Abstract
BrainInfo (http://braininfo.org) is a growing portal to neuroscientific information on the Web. It is indexed by NeuroNames, an ontology designed to compensate for ambiguities in neuroanatomical nomenclature. The 20-year old ontology continues to evolve toward the ideal of recognizing all names of neuroanatomical entities and accommodating all structural concepts about which neuroscientists communicate, including multiple concepts of entities for which neuroanatomists have yet to determine the best or ‘true’ conceptualization. To make the definitions of structural concepts unambiguous and terminologically consistent we created a ‘default vocabulary’ of unique structure names selected from existing terminology. We selected standard names by criteria designed to maximize practicality for use in verbal communication as well as computerized knowledge management. The ontology of NeuroNames accommodates synonyms and homonyms of the standard terms in many languages. It defines complex structures as models composed of primary structures, which are defined in unambiguous operational terms. NeuroNames currently relates more than 16,000 names in eight languages to some 2,500 neuroanatomical concepts. The ontology is maintained in a relational database with three core tables: Names, Concepts and Models. BrainInfo uses NeuroNames to index information by structure, to interpret users’ queries and to clarify terminology on remote web pages. NeuroNames is a resource vocabulary of the NLM’s Unified Medical Language System (UMLS, 2011) and the basis for the brain regions component of NIFSTD (NeuroLex, 2011). The current version has been downloaded to hundreds of laboratories for indexing data and linking to BrainInfo, which attracts some 400 visitors/day, downloading 2,000 pages/day.
Keywords: NeuroNames, BrainInfo, ontology, web portal, neuroanatomical nomenclature, web search
Introduction
The Internet, the World Wide Web and search engines such as Google, Bing, and Yahoo have brought about the greatest advance in human communication since invention of the printing press. While textbooks have long provided entrée to scientific knowledge for individuals who have access to them, all of human knowledge is rapidly becoming accessible at minimal expense to every person on the planet. The next challenge is to develop technologies that will enable readers to find specific information on the Web (Berners-Lee, 2001). Our efforts with NeuroNames and BrainInfo have been directed at meeting that further challenge.
Limitations of conventional textbooks, search engines and existing knowledge bases
The greatest limitations of textbooks as a means of communicating scientific knowledge are that they are expensive, limited in the amount of text, raw data and illustrations they can devote to a given topic, and they can only be updated every several years. The Web has largely eliminated those limitations on knowledge transfer.
The Web, however, poses its own challenges in terms of indexing and quality control. Indexes to the world’s documents, such as Google and PubMed, have grown so large that in some knowledge domains searchers cannot find the information they seek in a reasonable time, and many web pages, particularly those devoted to scientific topics, have not been sufficiently edited for completeness, accuracy or balance to assure users unfamiliar with the area that they are trustworthy.
Our quest for an ontology grew out of the wish to produce a retrieval engine for neuroscientific information on the Web that would address the challenges of incompleteness, inaccuracy and bias more effectively than Google (2011), PubMed (2009), Wikipedia (2011) and the hundreds of websites dedicated to specific aspects of neuroscience. Search results provided by Google and PubMed are exhaustive, but in some subject areas, such as neuroscience, their multipage lists of citations are dominated by documents that do not answer the query fully, clearly and accurately, and the most pertinent citations may appear so deep in the listing that the user fails to find them. Information in Wikipedia tends to be limited to topics that are of sufficient current interest that individual scientists are motivated to contribute information on them; the contributions lack a consistent terminology, are often incomplete and contain a small but significant amount of erroneous content. Hundreds of individual websites contain excellent information on specific topics but are difficult to find, complex to navigate and may use unfamiliar terminology. We wanted to develop a system that would eliminate terminology, semantic ambiguity and navigational complexity as obstacles to accurate and efficient retrieval of neuroscientific information.
A web portal should enable scientists, clinicians, students and the public to obtain up-to-date information at any desired level of detail. The system we envisioned would initially serve as a smart index to the Web and as a distributed textbook and database of neuroscientific information. It would provide answers to queries within a couple of minutes and a few mouse clicks. If it could not provide an answer it would inform the user and offer to compose a ‘smart query’ for the user to submit to other resources, such as PubMed. Basic information, such as the names and definitions of brain structures would reside on the system’s own server, where they could be presented in a standard, internally consistent terminology. It would use its own database to clarify terminological equivalents across sources and across species, provide detailed English descriptions of structures reported in foreign or out of print publications, and provide a much richer variety of illustrations than is feasible in a conventional textbook. Most further information would be retrieved by linking users to pages at other websites. By recognizing all names for a given structure, the system would remove differences in terminology as a cause of false negative returns, i.e., failure to return existing relevant information. By linking only to the most complete, clear and accurate web pages it would avoid returning thousands of false positive documents and unclear, incomplete or erroneous displays. By enabling users to navigate according to the logic of the discipline, i.e., from topic to related topic, as in the latest version of the Neuroscience Information Framework (NIF, 2011), rather than the more common logic of a resource-oriented system, i.e., from portal to website to site-map or menu to web-page of possible but not assured relevance, it would free the user from frustrating details of intra-site navigation.
