Abstract
Urinary metabolites of the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) and its glucuronides, termed total NNAL, have recently been shown to be good predictors of lung cancer risk, years prior to diagnosis. We sought to determine the contribution of several genetic polymorphisms to total NNAL output and inter-individual variability. The study subjects were derived from the Harvard/Massachusetts General Hospital Lung cancer case-control study. We analyzed 87 self-described smokers (35 lung cancer cases and 52 controls), with urine samples collected at time of diagnosis and (1992–1996). We tested 82 tagging SNPs in 16 genes related to the metabolism of NNK to total NNAL. Using weighted case status least squares regression, we tested for the association of each SNP with square-root (sqrt) transformed total NNAL (pmol per mg creatinine), controlling for age, sex, sqrt packyears and sqrt nicotine (ng per mg creatinine). After a sqrt transformation, nicotine significantly predicted a 0.018 (0.014, 0.023) pmol/mg creatinine unit increase in total NNAL for every ng/mg creatinine increase in nicotine at p<10E-16. Three HSD11B1 SNPs and AKR1C4 rs7083869 were significantly associated with decreasing total NNAL levels: HSD11B1 rs2235543 (p= 4.84E-08) and rs3753519 (p= 0.0017) passed multiple testing adjustment at FDR q=1.13E-05 and 0.07 respectively, AKR1C4 rs7083869 (p=0.019) did not, FDR q=0.51. HSD11B1 and AKR1C4 enzymes are carbonyl reductases directly involved in the single step reduction of NNK to NNAL. The HSD11B1 SNPs may be correlated with the functional variant rs13306401 and the AKR1C4 SNP is correlated with the enzyme activity reducing variant rs17134592, L311V.
Keywords: NNK, NNAL, tobacco specific nitrosamine, genetic polymorphism, HSD11B1
Introduction
Lung cancer is the leading cause of cancer-related mortality and has the third leading incidence rate of cancer in the United States.1 Long-term inhalation of tobacco smoke is the main cause of lung cancer with a relative risk of approximately 20 for smokers compared to non-smokers and a 30 percent increased risk for those exposed to second-hand smoke.2 Yet not all smokers develop lung cancer; the cumulative risk by age 75 for smokers is an estimated 10–20%,3, 4 which may indicate that individual susceptibility to tobacco smoke carcinogens is modified by other factors such as genetics. Tobacco-specific nitrosamines (TSNA) such as NNK (4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone) and polycyclic aromatic hydrocarbons are considered to be important carcinogens for lung cancer development. Formed from nicotine in the tobacco curing process, NNK is one of the most carcinogenic TSNAs in unburned tobacco and its smoke. 5 The increase in nitrate levels in cigarettes to enhance more complete combustion of tobacco has been postulated as one factor leading to the increase in lung adenocarcinoma due to the increased formation of TSNAs.6 NNK induces lung adenocarcinomas in various laboratory rodents, regardless of the route of administration.7 After humans smoke or otherwise consume tobacco, NNK is rapidly metabolized to NNAL (4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol) and NNAL-Gluc.8, 9 NNK and NNAL form DNA adducts after α-hydroxylation and are among the most potent carcinogenic TNSAs for lung tumorigenesis studied in mice.7 NNK is involved in multiple mechanisms in carcinogenesis. NNK induces signaling pathways that promote cell survival and proliferation10 and anti-inflammation in part through its role as a high affinity agonist of β-adrenergic and α7-nicotinic acetylcholine receptors.11, 12 This altered signaling not only affects neoplastic cells in various organs (lung, nose, colon, kidney, pancreas, liver) but also appears to affect pulmonary neuroendocrine cells leading to pulmonary disorders such as asthma and sudden infant death syndrome in infants of smoking mothers.13
Urinary levels of total NNAL (NNAL plus NNAL-Gluc) have recently been shown to be associated with lung cancer in a dose-dependent manner in cigarette smokers in two separate nested case-control studies.14 The highest tertile of total NNAL conferred an odds ratio of 2.11 (1.25–3.54) after controlling for measures of cigarette smoking and cotinine levels. Similarly, total NNAL levels have been associated with oral leukoplakia in smokeless tobacco users.15 Levels of total NNAL increase with an increase in the number of cigarettes smoked per day (CPD), but the rate decreases over 15 cigarettes per day and there is considerable variability.16, 17 Inter-individual variability in NNK metabolism was directly demonstrated by incubating freshly isolated human lung non-tumor cells with NNK and measuring metabolites of NNK bio-activation and transformation to NNAL, with wide metabolite ranges.18 Genetic polymorphisms in the genes affecting NNK metabolism may contribute to this variability. SNPs in UGT2B10 and CYP2A13 have been shown to alter NNAL-Gluc formation and NNK α-hydroxylation activity respectively in vitro.19, 20 There have been no prior epidemiological studies, however, examining the associations of these genes with total NNAL levels.
