 0.17611 (3) | 0.62056 (10) | 0.48355 (5) | 0.01523 (12) | |
| C9 | 0.12450 (3) | 0.70822 (10) | 0.46399 (5) | 0.01629 (12) | |
| C10 | 0.11223 (3) | 0.66071 (10) | 0.31348 (5) | 0.01553 (12) | |
| C11 | 0.03308 (3) | 0.71217 (13) | 0.16554 (6) | 0.02211 (15) | |
| H11A | 0.0227 | 0.7796 | 0.2093 | 0.027* | |
| H11B | 0.0114 | 0.6027 | 0.1450 | 0.027* | |
| C12 | 0.02289 (3) | 0.82567 (13) | 0.07750 (6) | 0.02274 (15) | |
| H12A | 0.0392 | 0.9444 | 0.0988 | 0.027* | |
| H12B | −0.0153 | 0.8434 | 0.0394 | 0.027* | |
| C13 | 0.04478 (3) | 0.73902 (13) | 0.01359 (6) | 0.02228 (15) | |
| H13A | 0.0257 | 0.6267 | −0.0141 | 0.027* | |
| H13B | 0.0398 | 0.8202 | −0.0403 | 0.027* | |
| C14 | 0.10279 (3) | 0.69973 (12) | 0.07298 (6) | 0.01982 (14) | |
| H14A | 0.1221 | 0.8133 | 0.0961 | 0.024* | |
| H14B | 0.1165 | 0.6393 | 0.0320 | 0.024* | |
| C15 | 0.11176 (4) | 0.58124 (12) | 0.15908 (6) | 0.02219 (15) | |
| H15A | 0.0958 | 0.4627 | 0.1359 | 0.027* | |
| H15B | 0.1498 | 0.5640 | 0.1991 | 0.027* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.01844 (8) | 0.02974 (10) | 0.02435 (9) | 0.00495 (7) | 0.01001 (7) | 0.00259 (7) |
| Cl2 | 0.01920 (9) | 0.04116 (12) | 0.01293 (7) | 0.00453 (7) | 0.00879 (6) | 0.00332 (7) |
| S1 | 0.01586 (8) | 0.02028 (8) | 0.01193 (7) | 0.00101 (6) | 0.00634 (6) | −0.00006 (6) |
| O1 | 0.0210 (3) | 0.0317 (3) | 0.0141 (2) | 0.0032 (2) | 0.0092 (2) | −0.0011 (2) |
| N1 | 0.0159 (3) | 0.0229 (3) | 0.0124 (2) | 0.0001 (2) | 0.0069 (2) | 0.0001 (2) |
| N2 | 0.0163 (3) | 0.0272 (3) | 0.0112 (2) | 0.0023 (2) | 0.0060 (2) | 0.0005 (2) |
| C1 | 0.0199 (3) | 0.0215 (3) | 0.0134 (3) | 0.0007 (3) | 0.0083 (2) | 0.0006 (2) |
| C2 | 0.0208 (3) | 0.0212 (3) | 0.0169 (3) | 0.0014 (3) | 0.0105 (3) | 0.0005 (2) |
| C3 | 0.0165 (3) | 0.0178 (3) | 0.0180 (3) | 0.0004 (2) | 0.0081 (2) | 0.0008 (2) |
| C4 | 0.0167 (3) | 0.0204 (3) | 0.0146 (3) | −0.0003 (2) | 0.0064 (2) | 0.0014 (2) |
| C5 | 0.0164 (3) | 0.0203 (3) | 0.0125 (3) | −0.0014 (2) | 0.0075 (2) | 0.0005 (2) |
| C6 | 0.0158 (3) | 0.0175 (3) | 0.0126 (3) | −0.0016 (2) | 0.0066 (2) | 0.0004 (2) |
| C7 | 0.0166 (3) | 0.0201 (3) | 0.0127 (3) | −0.0010 (2) | 0.0068 (2) | 0.0003 (2) |
| C8 | 0.0159 (3) | 0.0178 (3) | 0.0126 (3) | −0.0014 (2) | 0.0068 (2) | −0.0002 (2) |
| C9 | 0.0164 (3) | 0.0196 (3) | 0.0133 (3) | −0.0010 (2) | 0.0070 (2) | 0.0001 (2) |
| C10 | 0.0150 (3) | 0.0189 (3) | 0.0125 (3) | −0.0008 (2) | 0.0060 (2) | 0.0003 (2) |
| C11 | 0.0155 (3) | 0.0365 (4) | 0.0139 (3) | 0.0009 (3) | 0.0061 (2) | 0.0022 (3) |
| C12 | 0.0190 (3) | 0.0339 (4) | 0.0148 (3) | 0.0050 (3) | 0.0070 (3) | 0.0033 (3) |
| C13 | 0.0230 (4) | 0.0303 (4) | 0.0133 (3) | 0.0015 (3) | 0.0078 (3) | 0.0010 (3) |
| C14 | 0.0220 (3) | 0.0245 (4) | 0.0159 (3) | 0.0008 (3) | 0.0111 (3) | −0.0006 (3) |