A web portal as communication channel
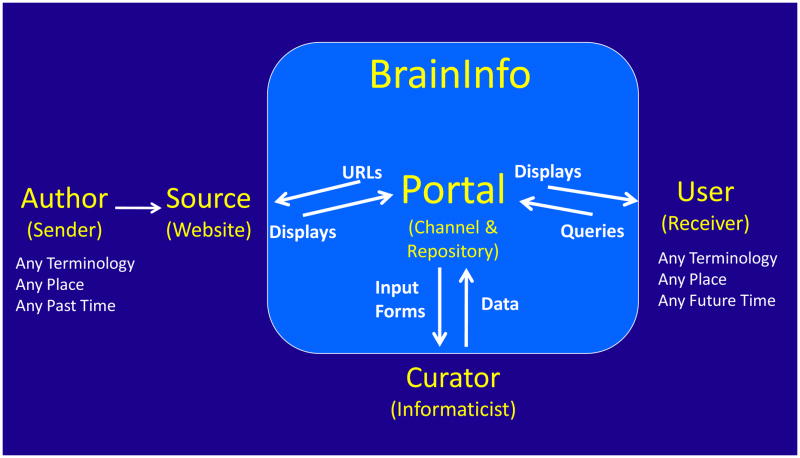
The web portal that we envisioned is a link in an intelligent communication channel (Fig. 1). It differs from pre-web communication channels, such as telephone and radio systems, in that it enables the sender (an author) to transmit information to the receiver (a user) anywhere in the world without regard to distance, time, or terminology. The Web itself compensates for distance by electronic transmission and compensates for time by storing the information in website server repositories where it is continuously accessible over the internet. The portal compensates for differences and ambiguities in terminology by processing queries through an ontology that is capable of handling synonyms, homonyms and multiple concepts of the same entity.
Figure 1.
The BrainInfo Portal, as a communication channel, is similar to conventional portals in that it has a repository of links to documents on the Web. It differs from conventional portals in that: 1) information in the repository is organized according to an ontology that allows indexing of documents based on the concepts rather than the words therein; 2) the portal interacts with users to interpret and disambiguate queries; and 3) a curator continuously updates the portal ontology with new terms, new concepts and the addresses of web pages that define and illustrate them.
An ontology for human communication via the Web
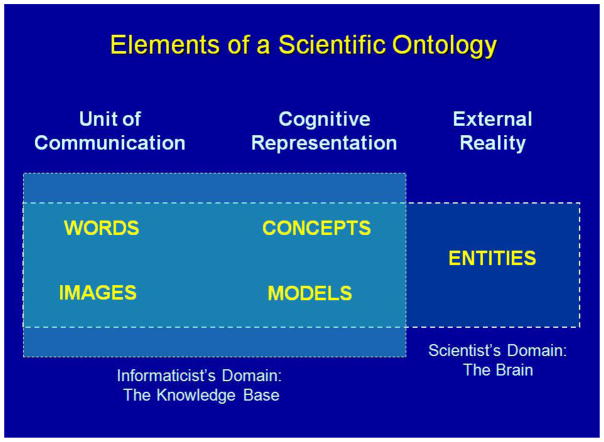
At the most general level, a scientific ontology is a conceptual model that represents in word definitions the relations between words in a vocabulary, cognitive constructs in subjective reality and entities in physical reality (Fig. 2). In NeuroNames at its current state of development words are names associated with neuroanatomical concepts, models and entities. Names are expressed in character strings; concepts and models are expressed in text definitions, lists and hierarchies and entities are structures in the central nervous system.
Figure 2. Elements of a Scientific Ontology Include Words, Concepts, Models and Entities.
Words are character strings, which are names representing concepts, models and the entities they represent. Names are linked to definitions of concepts and models. The contents of models are combinations of concepts and of smaller models represented by their names in lists, hierarchies and system diagrams. Entities are the objects and combinations of objects represented by concepts and models. (Images are represented here because, while they are not considered components of an ontology, they are equally or more important than words for communication of anatomical concepts.)
Scientific concepts differ from some other kinds of concept in that they are ultimately based on operational definitions of entities. Unlike some concepts in other knowledge domains their validity can be tested by experiment. In the on-going development of a scientific ontology the scientists’ role is to grapple with nature and to develop concepts and models of entities that they detect in external reality. Their role is to conceptualize, define and name entities, to relate them to existing knowledge and to communicate them to fellow scientists. The role of the informaticist in a communication system is to codify the relations of names, concepts, models and entities in a logical, internally consistent framework that aids efficient, unambiguous transmission of concepts from scientists to other scientists, students and the public at large.
To develop the ontology of a particular scientific domain the informaticist must understand the knowledge base of the domain as well as scientists working there, but his role is different. The informaticist does not create an ontology so much as codify and systematize the ontology of relations among names, concepts and entities that is embedded in the language and knowledge base of the domain. In our project, part of that task is to compile a standard vocabulary of unique names for concepts and models that is as suitable for unambiguous communication between humans as uniform identification numbers are for interoperability between computerized systems. A scientist can, of course, perform the role of an informaticist, but it is not always an easy fit. Most scientists identify with a particular model of reality and corresponding terminology. Only a few scientists are interested in expending significant time and effort on detailed analysis of how their terminology relates to the terminologies of other models. The role of informaticist in ontology development is more that of a textbook writer who organizes and describes the knowledge base of a domain in a consistent terminology than of a scientist who generates facts and models for a subdomain of the knowledge base.