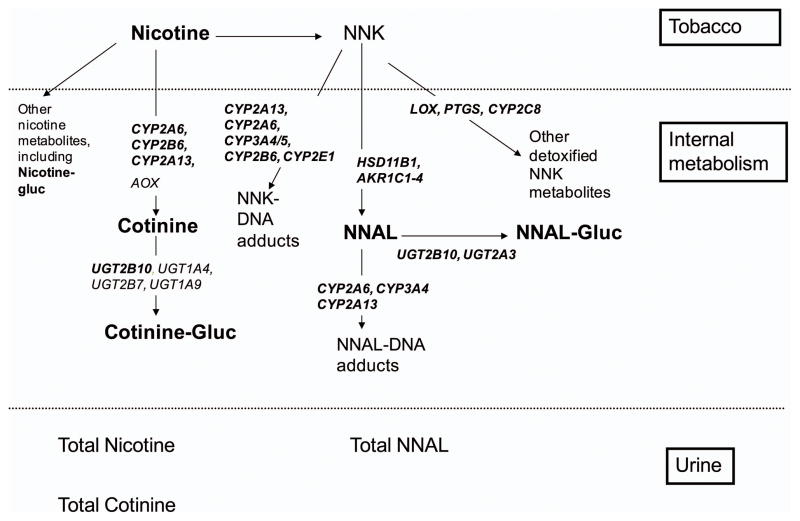
Carbonyl reductases and UDP-glucuronosyl transferases are involved in conversion of NNK to NNAL and detoxification to NNAL-Gluc. HSD11B1 codes for 11-Beta-Hydroxysteroid Dehydrogenase, Type I, located in lung and liver, which converts NNK to (R)-NNAL and shows a range of variability in mRNA expression and activity.21 AKR1C1, AKR1C2 and AKR1C4 code for aldo-keto reductases that convert NNK to (S)-NNAL.22 AKR1C1 has been shown to be highly over-expressed in non-small cell lung carcinoma (NSCLC) and small cell lung carcinoma, bronchial epithelial cells of NSCLC, and oral cancer cells by Affymetrix microarray.23 AKR1C4*5 (L311V rs17134592) is found on the C-terminal loop and affects steroid substrate specificity and Kcat.23 UGT2B10 codes for the UDP-glucuronosyl transferases that converts NNAL to NNAL-Gluc. In microsomes from human embryonic kidney (HEK)-293 cells over-expressing the UGT2B10 variant of SNP rs4657958 Asp67Tyr exhibited minimal glucuronide formation activity from NNAL or other TSNAs tested in vitro.19 Other pathways reduce the availability of NNK to be converted to NNAL. Cytochrome P450s are involved in toxification, converting NNK to intermediates that produce DNA adducts. Relevant genes are CYP2A13, CYP2A6, CYP3A4/5, CYP2B6, and CYP2E1. CYP2A13 is mainly expressed in nasal epithelium, trachea and lung and is primarily responsible for α-hydroxylation of NNK.24 CYP2A13 R257C (rs8192789), D158E, and V323L (rs3885816), had two- to three-fold decreased catalytic efficiency for NNK α-hydroxylation20 although allele frequencies in Caucasians are less than 3% for these SNPs. The CYP2A13 R257C variant carrier was associated with substantially reduced risk for lung adenocarcinoma [odds ratio= 0.41 (0.23–0.71)].25 The enzyme CYP2A6 is also involved in NNK metabolism, primarily in the liver, but also metabolizes nicotine to cotinine. Pyridine-N-oxidation detoxifies both NNK and NNAL. Other genes relevant for NNK metabolism are CYP2C8, LOX (lysyl oxidase), PTGS (prostaglandin endoperoxide synthase).26 In summary, candidate genes in this study include HSD11B1, AKR1C1/2/34, UGT2B10, CYP2A13, CYP2A6, CYP3A4/5, CYP2B6, CYP2E1, CYP2C8, LOX, PTGS1/2 and UGT2A3 (Figure 1).
Figure 1. Selected genes of NNK to NNAL and of nicotine to cotinine metabolism.
selected genes or measured compounds are in bold; Total NNAL, nicotine or cotinine includes their glucuronides
The total NNAL biomarker endpoint is narrowly defined compared to the broader phenotype of lung cancer although urinary measurements are representative of a relatively short time window of exposure prior to the time of collection. We hypothesized inter-individual variability in total NNAL levels is due to in part to genetic variation in specific metabolic genes of NNK metabolism which could help explain genetic susceptibility of smokers to lung cancer. We aimed to examine the roles of polymorphisms in 17 genes involved in NNK metabolism in the large Harvard/Massachusetts General Hospital (MGH) Lung Cancer Study from which study participants had been genotyped in a genome-wide association study (GWAS) and for whom there were also frozen urine samples.
Materials and Methods
Study population
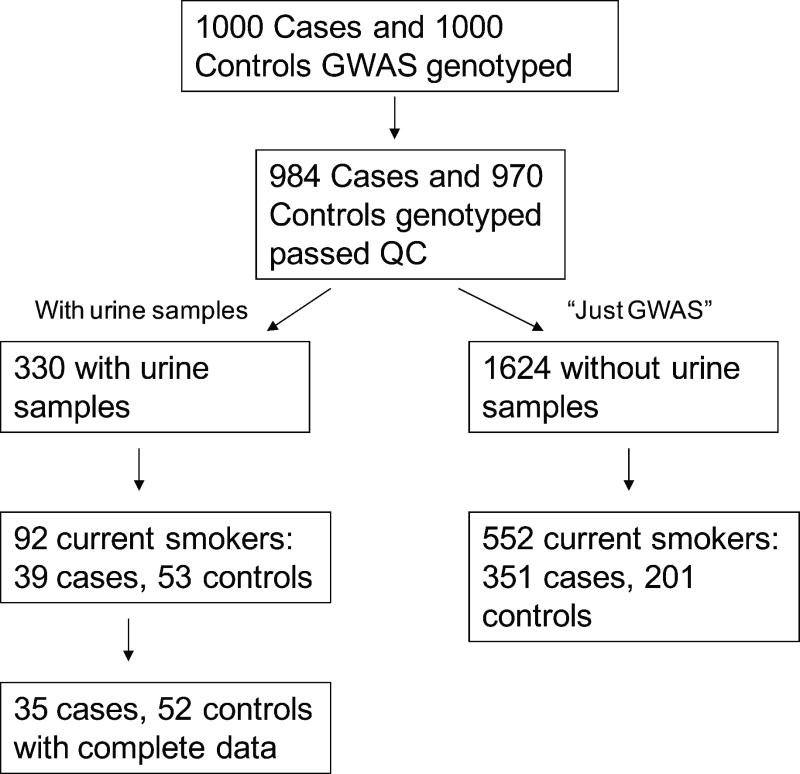
Samples were drawn from the Harvard/MGH Lung Cancer Study conducted from 1992 to the present. Interviewer-administered health questionnaires adapted from American Thoracic Society questionnaire27 provided information on demographics (such as age, sex) and detailed smoking histories from each subject. Current smokers were classified as those who reported smoking more than 100 cigarettes during their lifetime and who were smoking less than a 1 year prior to diagnosis or enrollment. Figure 2 outlines our sampling procedure. One thousand cases and one thousand controls, all self-reported Caucasian, were genotyped using the Illumina Human 610-Quad BeadChip for a GWAS on lung cancer risk and survival. 984 cases and 970 controls remained after applying quality control on ambiguous gender (genotype did not agree with the questionnaire), possible relatives and population outliers identified by a principle components analysis.
Figure 2.
Sampling flowchart from study participants genotyped for a Genome Wide Association Study (GWAS) and from those that also had urine samples
Urine samples had been collected early in the study (1992–1996) on 699 subjects at the time of diagnosis for the cases. The collected samples were frozen immediately and stored at −20°C for 5 years and −80°C for 10 years. Cases were requested to cease smoking prior to tumor resection and sample collection. Three hundred and thirty genotyped subjects also had urine samples (5cc) which could be used for this study of which 92 were current smokers. We limited our analysis of total NNAL, total nicotine and total cotinine in urine samples to the current smokers only, to maximize the probability of NNK exposure from mainstream tobacco smoking. There were 39 lung cancer cases and 53 controls.
Analysis of biomarkers
The outcome, total NNAL excretion (pmol per mg creatinine of NNAL plus NNAL-Gluc) in human urine was measured by gas chromatography with nitrosamine selective detection (GC-TEA).28 Total NNAL LOD was set at < 0.15 pmol/ml urine. Total NNAL LOD values were replaced with 0.07 pmol/ml urine. Total nicotine was measured as an alternative measure of exposure to NNK, which is undetectable in urine by GC-TEA. Total cotinine was measured as a potential confounder. Total nicotine per mg creatinine and total cotinine per mg creatinine (Free nicotine or cotinine plus their N-glucuronides) were analyzed by gas chromatography-mass spectrometry.29 Urinary creatinine was assayed with VITROS chemistry products CREA slides from Ortho Clinical Diagnostics (Raritan, NJ).