| C15 | 0.0252 (4) | 0.0291 (4) | 0.0138 (3) | 0.0072 (3) | 0.0100 (3) | 0.0015 (3) |
Geometric parameters (Å, °)
| Cl1—C3 | 1.7357 (8) | C6—C7 | 1.4560 (11) |
| Cl2—C5 | 1.7397 (7) | C7—C8 | 1.3498 (10) |
| S1—C8 | 1.7402 (7) | C7—H7A | 0.9500 |
| S1—C10 | 1.7839 (8) | C8—C9 | 1.5148 (11) |
| O1—C9 | 1.2265 (9) | C11—C12 | 1.5207 (12) |
| N1—C10 | 1.3186 (10) | C11—H11A | 0.9900 |
| N1—C9 | 1.3722 (10) | C11—H11B | 0.9900 |
| N2—C10 | 1.3261 (9) | C12—C13 | 1.5296 (12) |
| N2—C15 | 1.4690 (10) | C12—H12A | 0.9900 |
| N2—C11 | 1.4743 (11) | C12—H12B | 0.9900 |
| C1—C2 | 1.3851 (11) | C13—C14 | 1.5223 (12) |
| C1—C6 | 1.4075 (10) | C13—H13A | 0.9900 |
| C1—H1A | 0.9500 | C13—H13B | 0.9900 |
| C2—C3 | 1.3900 (11) | C14—C15 | 1.5252 (12) |
| C2—H2A | 0.9500 | C14—H14A | 0.9900 |
| C3—C4 | 1.3880 (11) | C14—H14B | 0.9900 |
| C4—C5 | 1.3893 (11) | C15—H15A | 0.9900 |
| C4—H4A | 0.9500 | C15—H15B | 0.9900 |
| C5—C6 | 1.4102 (10) | ||
| C8—S1—C10 | 88.74 (3) | N1—C10—S1 | 117.13 (5) |
| C10—N1—C9 | 111.56 (6) | N2—C10—S1 | 118.84 (6) |
| C10—N2—C15 | 122.99 (7) | N2—C11—C12 | 111.35 (7) |
| C10—N2—C11 | 120.09 (6) | N2—C11—H11A | 109.4 |
| C15—N2—C11 | 115.73 (6) | C12—C11—H11A | 109.4 |
| C2—C1—C6 | 122.55 (7) | N2—C11—H11B | 109.4 |
| C2—C1—H1A | 118.7 | C12—C11—H11B | 109.4 |
| C6—C1—H1A | 118.7 | H11A—C11—H11B | 108.0 |
| C1—C2—C3 | 118.91 (7) | C11—C12—C13 | 111.84 (7) |
| C1—C2—H2A | 120.5 | C11—C12—H12A | 109.2 |
| C3—C2—H2A | 120.5 | C13—C12—H12A | 109.2 |
| C4—C3—C2 | 121.44 (7) | C11—C12—H12B | 109.2 |
| C4—C3—Cl1 | 119.28 (6) | C13—C12—H12B | 109.2 |
| C2—C3—Cl1 | 119.29 (6) | H12A—C12—H12B | 107.9 |
| C3—C4—C5 | 118.17 (7) | C14—C13—C12 | 109.79 (6) |
| C3—C4—H4A | 120.9 | C14—C13—H13A | 109.7 |
| C5—C4—H4A | 120.9 | C12—C13—H13A | 109.7 |
| C4—C5—C6 | 123.08 (7) | C14—C13—H13B | 109.7 |
| C4—C5—Cl2 | 116.79 (5) | C12—C13—H13B | 109.7 |
| C6—C5—Cl2 | 120.12 (6) | H13A—C13—H13B | 108.2 |
| C1—C6—C5 | 115.83 (7) | C13—C14—C15 | 110.80 (7) |
| C1—C6—C7 | 123.46 (7) | C13—C14—H14A | 109.5 |
| C5—C6—C7 | 120.64 (7) | C15—C14—H14A | 109.5 |
| C8—C7—C6 | 130.21 (7) | C13—C14—H14B | 109.5 |
| C8—C7—H7A | 114.9 | C15—C14—H14B | 109.5 |
| C6—C7—H7A | 114.9 | H14A—C14—H14B | 108.1 |
| C7—C8—C9 | 121.03 (7) | N2—C15—C14 | 110.69 (7) |
| C7—C8—S1 | 129.85 (6) | N2—C15—H15A | 109.5 |
| C9—C8—S1 | 109.10 (5) | C14—C15—H15A | 109.5 |
| O1—C9—N1 | 124.51 (7) | N2—C15—H15B | 109.5 |
| O1—C9—C8 | 122.08 (7) | C14—C15—H15B | 109.5 |
| N1—C9—C8 | 113.41 (6) | H15A—C15—H15B | 108.1 |
| N1—C10—N2 | 124.03 (7) | ||
| C6—C1—C2—C3 | 1.25 (12) | C7—C8—C9—O1 | 3.74 (12) |
| C1—C2—C3—C4 | 0.26 (12) | S1—C8—C9—O1 | −177.51 (7) |
| C1—C2—C3—Cl1 | −179.63 (6) | C7—C8—C9—N1 | −175.99 (7) |
| C2—C3—C4—C5 | −1.49 (12) | S1—C8—C9—N1 | 2.77 (8) |