Indexing by concept vs. character string
One reason existing search engines cannot reliably retrieve certain kinds of information is that the information resides not in an html or xml document but in a database where it can only be retrieved by calling local routines that retrieve and format data on the fly. A more integral limitation is that conventional portals index documents by the words (character-strings), not by the information (concepts) they contain. This is not a problem for informational domains where a one-to-one relation of words to concepts and entities is enforced. Institutions pay millions of dollars to protect the words they use as brands and trademarks. That is why, if one searches by Google for ‘bolt for a Whirlpool washer’, the first page of citations lists nine different web-searchable parts lists, any one of which is likely to provide the precise information one seeks. Non-unique relations between words, concepts and entities, however, pose a great obstacle to search in a scientific domain, such as neuroscience. Different words are used for the same concept, the same word is used for different concepts, and authors and readers of messages can have different concepts of the same entity, reinforced by differences in belief as to which concept ‘truly’ represents the entity
Ambiguity: the greatest obstacle to accurate communication
The ambiguities caused by non-unique relations between words, concepts and entities often cannot be resolved by analysis of the message itself. Only authors know what they have in mind when they write ambiguous messages; and only users know what they have in mind when they submit ambiguous queries. Automated resolution of ambiguities requires an ontology that enables curators to index messages accurately with regard to the concepts of the authors and that enables the portal’s server to interact with users to determine accurately their concepts of interest. Ultimately the ontology database should contain: 1) all of the character strings used as names in the knowledge domain of interest; 2) all of the concepts that the names symbolize, and 3) descriptions of how authors believe the entities they define relate to entities defined by others.
In the late 1990s, unable to find an ontologic approach and communications software that addressed the challenges inherent in indexing systems and queries posed in natural language, we set out to design an efficient system for codifying the multiple relations of words to concepts and entities in the domain of neuroscience. To assure that, from the beginning, the system would be useful to a maximal portion of the neuroscientific community we focused on the subdomain of neuroanatomy, the common interface of all subdisciplines of neuroscience. As the oldest subdiscipline, neuroanatomy is unusually plagued by terminological and conceptual confusion. It represented a large but manageable subdomain of knowledge to serve as a test-bed for demonstrating feasibility and utility of the approach. If it proved successful for structures of the nervous system it might be extensible to functions of the nervous system and to other domains of biology.
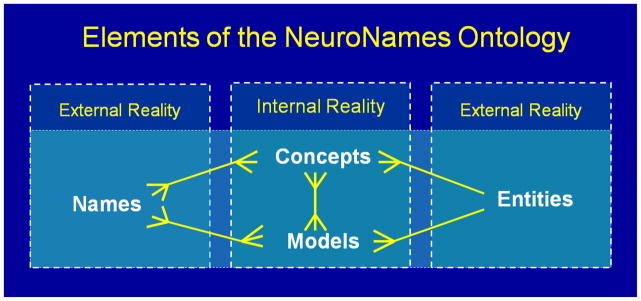
Ontological ambiguity comes in several forms. A word, such as ‘putamen’, can mean any of three things: the physical structure in a brain, a person’s concept of that structure, or simply itself, the character string ‘p-u-t-a-m-e-n’. The word-meaning-itself is important for the informaticist developing a standard nomenclature, but it is not likely to be an obstacle to accurate communication between authors and users of a portal. The bigger challenge is that the same and different words can represent the same and different concepts or entities. The ontology emerging from our project (Fig. 3) is designed to resolve semantic ambiguities that make queries uninterpretable on the basis of character strings alone.
Figure 3. NeuroNames is an informatics tool.
for resolving ambiguities in exchanges of information. In neuroanatomy the same structural concept, or model composed of structural concepts, can have multiple names and the same name can represent multiple concepts. A given model includes multiple concepts and the same concept can occur in multiple models. Different scientists can hold different concepts of the same structural entity and can model a complex entity in different ways.
Methods
The basic method for developing the NeuroNames ontology was to compile terminology from one book or review article at a time and to incorporate all terms that appeared to be relevant to the domain of neuroanatomy. The meaning of each term, i.e., the definition of the concept it represented was determined by consulting the text and illustrations in the source. If the concept was not already in the database, the term, the concept and the species to which they applied were added to the ontology, and the term was tagged as the tentative standard name for the concept. If the term and concept were already in the database but were attributed to a different species, the term was reentered into the database as applying to the new species. If the concept was there but the term was not, the term was added as a synonym for the concept. If the term was there but represented a different concept the term was added as a homonym of the new concept and a distinctive name was composed (attributed to NeuroNames as source) and added to the database as the tentative standard term. Once we had compiled the terminologies and concepts from the Nomina Anatomica, seven major textbooks and atlases and a number of neuroanatomical research articles and book chapters, we reviewed the names representing each concept and selected one as the permanent standard name (see Results and Discussion for selection criteria). We designed input forms and software of the system to maximize the efficiency of populating the ontology database.
We searched the web using Google to locate sources and pages with informative text and images. The informational value of web pages was judged on the basis of comprehensiveness, accuracy and intelligibility compared to pages already in the system. For example, an ideal page of text describing the connections of a structure would list all of its connections in a readily understood terminology and would cite a peer-reviewed source for each connection. An ideal image would show accurate labels and boundaries of all structures in the vicinity of the structure of interest and would be consistent with the text definitions of structures in NeuroNames.