Selection of candidate genes and genotyping methods
Genotyping had been completed with the Illumina Human 610-Quad BeadChip from which tag SNPs could be extracted on selected genes. Seventeen candidate genes for metabolic enzymes with good evidence that they use NNK as a substrate (Table 1) were selected from the literature9, 19, 20, 22, 23, 25, 26, 30 and the bioinformatic databases GeneCards (http://www.genecards.org/index.shtml) and PharmGKB (http://www.pharmgkb.org/). 141 tag SNPs from Illumina Human 610-Quad BeadChip within 2 kb of the gene were chosen where gene start and stop endpoints were used as defined by National Center for Biotechnology Information (NCBI) build 36, the same build that the Illumina 610 BeadChip was based on. Call rates of genotyping were all greater than 95%. 126 SNPs passed the Hardy-Weinberg test of equilibrium at p≥0.05 in the original GWAS 970 controls. One per pair of SNPs in perfect LD (r2=1) were removed. The final 82 SNPs analyzed (Appendix 1) had sufficiently large minor allele frequencies (MAFs) and representative genotype frequencies in our subset of subjects with urine samples, using the following criteria. Recall that there were a total of 984 cases and 970 controls GWAS genotyped and passed quality control; of these there were 92 current smokers with urine samples and 552 current smokers without urine samples (referred to as the just GWAS sample) (Figure 2). The SNPs had an MAF≥0.05 in all 254 current smoker controls. We compared genotype frequencies in all 92 subjects analyzed in this study to those in the 552 just GWAS current smokers. We also compared the genotype frequencies in 53 controls analyzed in this study to the 201 just GWAS current smoker controls. To ensure our sample is similar to the GWAS sample, we retained SNPs where the genotype frequencies did not differ by the Fisher exact test (alpha=0.05) in the two comparisons.
Table 1.
Candidate genes in NNK metabolism
| Chromosome | Gene | number of SNPs |
|---|---|---|
| 10 | AKR1C1 | 2 |
| 10 | AKR1C2 | 3 |
| 10 | AKR1C3 | 3 |
| 10 | AKR1C4 | 9 |
| 19 | CYP2A13 | 3 |
| 19 | CYP2A6 | 0 |
| 19 | CYP2B6 | 12 |
| 10 | CYP2C8 | 11 |
| 10 | CYP2E1 | 9 |
| 7 | CYP3A4 | 2 |
| 7 | CYP3A5 | 6 |
| 1 | HSD11B1 | 5 |
| 5 | LOX | 2 |
| 9 | PTGS1 | 7 |
| 1 | PTGS2 | 2 |
| 4 | UGT2A3 | 4 |
| 4 | UGT2B10 | 2 |
Statistical analysis
We used SAS/Genetics software (ver. 9.1.3; SAS Institute, Cary, NC) and PLINK (ver.1.06)31 http://pngu.mgh.harvard.edu/purcell/plink/ to perform the analyses. We determined allele frequencies in cases and controls separately. To check for genotyping error, we examined departure from Hardy Weinberg Equilibrium in controls, using a χ2 test. All statistical testing was done at the two-sided 0.05 level for the p-value and 20% for the Benjamini-Hochberg false discovery rate (FDR) q-value.32
We examined the role of each genetic polymorphism in predicting urinary total NNAL levels using modified linear regression models. Since we performed linear regression on a secondary outcome from data derived from a case-control study we had to take into account that cases are oversampled in case-control designs. We used a weighted least squares regression, weighting cases by (the prevalence of lung cancer in the Massachusetts general population (=0.000745))/(the proportion of cases in the dataset) and weighting controls by (1- the lung cancer prevalence)/(the proportion of controls in the dataset).33 We also tested associations in the control group alone. Biomarker results and packyears34 were square-root transformed prior to analysis to normalize. Variables in the model included age, sex, square-root packyears and the concentration of urinary total nicotine (ng/mg creatinine) as potential confounding variables. The associations between the polymorphisms and total NNAL levels are reported as the change in total NNAL levels for each additional increase in the number of variant alleles (additive genetic model), and variant carriers vs. non-carriers (dominant model) and their corresponding 95% confidence intervals (95% CI) and p-values. We used the SAS macro, Happy, http://www.hsph.harvard.edu/faculty/kraft/soft.htm,35 to analyze haplotype associations for significant genes using the additive model. Haplotypes with frequencies less than 5% were combined into a single group. We also tested for interactions among significant SNPs in different genes with a cross product term.
We tested whether cotinine could be a confounder of the SNP-NNAL association, (since some of the same enzymes (CYP2A6, UGT2B10, and CYP2A13) also metabolize nicotine to cotinine). We tested the associations of cotinine with each of the SNPs controlling for nicotine and packyears, and tested the association of cotinine with NNAL, using all subjects and in controls alone. Biomarkers were log-transformed to meet the normality assumption with cotinine as the outcome in this analysis alone. For the SNPs significantly associated with cotinine, we included sqrt-cotinine in the model (with sqrt-nicotine and other variables previously mentioned) as a confounder of total NNAL outcome.
Results
Ninety-two Caucasian samples were analyzed for total NNAL, total nicotine, total cotinine and for creatinine. Creatinine was determined for all samples. For total NNAL, 4 samples (all cases) were not determined due to co-eluting peaks and 14 samples (12 cases) were at the LOD and so were replaced with 0.7 pmol/ml. For total nicotine only one sample, a control, was not determined due to co-eluting peaks. The same sample was not determined for cotinine, in addition to another case. Since the undetermined samples for nicotine and cotinine did not overlap for NNAL, our effective sample size controlling for nicotine was 87 (35 lung cancer cases and 52 controls) and for our cotinine analysis was 86. Sqrt –total NNAL was correlated with sqrt-cotinine (spearman ρ= 0.82 in controls, 0.37 in cases) and sqrt-nicotine (spearman ρ=0.74 in controls, 0.14 in cases).