| Cl1—C3—C4—C5 | 178.41 (6) | C9—N1—C10—N2 | −178.19 (8) |
| C3—C4—C5—C6 | 1.30 (12) | C9—N1—C10—S1 | 1.36 (9) |
| C3—C4—C5—Cl2 | −177.86 (6) | C15—N2—C10—N1 | −175.15 (8) |
| C2—C1—C6—C5 | −1.41 (12) | C11—N2—C10—N1 | −8.17 (12) |
| C2—C1—C6—C7 | 175.53 (8) | C15—N2—C10—S1 | 5.31 (11) |
| C4—C5—C6—C1 | 0.10 (11) | C11—N2—C10—S1 | 172.29 (6) |
| Cl2—C5—C6—C1 | 179.24 (6) | C8—S1—C10—N1 | 0.25 (7) |
| C4—C5—C6—C7 | −176.93 (7) | C8—S1—C10—N2 | 179.82 (7) |
| Cl2—C5—C6—C7 | 2.21 (10) | C10—N2—C11—C12 | 140.61 (8) |
| C1—C6—C7—C8 | 7.99 (13) | C15—N2—C11—C12 | −51.50 (10) |
| C5—C6—C7—C8 | −175.21 (8) | N2—C11—C12—C13 | 51.51 (10) |
| C6—C7—C8—C9 | −179.51 (7) | C11—C12—C13—C14 | −55.21 (10) |
| C6—C7—C8—S1 | 2.03 (13) | C12—C13—C14—C15 | 56.91 (10) |
| C10—S1—C8—C7 | 177.01 (8) | C10—N2—C15—C14 | −139.08 (8) |
| C10—S1—C8—C9 | −1.60 (5) | C11—N2—C15—C14 | 53.41 (10) |
| C10—N1—C9—O1 | 177.67 (8) | C13—C14—C15—N2 | −55.37 (9) |
| C10—N1—C9—C8 | −2.61 (9) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1A···S1 | 0.95 | 2.49 | 3.2260 (8) | 134 |
| C4—H4A···O1i | 0.95 | 2.40 | 3.3080 (9) | 160 |
| C15—H15A···O1ii | 0.99 | 2.57 | 3.2778 (11) | 129 |
Symmetry codes: (i) −x+1/2, y−1/2, −z+3/2; (ii) x, −y+1, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB6435).
References
- Ahn, J. H., Kim, S. J., Park, W. S., Cho, S. Y., Ha, J. D., Kim, S. S., Kang, S. K., Jeong, D. G., Jung, S. K., Lee, S. H., Kim, H. M., Park, S. K., Lee, K. H., Lee, C. W., Ryu, S. E. & Choi, J. K. (2006). Bioorg. Med. Chem. Lett. 16, 2996–2999.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2009). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Cosier, J. & Glazer, A. M. (1986). J. Appl. Cryst. 19, 105–107.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Geronikaki, A., Eleftheriou, P., Vicini, P., Alam, I., Dixit, A. & Saxena, A. K. (2008). J. Med. Chem. 51, 5221–5228. [DOI] [PubMed]
- Havrylyuk, D., Zimenkovsky, B., Vasylenko, O., Zaprutko, L., Gzella, A. & Lesyk, R. (2009). Eur. J. Med. Chem. 44, 1396–1404. [DOI] [PubMed]
- Lesyk, R., Vladzimirska, O., Holota, S., Zaprutko, L. & Gzella, A. (2007). Eur. J. Med. Chem. 42, 641–648. [DOI] [PubMed]
- Lesyk, R. & Zimenkovsky, B. (2004). Curr. Org. Chem. 8, 1547–1577.
- Park, H., Jung, S. K., Jeong, D. G., Ryu, S. E. & Kim, S. J. (2008). Bioorg. Med. Chem. Lett. 18, 2250–2255. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Zimenkovsky, B., Kazmirchuk, G., Zaprutko, L., Paraskiewicz, A., Melzer, E. & Lesyk, R. (2005). Pol. Chem. Soc. 1, 69–72.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811040785/hb6435sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811040785/hb6435Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811040785/hb6435Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