Defining the domain
The creation of a portal to all of neuroscience on the Web promised to be a multidecade project. Wishing to make it useful from the beginning we had to decide whether to start with a high level ontology that would cover the entire domain of neuroscience superficially or to cover a portion of the domain thoroughly. We concluded that starting with a high level ontology would only make available information that is easily accessible in textbooks, whereas populating the database comprehensively in a limited domain would make accessible a collation of detailed information not currently available from any one source. Thus, while we designed the schema of the ontology to be extensible to any domain of information communicated by human language, we began by populating only the portion related to the anatomy of the primate brain (Bowden and Martin, 1995). Subsequently we extended the domain considerably to include all structures in the brain and spinal cord of the four genera most studied by neuroscientists: human, macaque, rat and mouse (BrainInfo, 2010c).
Another consideration in defining the domain was whether to strive for historical completeness or to include only names, concepts and models that play a role in current neuroanatomical discourse. For example, since publication of the first version of the Nomina Anatomica more than a century ago most anatomists have adopted a model of the brain as composed of forebrain, midbrain, and hindbrain with the forebrain composed of two parts and the hindbrain of three parts. But that was not always true. At least seven different concepts of the brain as sum-of-parts have dominated in one scholarly tradition or another (Swanson, 2000). Most of those models are now obsolete and need not be included in an ontology for use now or in the future.
Elements of the NeuroNames ontology
In developing an ontology for neuroanatomy we found it useful to distinguish two kinds of cognitive element: concepts and models (Figs. 2 and 3). A concept is a cognitive representation of an entity believed to exist in objective reality. The concept is represented in the ontology by a written definition. Models are combinations of concepts grouped on the basis of specified relations, most commonly partitive or categorical relations. Inasmuch as models are units of cognition and communication they are themselves concepts. As combinations of concepts, however, they are better judged in terms of utility for a given purpose than as true or untrue representations of entities. They can play different roles from simple concepts in logical processing; we found it useful to distinguish them from simple concepts by designating them ‘models’.
We distinguish three categories of model: lists, hierarchies and systems (Bowden and Dubach, 2004). List models are exemplified by the alphabetical indexes to structures in brain atlases. The most comprehensive hierarchical models in the neuroanatomical domain represent groupings of structures based on different kinds of relationship, such as the brain hierarchy in the Nomina Anatomica (1983), where structures are grouped by proximity on the basis of dissection, and the brain hierarchy of Swanson (2004), where structures are grouped largely based on function. Systems models, such as those summarized in Arbib and Amari’s (1988) Dynamic Interactions in Neural Networks: Models and Data, represent more complex relationships among structures than lists and hierarchies. Neuroanatomical circuits, systems and networks are usually illustrated by diagrams that include such relations as reciprocal, excitatory and inhibitory connections.
In our ontology, hierarchical models can be either categorical or partitive, and each of those can be open or closed. In a closed partitive hierarchy, such as that of the comprehensively segmented macaque brain of Martin and Bowden (2000), the children of a given parent structure constitute a complete set of mutually exclusive parts of the parent structure. In an MRI atlas segmented according to that model (BrainInfo, 2010a), every voxel of the brain is identified with one and only one primary structure. No substructure can be added and no set of voxels representing one of the substructures in the canonical atlas can be changed without changing the voxel set of one or more of the others. The fact that the parts are exhaustive and mutually exclusive means that data mapped to them is indexed to a closed partitive hierarchy and suitable for parametric analysis by fixed-effects statistical models (Bowden, 2000).
An open partitive hierarchy does not require specification of a complete set of mutually exclusive subparts. The classical Hierarchy of Brain Structures of NeuroNames (Bowden and Martin, 1995; NeuroNames, 2010) is such a hierarchy. The children of a given parent include all of the parts of the structure that exist in either the human or the macaque. Since the structures that make up a superstructure, such as the frontal lobe, differ in the two species, the total number of children in the primate brain hierarchy, which includes both human and macaque structures, is greater than the number in either the human hierarchy or the macaque hierarchy alone. Such a hierarchy is useful for indicating equivalent and non-equivalent structures between species, for categorical indexing, for inferential logic and nonparametric statistical analyses of neuroanatomical data, but not for parametric, quantitative neuroanatomical analyses.
Structure of the NeuroNames ontology
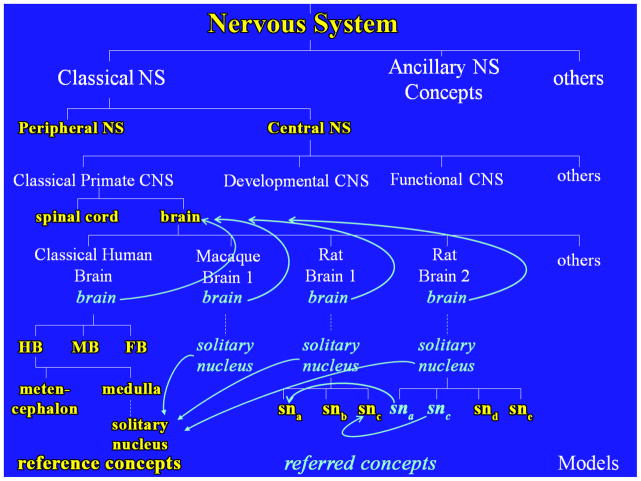
To interpret and resolve ambiguities in queries submitted to BrainInfo, the NeuroNames ontology had to codify the relations among all names, concepts and models in the domain of discourse. Those relations are illustrated in Figure 4.