For the 87 samples analyzed with complete total NNAL and nicotine (Table 2), there were 48 females and 39 males with ages ranging from 33 to 77 with a median of 62 years. Participants smoked 4 to 60 cigarettes per day (CPD) with a median of 20. There were no missing values for age, sex or CPD. Age, gender and sqrt-packyears were not significant variables predicting urinary sqrt-total NNAL when sqrt-nicotine was included. Sqrt-nicotine was highly significant predicting a 0.018 (0.014, 0.023) pmol/mg creatinine unit increase in sqrt total NNAL for every ng/mg creatinine increase in sqrt-nicotine at p<10E-16. Removing the two outliers, one in cases and controls each (the highest value of total nicotine), did not significantly change the results of the association with sqrt-nicotine and sqrt-total NNAL (beta =0.0188 (0.014, 0.023), p=3.11E-15), therefore outliers remained in the analysis. For controls only, only sqrt-nicotine was significant at beta=0.0185 (0.013, 0.024), p=5.52E-09. Four SNPs in HSD11B1 and AKR1C4 were associated with sqrt NNAL under the additive model (Table 3; Appendix 2 shows all SNP results under the additive model). One SNP in CYP2E1 was also significant under the dominant model.
Table 2.
Descriptive characteristics of the study populationa
| Characteristics | Total (n=87) | Lung cancer cases (n= 35) | Controls (n=52) |
|---|---|---|---|
| Ageb | 62 (33–77) | 65 (35–77) | 59 (33–76) |
| Gender | |||
| Males | 39 (44.8%) | 20 (57.1%) | 19 (36.5%) |
| Females | 48 (55.2%) | 15 (42.9%) | 33 (63.5%) |
| Pack yearsb | 45.5 (8.3–172.5) | 58 (8.3–172.5) | 36.1 (8.7–89.9) |
| Sqrt (Packyrs) | 6.67 (2.88–13.13) | 7.62 (2.88–13.13)n | 6.0 (2.94–9.48)n |
| Cigarettes/dayb | 20 (4–60) | 20 (4–60) | 20 (4–50) |
| Time smoking (yrs)b | 42.8 (17.3–61.7) | 49 (20–58) | 41.5 (17.3–61.7) |
| Total NNAL (pmol/mg creatinine)c | 1.41 (0.04–9.83) | 0.44 (0.04–7.62) | 2.42 (0.09–9.83) |
| Sqrt(NNAL) | 1.19 (0.2–3.1) | 0.66 (0.2–2.76) | 1.55 (0.2–3.13)n |
| LODc | 14 | 12 | 2 |
| Nicotine (ng/mg creatinine) | 740.57 (2.66–10933.8) | 31.1 (2.66–2139.9) | 2034.1(12.74–10933.8) |
| Sqrt(nicotine) | 27.2 (1.63–104.56) | 5.58 (1.63–46.26) | 45.10 (3.57–104.56)n |
| Cotinine (ng/mg creatinine) | 1714.96 (4.53–10939.1)d | 129 (4.53–5314.4)d | 3259.9 (22.2–10939.1) |
| Sqrt(cotinine) | 41.4 (2.13–104.59) | 11.3 (2.13–72.89) | 57.09 (4.72–104.59)n |
| Creatinine (mg/ml) | 0.97 (0.15–5.54) | 0.80 (0.15–5.54) | 1.10 (0.35–3.32) |
all Caucasian smokers, restricted to complete measurements on total NNAL and nicotine
median (range)
Total NNAL pmol/ml LOD ~= < 0.15 pmol/ml urine, (LOD values replaced with 0.07 pmol/ml urine)
Total N=86, Cases N=34
normally distributed
Table 3.
Association of significant SNPs with square-root total NNAL
| Gene | SNP | All subjects | Controls only | Case genotype countsb | Control genotype counts b | Case minor allele frequency | Control minor allele frequency | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate (95%CI)a | p-value | fdr q- value | Estimate (95%CI)a | p- value | fdr q- value | ||||||
| HSD11B1 | rs2235543 | −0.38 (−0.52, −0.27) | 1.37E-07 | 1.07E-05 | −0.38 (−0.59, −0.16) | 0.0013 | 0.10 | 2/5/28 | 1/8/43 | 0.13 | 0.10 |
| HSD11B1 | rs3753519 | −0.42 (−0.68, −0.16) | 0.0017 | 0.07 | −0.42 (−0.73, −0.12) | 0.0076 | 0.31 | 2/5/28 | 0/7/45 | 0.13 | 0.07 |
| AKR1C4 | rs7083869 | −0.21 (−0.38, −0.04) | 0.019 | 0.51 | −0.21 (−0.43, 0.013) | 0.06 | 0.99 | 2/14/19 | 0/19/33 | 0.26 | 0.18 |
| HSD11B1 | rs10863782 | −0.26 (−0.51, −0.02) | 0.035 | 0.72 | −0.26 (−0.52, −0.01) | 0.039 | 0.99 | 2/7/26 | 0/12/40 | 0.16 | 0.12 |
additive genetic model, controlling for age, sex, square-root packyears, square-root nicotine
homozygous variant/heterozygous/homozygous wild type
Three HSD11B1 SNPs, located in a 14kb region flanked by rs2235543 and rs3753519 in intron 1 with strong linkage disequilibrium, were significantly associated with decreased total NNAL levels (Table 3). Using the whole data set, two of these SNPs passed multiple testing adjustment: rs2235543 C>T (padditive= 1.37E-07, FDR qadditive=1.13E-05; pdominant=7.61E-05, FDR qdominant=0.0062) and rs3753519 G>A (padditive/dominant= 0.0017, FDR qadditive/dominant= 0.07). HSD11B1 rs10863782 was also significant at p=0.035 under both models, but did not pass multiple testing adjustment. Similar although slightly attenuated results were seen in controls alone. The same three HSD11B1 SNPs were significant and rs2235543 still passed a multiple testing adjustment with padditive=0.0013, FDR qadditive=0.10; pdominant=0.004 FDR qdominant=0.31. A haplotype analysis of all 5 SNPs included for HSD11B1 showed that one haplotype that contained a minor allele of the 3 SNPs rs2235543, rs10863782 and rs3753519 and major allele of the 2 others had a frequency of 5% in our population (3.6 in controls and 7% in cases) and was associated with decreasing NNAL levels, beta = −0.59 (−0.87, −0.31) with a p-value of 3.41E-05 using all subjects and 0.0098 in controls only. None of the other haplotypes were significantly associated.
AKR1C4 rs7083869 G>A, located in the second intron, was also associated with sqrt-total NNAL at p=0.019 under both models, using all subjects, but did not pass multiple testing adjustment, FDR q=0.51. In controls alone, this SNP was borderline not significant at p=0.06, FDR q=0.99 under both models. A haplotype analysis of all 9 SNPs selected in AKR1C4 showed that the minor allele of this SNP was located only on a single common haplotype, along with minor alleles of rs11253042 and rs6601927. This haplotype had a frequency of 14.4% in our population (15.4% in controls and 12.9% in cases). The haplotype was associated with decreasing NNAL levels, beta = −0.17(−0.44, 0.09) with a p-value of 0.21 using all subjects and 0.27 in controls only. The other haplotypes were also not significantly associated. There were no significant interactions of AKR1C4 rs7083869 with each of the three significant HSD11B1 SNPs.
One SNP in CYP2E1 rs915907 C>A had a borderline significant result under the dominant genetic model, regression coefficient = −0.19 (−0.37, −0.0009), p=0.048, FDR q=0.75, but not under the additive model, p=0.12, FDR q=0.73, using all subjects. This SNP was also not significant for controls only under either genetic model, p=0.19, FDR q=0.99.
Cotinine was a statistically significant confounder of the association with total NNAL for SNPs in genes not known to be involved in nicotine to cotinine metabolism except for UGT2B10. Cotinine was significantly associated with total NNAL after controlling for packyears and nicotine (with a log transformation of all variables, beta=1.23 (0.84. 1.62) p<5E-05). Cotinine was significantly associated (although not after multiple testing adjustment), with similar results under both genetic models, with the following SNPs after controlling for age, sex, log-pack-years and log-nicotine using all subjects: under the additive model, HSD11B1 rs3753519 with p= 0.004 FDR q=0.24, CYP3A4 rs4646437 at p=0.006 FDR q=0.24, and HSD11B1 rs10863782 at p=0.026, and HSD11B1 rs2235543 at p=0.028, both FDR q=0.57. In controls alone, the same SNPs and UGT2B10 rs861340 at p=0.03 and UGT2B10 rs835316 at p=0.04, both FDR q=0.55, were nominally significant under both genetic models.
For SNPs that appeared to be confounded with cotinine levels, we reanalyzed the NNAL associations including sqrt-cotinine in the model. In all subjects, the SNPs HSD11B1 rs2235543 and rs3753519 remained significant at p=4.86E-06 and p= 0.014 respectively, although the regression coefficients were decreased by about half to −0.27 (−0.39, −0.16) and to −0.23 (−0.42, −0.047) respectively. HSD11B1 rs10863782 was no longer significant at p=0.18 and the UGTB10 SNP remained unassociated with total NNAL levels after adjusting for cotinine.
Discussion
After adjusting for multiple testing, genetic polymorphisms HSD11B1 rs2235543 and rs3753519 appear to be associated with total NNAL metabolite levels with the minor alleles associated with decreasing levels. Variants HSD11B1 rs10863782, AKR1C4 rs7083869 and CYP2E1 rs915907 may also be associated with decreasing total NNAL metabolite levels but these SNPs did not pass a multiple testing adjustment, possibly due to sample size. Both enzymes HSD11B1 and AKR1C4 are carbonyl reductases directly involved in the single step reduction of NNK to NNAL.
The enzyme HSD11B1 is primarily known for its ability to reversibly oxidize glucocorticoids at carbon 11 such as cortisol to cortisone, but its ability to catalyze the carbonyl reduction of NNK and other non steroidal carbonyl compounds has recently been discovered.36 The enzyme 11β-hydroxysteroid dehydrogenase shows high inter-individual variation both in the mRNA expression and activity of NNAL formation.21 HSD11B1 has 2 mRNA transcripts, 48.7kb and 30.1kb. One potentially functional genetic variant G>A, in HSD11B1 has been reported, rs13306421, located two nucleotides 5′ to the translation initiation site of the smaller transcript. Compared with the common G allele, the A allele in vitro was translated at higher levels and resulted in higher enzyme expression and activity.37 However this variant has a very low frequency in the Caucasian population. Another study reported a 20% reduction in luciferase activity in a reporter-gene assay with a rare HSD11B1 haplotype including rs846911 and rs860185 which indicated altered transcription.38 SNPs rs846911 and rs860185 are only 2kb 5′ to rs1330621 and only 0.73kb downstream of the significant SNP rs3753519. The SNPs we analyzed encompass this 2kb area around the second transcription start site. Similar to HapMap CEU (CEPH (Centre d’Etude du Polymorphisme Humain) Utah residents with ancestry from northern and western Europe) data (release 24),39 all the selected SNPs in HSD11B1 had a pairwise r2<0.80 but strong LD as measured by D′. The single haplotype containing the minor alleles of the 3 significant HSD11B1 SNPs was associated with a stronger regression coefficient than the single SNPs alone. It is possible that these 3 SNPs, which are 5′ to these reported variants close to the initiation site for the smaller HSD11B1 transcript, serve as a marker for the causal SNP or haplotype producing reduced enzyme activity.
AKR1C4 is an aldo-keto reductase, along with AKR1C1 and AKR1C2, involved in formation of (S)-NNAL, the NNAL enantiomer that is less readily glucuronidated in rats.22, 23, 40 (S)-NNAL can be oxidized back to NNK. In contrast, 11β-HSD catalyzes formation of (R)-NNAL which, in rats, is further glucuronidated and excreted.23, 40 This enzymatic stereospecificity of AKR1C4 to a less excretable NNAL form may explain the lower point estimate of the association of the AKR1C4 SNP compared to the HSD11B1 SNPs. A number of studies have reported that AKR1C4 L311V (encoded by rs17134592) lowers enzyme activity by as much as 3–5 fold.41 The SNP rs17134592 has a 15% MAF in Caucasians (dbSNP) and is in strong LD (D′ of 1 with HapMap CEU data) with our significant SNP rs7083869, but is also correlated with the other non-significant SNPs. There were no significant haplotypes associated with NNAL values although the regression coefficient of the single haplotype containing the minor allele rs7083869 was very similar to the single SNP alone. It is possible that the common minor allele of rs11253042, located in this haplotype and also other haplotypes, is attenuating the signal.
CYP2E1 oxidizes NNK to intermediates that can form DNA adducts. This enzyme also metabolizes a wide variety of other compounds and has a number of inducers (such as ethanol) and inhibitors. The gene has 2 known functional polymorphisms, more common in Asian populations than Caucasian, commonly known by their restriction sites, RsaI (−1053C>T, rs2031920) reducing inducibility, and DraI restriction fragment in intron 6 (7632T>A, rs6413432), with reduced in vivo enzyme activity. Various cancer associations have been reported with these polymorphisms in Asian populations compared to mostly non-significant results reported in Caucasian populations.42 The SNP we found to have borderline significant nominal p-value in all subjects, CYP2E1 rs915907 is 7kb downstream of RsaI and 1.25kb downstream of DraI and is linked to these functional polymorphisms by D′. However, it is not highly correlated with them since it is more frequent in Caucasians, (HapMap CEU MAF 0.18, compared to 0.06 and 0.02 for RsaI and DraI respectively). RsaI or rs2031920 was dropped from our original analysis due to a low minor allele frequency in our data, but testing this SNP’s associations with total NNAL levels using both additive and dominant genetic models did not produce significant results, (p=0.35). The association of rs915907 with the dominant model is suggestive but further evidence is needed to identify the role of this SNP in NNK metabolism.