Figure 4. NeuroNames Ontology of the Nervous System.
A hypothetical set of brain models (one human, one macaque and two rat models) are integrated with a subset of concepts from the NeuroNames ontology to illustrate relations among reference concepts (embossed or yellow), referred concepts (italicized or light blue) and models (capitalized or white). CNS: central nervous system; HB: hindbrain; MB: midbrain; FB: forebrain; NS: nervous system; sna-e: subnuclei of solitary nucleus in two hypothetical atlases of the rat brain. Dashed lines indicated intervening levels of hierarchy. Arrows indicate identity of referred concepts in models with their reference concepts. All instances of ‘brain’ in italicized or light blue share the standard name and definition of the reference concept ‘brain’ in embossed or yellow letters. The coding of subnuclei sna-c in Rat Brain 1 as reference concepts and of sna and snc as referred concepts in Rat Brain 2 indicates that the concepts were first found, coded and defined in the ontology from the Rat Brain 1 atlas.
Definitions of Concepts and Models
Concepts are defined by features that distinguish them from all other concepts. A scientific concept is represented by an operational definition, a text description of distinguishing features of an entity, the procedures or operations by which those features are revealed and the source of the concept. Neuroanatomical structures are defined on the basis of several methods including dissection and inspection, histological staining, electrical recording, x-ray and others. For example, an operational definition of the lateral medullary lamina takes the form: “The lateral medullary lamina is a thin myelinated structure located in the basal ganglia of the primate brain. Bounded laterally by the putamen and medially by the globus pallidus, it is defined on the basis of Nissl or myelin stain (Carpenter and Sutin, 1983).” The definition is logically unique in the sense that no two physical objects can fill the same space. It is true on the condition that one looks for the entity using the operations described in the definition: microscopic examination of Nissl or myelin stained sections in the primate brain. In our view, a definition that omits methodology and source is satisfactory for teaching the current state of knowledge. But an ontology intended to serve the needs of scientists experimenting at the unstable conceptual frontier of knowledge must include the methodology and source of the definition. A scientist who needs to resolve a conflict between his findings and the findings predicted on the basis of previous work needs to know the source and methodology of the earlier reports.
The definition of an anatomical model consists of a list, hierarchy or system of concepts that are the content of the model. Thus, the definition of the hypothetical model ‘Rat Brain 1’ in Figure 4 is the hierarchical list of structures illustrated in that particular atlas of the rat brain.
An important feature of hierarchical models is that the concept of the model itself is different from the top concept in the hierarchy, which is identical to the parent concept of the model. Figure 4 shows small corresponding parts of four models of the entity ‘brain’: the classical human brain hierarchy and hypothetical models of a macaque brain and two rat brains. Each model represents a somewhat different concept of the brain as a composite structure. The same entity, viz., the brain as cranial component of the central nervous system, appears as the parent and first child of each model. The common identity of the models is represented by their belonging to the same category, ‘brain’, and by the fact that the top concept in the model hierarchy is the same ‘brain’ for all. Their differences are represented in the unique names of the models and the composition and organization of parts below the top concept, ‘brain’.
This organization of the ontology accommodates the senses in which each model is the same and different from the others. One can think of each referred concept at a node in a partitive hierarchical model of neuroanatomical structure as meaning two things, i.e., as having two definitions: a categorical definition and a partitive definition. Consider the solitary nucleus, which appears in four models in Figure 4, two of which are in Rat Brain 1 and Rat Brain 2. Because the definitions of the solitary nuclei in topological terms are identical, they belong to the same category as the solitary nucleus defined in the Classical Human Brain. Top down they fit the category of ‘solitary nucleus’ based on the operational definition recorded there. Bottom up, however, the partitive definitions are different. The definition of the solitary nucleus in the Rat Brain 1 model contains a different set of parts from that of the Rat Brain 2 model. Thus, the partitive definitions of the solitary nucleus are different depending on the context, i.e., the model to which it belongs. The ontology codifies the identities and differences in concept definitions by specifying the models where they appear, e.g., in Rat Brain 1 or Rat Brain 2. This integrated codification of relationships between reference and model concepts allows an automated system to interact with the user to resolve query ambiguities that are based on multiple concepts and models of the same entity.
The NeuroNames ontology addresses the synonyms and homonyms problems by linking all names (including the standard name) for a concept, its text definition, and other information common to all instances of the concept to one instance of the concept in the hierarchy, the reference concept (Fig. 4). All subsequent instances of the same concept in other parts of the hierarchy are identified as referred concepts.
The first and largest sources of reference concepts in NeuroNames (Bowden and Martin 1995; BrainInfo 2010c) were the Encephalon division of the Nomina Anatomica (IANC, 1983) and two textbooks that most systematically extended the Nomina-style hierarchy to smaller structures (Crosby, 1962; Carpenter, 1983). Thus, the reference concepts illustrated in Figure 4 are indexed to the hierarchical model Classical Human CNS, which contains some 800 concepts. The majority of NeuroNames concepts, however, are from other sources and are indexed not as classical structures, but as ancillary structures, structures whose location in the brain is defined by neuroanatomists in relation to the classical structures. The Ancillary NS Concepts model (Fig. 4, top right) is a non-hierarchical, open list model. It includes more than 1600 structures that overlap spatially but do not coincide exactly with structures in the classical hierarchy. It includes, for example, combinations of classical structures grouped differently, such as the combination of the putamen and globus pallidus known as the lenticular nucleus; the catecholaminergic nuclei as defined on the basis of markers other than the classical Nissl and myelin stains; cortical areas defined on the basis of internal, architectonic structure rather than classical sulcal landmarks, e.g., Brodmann’s areas; and structures only found in certain strains or species, such as the hypothetical subnuclei of the rat solitary nucleus shown in Figure 4. The flat structure of list models makes them of little use in classifying data for inferential logic or statistical analyses. Nothing can be inferred about individual ancillary concepts in a list beyond the information contained in their definitions and the fact that they represent entities in the nervous system. Thus, while structures are in the ancillary model no less important than structures in the classical model, a list is the least informational type of model in which to represent them, because the relational information implicit in the hierarchical and systems models, where they also appear, is lacking. To avoid giving users the misimpression that ancillary structures are less important than structures in the classical hierarchy we avoid use of the term ‘ancillary’ in BrainInfo graphic user interfaces.
In summary, models appear in the ontology as children of the concepts they represent. All structures in NeuroNames are categorized as belonging either to the Classical NS model or to the Ancillary NS Concepts model. Concepts in either category can serve as reference con cepts and can appear as referred concepts in any number of other models.
BrainInfo as a portal to the Web
The BrainInfo Portal [http://braininfo.org] has three interfaces with the Web (Fig. 1): a user interface to the person in search of information; a source interface to the website repositories of information recorded by authors; and a curator interface to the person who creates the index database that links users to pages in websites. Since all three interfaces open to the Web, curators as well as authors and users can be located anywhere in the world with access to the internet.
The user interface is interactive. It receives and disambiguates queries by exchange of text messages, and it displays pages from websites that deliver information in text and image formats. The source interface submits URLs (Uniform Resource Locators, web addresses) to the Web and retrieves pages from websites, which the Portal displays to the user together with terminological clarification and identity of the source. The curator interface provides input forms that enable curators to record information in the database: text to build the ontology, links to website pages, text for messages to the user, and thumbnail images to illustrate to the user the kinds of image information available from specific sources.
The BrainInfo Portal is maintained by the Center for Research in Biological Systems (CRBS, 2010) at the University of California, San Diego. It consists of the NeuroNames ontology, concept directories and web links, which reside in an SQL relational database (Microsoft SQL 2005 Server) with input forms and web displays programmed in C# (.NET). The SQL server houses proprietary software (copyright University of Washington), including BrainInfo and log databases supported by a Windows 2003 Server. The BrainInfo database consists of more than 80 tables, 250 stored procedures, and 10 triggers. A separate Windows 2003 Web Server houses the BrainInfo website and custom Windows services to generate XML files. A Windows 2000 SVN server maintains files for source control: SQL Scripts for BrainInfo tables, stored procedures, triggers and source codes for ‘BI Input Forms’ and ‘BI Website’. All servers are equipped with Symantec LiveState Recovery Standard Server 6.0 and McAfee VirusScan Enterprise 8.0. Curator and programmer functions are supported by links into the system from standard PC and Apple Macintosh workstations.
Results and Discussion
Within the domain of neuroanatomy it was necessary to define limits to the sets of entities, concepts and names included in the ontology.
Entities
We limited the set of entities that we would cover comprehensively to structures in the mature central nervous system as defined or illustrated in the most widely used textbooks and brain atlases of the human, macaque, rat and mouse. Other entities were included if their names could be confused with the names of entities in the domain. The classical brain hierarchy of NeuroNames was designed to illustrate spatial relations among structures grouped by proximity. Thus, we only extended the hierarchy down to the level of structures that anatomists have seen fit to assign unique names, e.g., ‘solitary nucleus’, otherwise known as ‘nucleus of the solitary tract’. Structures below that level have generic names modified by the name of the parent structure, e.g., ‘medial subnucleus of the solitary tract’. Thus, in NeuroNames the classical hierarchy of brain structure extends from ‘brain’ down to the level of nuclei and gyri.
The entity domain includes hundreds of structures that are not part of the classical hierarchy. They include structures whose boundaries do not coincide with boundaries of the classical structures, such as cortical areas defined by internal architecture; alternate groupings of hierarchy structures, e.g., according to embryologic origin or function; and partial subdivisions of classical structures. Such structures are categorized as ‘ancillary structures’. Many of the structures defined in the Ancillary NS Concepts Model served subsequently as reference concepts when they were incorporated into more useful models, such as the Functional CNS model (a hierarchy of ~575 structures grouped by function) and the Classical Spinal Cord model (a hierarchy of ~70 structures grouped by proximity).
Concepts
The number of concepts of an entity is as great as the number of individuals who think about it. Some cognitive scientists, in fact, propose that the number is much greater. They observe that one’s concept of an entity undergoes revision into a slightly different concept every time it is drawn from long-term memory into working memory and rewritten to long-term memory. Thus, at any given time a scientist’s concept of an entity is based on a unique history of direct and indirect experience with it.