We observed that different SNPs were associated with total NNAL levels and with cotinine levels although there was some overlap with HSD11B1 rs3753519 G>A. Neither HSD11B1 nor CYP3A4 are known to be directly involved in nicotine to cotinine metabolism, although nicotine has been shown to induce CYP3A4 and other CYP enzyme expression through the nuclear receptor pregnane X receptor.43, 44 While both SNPs in both HSD11B1 and CYP3A4 did not pass a multiple testing adjustment for an association with cotinine levels, the raw p-value could also be the result of unknown uncontrolled confounders since both enzymes have broad substrate specificity.
Some inhibitors of HSD11B1 and AKRC enzymes could explain some of the interindividual variability seen in NNAL output. They could be potential uncontrolled confounders if related to the polymorphisms and if a significant proportion of the study population is exposed. Some known HSD11B1 enzyme inhibitors are endogenous steroids (glucocorticoids, estrogen, progesterone, cholesterol, bile acids), exogenous steroids (glucocorticoids, dexamethasone, oral contraceptives), drugs (carbenoxolone, furosemide, ethacrynic acid, gossypol, ketoconazole, metyrapone) and dietary compounds (naringenin (found in grapefruit), glycyrrhetinic acid (licorice), and flavonoids).45, 46 Alcohol is also an inhibitor of HSD11B1 and AKR1C1, 1C2, 1C4 enzymes. We had insufficient information on these compounds in our study. On the other hand, the SNP(s) may modify the inhibitors effects, in other words an inhibitor may have different effects in the presence of different forms of the HSD11B1 enzyme due to the polymorphism. Additional study on genetic interactions with these inhibitors could further highlight the importance of HSD11B1 in contributing to NNAL variability and by extension to lung cancer development in inhibitor exposed versus unexposed populations.
There a few limitations to this study. Coverage of some relevant genes may be insufficient in part due to coverage of the Illumina 610 chip (e.g. CYP2A6), and due to our own stringent criteria for SNPs that would sufficiently capture the genotype frequencies in the larger sample of smokers genotyped for the GWAS. The total sample size of 87 subjects may be insufficient to detect moderate associations or associations with low frequency SNPs, although two highly associated SNPs we found had MAF’s less than 10% in controls. Additionally, our SNP selection process tried to ensure that our small sample was comparable to the much larger sample of genotyped individuals that we did not have urine samples for. The controls are fairly representative of smokers in Eastern Massachusetts in terms of self reported smoking habits,47 and the lung cancer cases’ self reported smoking habits are significantly higher as expected. However, cases were requested to cease smoking prior to tumor resection and before urine sample collection which resulted in lower median nicotine and NNAL levels in the cases compared to controls that smoked, although higher levels than non-smokers or individuals exposed to second-hand smoke.48 Given the small sample size of cases, the request to stop smoking and possibly the motivation of a recent diagnosis, it is difficult to determine if the lung cancer disease condition itself had any impact on the inter-individual variability of NNAL.
We were able to detect NNAL in long term frozen urine samples with little apparent loss, as has been demonstrated in other studies.49 Time since smoking last cigarette to urine sample collection may have affected NNAL levels since the initial half-life of NNAL is 3 days.50 However, the nicotine, cotinine and NNAL ranges in our controls are representative of other smokers.16, 28 NNAL levels in the cases appear to be similar to people who stopped smoking two to three weeks prior to sample collection.50 The lower than expected NNAL levels in cases did not appear to have had much of an effect on our results since they were similar in all subjects combined compared to controls only.
To our knowledge, this study is the first to examine readily available, high quality genotyped data on a number of polymorphisms in NNK metabolism with total NNAL levels as a biomarker. The results show that the most significant SNPs that are associated with inter-individual variability of NNAL in a sample of smokers are also correlated with potentially functional variants in HSD11B1 and possibly AKR1C4 that have been associated with altered mRNA expression and enzyme activity in vitro. While confirmation in a larger sample of smokers is warranted, HSD11B1 and possibly AKR1C4 polymorphisms may provide part of the mechanism by which varying NNAL levels have been shown to be a useful prognostic factor for lung cancer.14
Supplementary Material
Acknowledgments
Grant Sponsors: Harvard- National Institute for Environmental Health Sciences (NIEHS) Center for Environmental Health, National Institutes of Health Office of Extramural Research (NIH-OER), National Cancer Institute (NCI)
We thank Salvatore Mucci for valuable contributions in data collection and management and Benjamin Church for sample management. M.T. was awarded the Harvard-NIEHS Center (ES00002) for Environmental Health pilot grant to conduct this work. Funding was also provided by NIH-OER Ruth L. Kirschstein-National Research Service Award (T32 ES 007069), and NCI grant CA074386
Abbreviations used
- AKR
aldo-keto reductases
- CEU
(CEPH (Centre d’Etude du Polymorphisme Humain) Utah residents with ancestry from northern and western Europe)
- CPD
cigarettes smoked per day
- CYP
cytochrome p450
- Gluc
glucuronide
- FDR
false discovery rate
- GWAS
genome-wide association study
- HSD11B1
11-hydroxy-steroid deydogenase type 1
- LOX
Lysyl oxidase, MAF, minor allele frequency
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNAL
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol
- PTGS
prostaglandin endoperoxide synthase
- TSNA
Tobacco-specific nitrosamines
- sqrt
square-root
- UGT
UDP-glucuronosyl transferases
Footnotes
Novelty and impact: This paper quantifies the associations of polymorphisms in several genes involved in the metabolism of a tobacco-specific nitrosamine NNK, a potent carcinogen, with its metabolite measured as total NNAL in urine. The paper provides evidence that genetic variability in NNK carbonyl reductases, HSD11B1 and AKR1C4, may contribute to the inter-individual variability seen in total NNAL, previously shown to be a predictor of lung cancer risk.