Fortunately, an ontology for communication does not have to accommodate all concepts. It only needs to codify the subset of concepts that people use to communicate about entities. Operationally, that means concepts that neuroscientists have found it useful to define and assign names. In the neuroanatomical domain we estimate the number of concepts of anatomical, as opposed to histological, cellular and smaller entities in the central nervous system, to be on the order of 3,000, i.e., a few hundred more than currently in NeuroNames. All should eventually be included in the NeuroNames ontology.
Models
The number of models, i.e., combinations of primary neuroanatomical concepts, is also very large. Most, however, need not be included in the ontology. We have identified only three hierarchical models of the central nervous system that are sufficiently comprehensive, systematic and embedded in the knowledge base of neuroanatomy to merit inclusion in NeuroNames. They differ in history, purpose and in the criteria by which concepts are grouped. The classical central nervous system (CNS) hierarchy, based on the Systema Nervosum section of the Nomina Anatomica (IANC, 1983) and extended in the first chapters of most neuroanatomy textbooks, structures are grouped by proximity to show which of them go together when the brain is dissected into successively smaller parts (Bowden and Martin, 1995). In Swanson’s (2004) Brain Maps: Structure of the Rat Brain, the same structures are grouped hierarchically, by embryonic origin at upper levels and by functional systems at lower levels, to indicate the structures involved in different behavioral and physiological functions. In the embryogenic, or developmental hierarchy of Paxinos et al. (2010) structures are grouped first as predominantly cellular, predominantly myelinated or ventricular; then predominantly cellular structures are grouped hierarchically to illustrate the rhombomeres of the embryonic nervous system from which they originate.
Most other hierarchies can be excluded because they are obsolete (Swanson, 2000), duplicative of parts of the classical, functional or developmental models, or idiosyncratic to a particular source and not used to communicate beyond that source. In many such models brain structures are organized hierarchically, but a lack of systematic criteria for inclusion, exclusion and grouping of items makes them unsuitable for communication or logical processing. For example, Wikipedia’s (2010) brain model, ‘List of regions in the human brain’, is an unstable, intermittently reorganized hierarchy designed to show associations of certain brain structures and functions as conceptualized by volunteer authors of variable interest and expertise. The National Library of Medicine’s MeSH vocabulary (NLM, 2010) is a hierarchy of names of brain structures with overlapping definitions that is designed to serve as a controlled vocabulary for indexing publications. Such models should not be included in a portal to information on the web, because the definitions implicit in the hierarchical relations of their structures often differ from conventional usage. Employed at face value in a portal, they would lead to false positive retrievals of information.
Names
The selection of names to include in the ontology was based on two goals: 1) to compile all names of structures that users might include in queries, and 2) to select a unique standard name for each anatomical concept in the domain of neuroanatomy. Work to achieve comprehensiveness was time-consuming but not difficult. We searched the text and indexes of the most widely used neuroanatomical books, atlases and journal articles for terms. If the source of the name was the publication where the concept was first defined in operational terms that publication was considered the expert source. If the name and concept were obtained from a textbook or review that cited original publications, the textbook or review was cited as the expert source. Terms lacking citation of an original source were assigned the author of the summary publi cation. Terms mean what people think they mean, and most scientists learn the meanings of neuroanatomical terms from textbooks and atlases. We regarded as expert sources textbooks that were comprehensive, clearly organized, internally consistent with regard to terminology, thoroughly referenced and widely used in universities and medical schools. Authoritative sources of the largest numbers of names and concepts included the Nomina Anatomica (IANC, 1983), which was the origin of the hierarchical organization of structures in most textbooks published since the late 1800s, Riley (1943), Crosby, Humphrey, and Lauer (1962), Stephan (1975), Carpenter and Sutin (1983), Paxinos and Mai (1990), Anthoney (1994), Swanson (2004), and Kahle and Frotscher (2001). The most widely disseminated textbooks have been translated into many languages. Thus, the most useful sources of terms in languages other than English, Latin or German were translations into other languages from widely used English or German textbooks and atlases. Structure names that were not defined or illustrated sufficiently to relate to known concepts were omitted. We continue to integrate new concepts and names into the NeuroNames database from original neuroanatomical reports and reviews; the number currently exceeds 450.
Selection of standard names from among synonyms
A more difficult challenge than the codification of names and concepts was to select a standard name for each concept that would be suitable for communication with users in a consistent terminology. The only absolute criterion for selection of a standard name was that it is used only in reference to the concept in question. A word means what most people think it means, so after uniqueness, the most important criterion is precedent. Neuroanatomists have little inclination to use the nomenclatures of other neuroanatomists. So, insofar as possible, we avoided the temptation to invent ‘better’ names than those existing in the natural language of the domain. The only situation in which we created a new term was the unusual case where every appropriate name is commonly used for more than one concept.