References
- 1.US Cancer Statistics Working Group. United States Cancer Statistics: 1999–2005 Incidence and Mortality Web-based Report. Atlanta: Department of Health and Human Services, Centers for Disease Control and Prevention, and National Cancer Institute; 2009. [Google Scholar]
- 2.IARC. Tobacco smoke and involuntary smoking. IARC Monogr Eval Carcinog Risks Hum. 2004;83:1–1438. [PMC free article] [PubMed] [Google Scholar]
- 3.Peto R, Darby S, Deo H, Silcocks P, Whitley E, Doll R. Smoking, smoking cessation, and lung cancer in the UK since 1950: combination of national statistics with two case-control studies. BMJ. 2000;321:323–9. doi: 10.1136/bmj.321.7257.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crispo A, Brennan P, Jockel KH, Schaffrath-Rosario A, Wichmann HE, Nyberg F, Simonato L, Merletti F, Forastiere F, Boffetta P, Darby S. The cumulative risk of lung cancer among current, ex- and never-smokers in European men. Br J Cancer. 2004;91:1280–6. doi: 10.1038/sj.bjc.6602078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stepanov I, Upadhyaya P, Carmella SG, Feuer R, Jensen J, Hatsukami DK, Hecht SS. Extensive metabolic activation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in smokers. Cancer Epidemiol Biomarkers Prev. 2008;17:1764–73. doi: 10.1158/1055-9965.EPI-07-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann D, Djordjevic MV, Hoffmann I. The changing cigarette. Prev Med. 1997;26:427–34. doi: 10.1006/pmed.1997.0183. [DOI] [PubMed] [Google Scholar]
- 7.Amin S, Desai D, Hecht SS, Hoffmann D. Synthesis of tobacco-specific N-nitrosamines and their metabolites and results of related bioassays. Crit Rev Toxicol. 1996;26:139–47. doi: 10.3109/10408449609017927. [DOI] [PubMed] [Google Scholar]
- 8.Maser E, Richter E, Friebertshauser J. The identification of 11 beta-hydroxysteroid dehydrogenase as carbonyl reductase of the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Eur J Biochem. 1996;238:484–9. doi: 10.1111/j.1432-1033.1996.0484z.x. [DOI] [PubMed] [Google Scholar]
- 9.Maser E. 11Beta-hydroxysteroid dehydrogenase responsible for carbonyl reduction of the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in mouse lung microsomes. Cancer Res. 1998;58:2996–3003. [PubMed] [Google Scholar]
- 10.Akopyan G, Bonavida B. Understanding tobacco smoke carcinogen NNK and lung tumorigenesis. Int J Oncol. 2006;29:745–52. [PubMed] [Google Scholar]
- 11.Wu WK, Wong HP, Luo SW, Chan K, Huang FY, Hui MK, Lam EK, Shin VY, Ye YN, Yang YH, Cho CH. 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone from cigarette smoke stimulates colon cancer growth via beta-adrenoceptors. Cancer Res. 2005;65:5272–7. doi: 10.1158/0008-5472.CAN-05-0205. [DOI] [PubMed] [Google Scholar]
- 12.Egleton RD, Brown KC, Dasgupta P. Nicotinic acetylcholine receptors in cancer: multiple roles in proliferation and inhibition of apoptosis. Trends Pharmacol Sci. 2008;29:151–8. doi: 10.1016/j.tips.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Schuller HM, Jull BA, Sheppard BJ, Plummer HK. Interaction of tobacco-specific toxicants with the neuronal alpha(7) nicotinic acetylcholine receptor and its associated mitogenic signal transduction pathway: potential role in lung carcinogenesis and pediatric lung disorders. Eur J Pharmacol. 2000;393:265–77. doi: 10.1016/s0014-2999(00)00094-7. [DOI] [PubMed] [Google Scholar]
- 14.Yuan JM, Koh WP, Murphy SE, Fan Y, Wang R, Carmella SG, Han S, Wickham K, Gao YT, Yu MC, Hecht SS. Urinary levels of tobacco-specific nitrosamine metabolites in relation to lung cancer development in two prospective cohorts of cigarette smokers. Cancer Res. 2009;69:2990–5. doi: 10.1158/0008-5472.CAN-08-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kresty LA, Carmella SG, Borukhova A, Akerkar SA, Gopalakrishnan R, Harris RE, Stoner GD, Hecht SS. Metabolites of a tobacco-specific nitrosamine, 4-(methylnitrosamino)- 1-(3-pyridyl)-1-butanone (NNK), in the urine of smokeless tobacco users: relationship between urinary biomarkers and oral leukoplakia. Cancer Epidemiol Biomarkers Prev. 1996;5:521–5. [PubMed] [Google Scholar]
- 16.Joseph AM, Hecht SS, Murphy SE, Carmella SG, Le CT, Zhang Y, Han S, Hatsukami DK. Relationships between cigarette consumption and biomarkers of tobacco toxin exposure. Cancer Epidemiol Biomarkers Prev. 2005;14:2963–8. doi: 10.1158/1055-9965.EPI-04-0768. [DOI] [PubMed] [Google Scholar]
- 17.Lubin JH, Caporaso N, Hatsukami DK, Joseph AM, Hecht SS. The association of a tobacco-specific biomarker and cigarette consumption and its dependence on host characteristics. Cancer Epidemiol Biomarkers Prev. 2007;16:1852–7. doi: 10.1158/1055-9965.EPI-07-0018. [DOI] [PubMed] [Google Scholar]
- 18.Smith GB, Castonguay A, Donnelly PJ, Reid KR, Petsikas D, Massey TE. Biotransformation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in freshly isolated human lung cells. Carcinogenesis. 1999;20:1809–18. doi: 10.1093/carcin/20.9.1809. [DOI] [PubMed] [Google Scholar]
- 19.Chen G, Dellinger RW, Gallagher CJ, Sun D, Lazarus P. Identification of a prevalent functional missense polymorphism in the UGT2B10 gene and its association with UGT2B10 inactivation against tobacco-specific nitrosamines. Pharmacogenet Genomics. 2008;18:181–91. doi: 10.1097/FPC.0b013e3282f4dbdd. [DOI] [PubMed] [Google Scholar]
- 20.Schlicht KE, Michno N, Smith BD, Scott EE, Murphy SE. Functional characterization of CYP2A13 polymorphisms. Xenobiotica. 2007;37:1439–49. doi: 10.1080/00498250701666265. [DOI] [PubMed] [Google Scholar]
- 21.Soldan M, Nagel G, Losekam M, Ernst M, Maser E. Interindividual variability in the expression and NNK carbonyl reductase activity of 11beta-hydroxysteroid dehydrogenase 1 in human lung. Cancer Lett. 1999;145:49–56. doi: 10.1016/s0304-3835(99)00216-5. [DOI] [PubMed] [Google Scholar]
- 22.Atalla A, Breyer-Pfaff U, Maser E. Purification and characterization of oxidoreductases-catalyzing carbonyl reduction of the tobacco-specific nitrosamine 4-methylnitrosamino-1-(3-pyridyl)-1-butanone (NNK) in human liver cytosol. Xenobiotica. 2000;30:755–69. doi: 10.1080/00498250050119826. [DOI] [PubMed] [Google Scholar]
- 23.Penning TM, Drury JE. Human aldo-keto reductases: Function, gene regulation, and single nucleotide polymorphisms. Arch Biochem Biophys. 2007;464:241–50. doi: 10.1016/j.abb.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossini A, de Almeida Simao T, Albano RM, Pinto LF. CYP2A6 polymorphisms and risk for tobacco-related cancers. Pharmacogenomics. 2008;9:1737–52. doi: 10.2217/14622416.9.11.1737. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Tan W, Hao B, Miao X, Zhou G, He F, Lin D. Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 2003;63:8057–61. [PubMed] [Google Scholar]
- 26.Smith GB, Bend JR, Bedard LL, Reid KR, Petsikas D, Massey TE. Biotransformation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in peripheral human lung microsomes. Drug Metab Dispos. 2003;31:1134–41. doi: 10.1124/dmd.31.9.1134. [DOI] [PubMed] [Google Scholar]
- 27.Ferris BG. Epidemiology Standardization Project (American Thoracic Society) Am Rev Respir Dis. 1978;118:1–120. [PubMed] [Google Scholar]
- 28.Carmella SG, Han S, Fristad A, Yang Y, Hecht SS. Analysis of total 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human urine. Cancer Epidemiol Biomarkers Prev. 2003;12:1257–61. [PubMed] [Google Scholar]
- 29.Hecht SS, Carmella SG, Chen M, Dor Koch JF, Miller AT, Murphy SE, Jensen JA, Zimmerman CL, Hatsukami DK. Quantitation of urinary metabolites of a tobacco-specific lung carcinogen after smoking cessation. Cancer Res. 1999;59:590–6. [PubMed] [Google Scholar]
- 30.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 33.Vanderweele TJ, Vansteelandt S. Odds ratios for mediation analysis for a dichotomous outcome. Am J Epidemiol. 2010;172:1339–48. doi: 10.1093/aje/kwq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thurston SW, Liu G, Miller DP, Christiani DC. Modeling lung cancer risk in case-control studies using a new dose metric of smoking. Cancer Epidemiol Biomarkers Prev. 2005;14:2296–302. doi: 10.1158/1055-9965.EPI-04-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraft P, Cox DG, Paynter RA, Hunter D, De Vivo I. Accounting for haplotype uncertainty in matched association studies: a comparison of simple and flexible techniques. Genet Epidemiol. 2005;28:261–72. doi: 10.1002/gepi.20061. [DOI] [PubMed] [Google Scholar]
- 36.Maser E, Friebertshauser J, Volker B. Purification, characterization and NNK carbonyl reductase activities of 11beta-hydroxysteroid dehydrogenase type 1 from human liver: enzyme cooperativity and significance in the detoxification of a tobacco-derived carcinogen. Chem Biol Interact. 2003;143–144:435–48. doi: 10.1016/s0009-2797(02)00180-1. [DOI] [PubMed] [Google Scholar]
- 37.Malavasi EL, Kelly V, Nath N, Gambineri A, Dakin RS, Pagotto U, Pasquali R, Walker BR, Chapman KE. Functional effects of polymorphisms in the human gene encoding 11 beta-hydroxysteroid dehydrogenase type 1 (11 beta-HSD1): a sequence variant at the translation start of 11 beta-HSD1 alters enzyme levels. Endocrinology. 2010;151:195–202. doi: 10.1210/en.2009-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Quervain DJ, Poirier R, Wollmer MA, Grimaldi LM, Tsolaki M, Streffer JR, Hock C, Nitsch RM, Mohajeri MH, Papassotiropoulos A. Glucocorticoid-related genetic susceptibility for Alzheimer’s disease. Hum Mol Genet. 2004;13:47–52. doi: 10.1093/hmg/ddg361. [DOI] [PubMed] [Google Scholar]
- 39.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breyer-Pfaff U, Martin HJ, Ernst M, Maser E. Enantioselectivity of carbonyl reduction of 4-methylnitrosamino-1-(3-pyridyl)-1-butanone by tissue fractions from human and rat and by enzymes isolated from human liver. Drug Metab Dispos. 2004;32:915–22. [PubMed] [Google Scholar]
- 41.Bains OS, Grigliatti TA, Reid RE, Riggs KW. Naturally occurring variants of human aldo-keto reductases with reduced in vitro metabolism of daunorubicin and doxorubicin. J Pharmacol Exp Ther. 2010;335:533–45. doi: 10.1124/jpet.110.173179. [DOI] [PubMed] [Google Scholar]
- 42.Neafsey P, Ginsberg G, Hattis D, Johns DO, Guyton KZ, Sonawane B. Genetic polymorphism in CYP2E1: Population distribution of CYP2E1 activity. J Toxicol Environ Health B Crit Rev. 2009;12:362–88. doi: 10.1080/10937400903158359. [DOI] [PubMed] [Google Scholar]
- 43.Lee MR, Kim YJ, Hwang DY, Kang TS, Hwang JH, Lim CH, Kang HK, Goo JS, Lim HJ, Ahn KS, Cho JS, Chae KR, et al. An in vitro bioassay for xenobiotics using the SXR-driven human CYP3A4/lacZ reporter gene. Int J Toxicol. 2003;22:207–13. doi: 10.1080/10915810305110. [DOI] [PubMed] [Google Scholar]
- 44.Lamba V, Yasuda K, Lamba JK, Assem M, Davila J, Strom S, Schuetz EG. PXR (NR1I2): splice variants in human tissues, including brain, and identification of neurosteroids and nicotine as PXR activators. Toxicol Appl Pharmacol. 2004;199:251–65. doi: 10.1016/j.taap.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 45.Maser E. Stress, hormonal changes, alcohol, food constituents and drugs: factors that advance the incidence of tobacco smoke-related cancer? Trends Pharmacol Sci. 1997;18:270–5. doi: 10.1016/s0165-6147(97)01090-0. [DOI] [PubMed] [Google Scholar]
- 46.Maser E. Significance of reductases in the detoxification of the tobacco-specific carcinogen NNK. Trends Pharmacol Sci. 2004;25:235–7. doi: 10.1016/j.tips.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Beiner L, Nyman AL, Roman AM, Flynn CA, Albers AB. Massachusetts Adult Tobacco Survey: Technical Report & Tables 1993–2000. Center for Survey Research. University of Massachusetts; Boston: 2001. [Google Scholar]
- 48.Hecht SS, Ye M, Carmella SG, Fredrickson A, Adgate JL, Greaves IA, Church TR, Ryan AD, Mongin SJ, Sexton K. Metabolites of a tobacco-specific lung carcinogen in the urine of elementary school-aged children. Cancer Epidemiol Biomarkers Prev. 2001;10:1109–16. [PubMed] [Google Scholar]
- 49.Kavvadias D, Scherer G, Urban M, Cheung F, Errington G, Shepperd J, McEwan M. Simultaneous determination of four tobacco-specific N-nitrosamines (TSNA) in human urine. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:1185–92. doi: 10.1016/j.jchromb.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Carmella SG, Chen M, Han S, Briggs A, Jensen J, Hatsukami DK, Hecht SS. Effects of smoking cessation on eight urinary tobacco carcinogen and toxicant biomarkers. Chem Res Toxicol. 2009;22:734–41. doi: 10.1021/tx800479s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.