We considered several indicators of precedent. When we began the project in the early1980s most brain atlases and textbooks listed anatomical terms in the native language of the author and in Latin. So our initial approach was to select Latin terms as standard names. From 1990, however, virtually all brain atlases, edited textbooks and journals with authors from language areas as diverse as Germany, Spain and Japan have been published with the neuroanatomical terminology in English (Paxinos, 1990; Ono et al., 1990). Thus, if a concept had English or anglicized Latin names we selected the standard name from among those. The second indicator of precedent was use frequency. We submitted each English and Latin term to PubMed and determined the number of abstracts in which it had appeared during the previous 25 years. All other things being equal, the term with the highest use frequency was made the standard name. For example, a structure in the subthalamus had nine legitimate English and Latin names. Of those, three were eliminated from consideration because they had not appeared in a single PubMed citation in 25 years. Five more had appeared in less than 25 citations. One, ‘field H’ appeared in 417 citations, so ‘field H’ was adopted as the standard name.
If multiple names appeared with equal frequencies we weighed several further principles of selection. Mnemonic terms, such as ‘basal forebrain nucleus’, were favored over eponyms, such as ‘basal nuclei of Meynert’. Common English spellings, such as ‘locus ceruleus’, were rated above awkward vowel combinations, such as ‘locus coeruleus’ or ‘locus caeruleus’. We avoided constructions that, while acceptable in written language, are awkward for spoken English. Thus, ‘transitional part of visual area 4’ was rated above ‘visual area 4, transitional part’. Short names and names consistent in format with the standard names of related structures were given preference.
The selection of standard names for models is less challenging than for simple concepts. The names of models are seldom used in thought, speech or writing. In NeuroNames the standard name for a model is created by the curator who adds the model to the ontology. The only criteria for model names are that they be unique, mnemonic and reasonably short, e.g., ‘Classical Primate CNS’, ‘Developmental CNS’, ’Functional CNS’ (Fig. 4).
A major purpose of a standard vocabulary is to minimize the number of names a user needs to know in order to understand text information in the domain of interest. Thus, we have attempted not to change the standard name of a concept without good reason. On occasion, however, we have found that the first name selected as standard was not in fact the best by NeuroNames criteria. For many years ‘nucleus of Darkschewitsche’ was the standard name for a structure in the midbrain tegmentum, because we had found no non-eponymic name and it appeared with by far the highest frequency in PubMed. The German spelling had led us to believe that the author was German. Later we learned that the author was Russian and that ‘Darkschewitsche’ was its German transliteration. The English transliteration better fit the English-spelling criterion and had in fact appeared in one U.S. textbook. So we changed the standard name to ‘nucleus of Darkshevich’.
Selecting a standard name when the best options are ambiguous
Ambiguity occurs when the same name can refer to different concepts, models or entities, i.e., represents the homonym problem. Avoidance of homonymy posed the single gr eatest challenge in selecting standard names. Homonymy gives rise to three kinds of ambiguity: resolvable and irresolvable conceptual ambiguities and entity ambiguity
Resolvable conceptual ambiguity arises when the same name can represent multiple concepts that domain experts agree are of different entities. The best strategy, if feasible, is to choose a different name from among existing synonyms for the less frequently used concept. We found this a good solution for part of the problem with ‘arcuate nucleus’. PubMed counts showed 20 times more abstracts for the query [“arcuate nucleus” AND hypothalamus] than for the query [“arcuate nucleus” AND thalamus]; and the thalamic nucleus had a frequently used synonym, ‘ventral posteromedial nucleus’. So we adopted ‘ventral posteromedial nucleus’ for the thalamic nucleus.
A second strategy is to append modifiers to the homonyms to distinguish their meanings. This is less desirable, because it violates the principle ‘short is better than long’. Nevertheless, for the hypothalamic and medullary arcuate nuclei it was necessary. The hypothalamic nucleus had a synonym, ‘infundibular nucleus’ that applied uniquely to it. But the PubMed count for [“infundibular nucleus” AND hypothalamus] was 30 times less than for [“arcuate nucleus” AND hypothalamus], indicating that the frequency with which authors use ‘arcuate nucleus’ for it is overwhelmingly greater than ‘infundibular nucleus’. The arcuate nucleus of the medulla had no acceptable synonym, so we adopted ‘arcuate nucleus of the hypothalamus’ and ‘arcuate nucleus of the medulla’ as standard names for the hypothalamic and medullary nuclei.
A third strategy is to assign the homonym to one of the concepts and create a name for the other concept that includes the homonym and a mnemonic qualifier for a feature that distinguishes it from the first concept. We took this approach with the basal ganglia. The name ‘basal ganglia’ is commonly associated with several concepts that differ with regard to the set of subcortical structures they include (Anthoney, 1994). We assigned the term ‘basal ganglia’ to the concept defined in the most long-standing authoritative source, Nomina Anatomica (IANC, 1983). For the next most common concept, which includes additional nuclei with connections to those in the classical concept, we created the name ‘basal ganglia circuit’. Compared with possible synonyms from the literature, viz., ‘basal ganglia (clinical)’ or ‘basal ganglia-2’, this name was mnemonic and pronounceable. For more idiosyncratic definitions of ‘basal ganglia’, which merited inclusion because of the authoritative status of the sources but which are seldom if ever encountered in discourse, we created unique names by referencing the authors, viz., ‘basal ganglia of Crosby’ and ‘basal ganglia of Carpenter’. Creation of the term ‘basal ganglia circuit’ violated the principle not to invent ‘better terms’, but it solved the ambiguity problem and it resulted in a mnemonic term more likely to be used in oral and written discourse than any alternative.
Irresolvable conceptual ambiguity
Very occasionally we came upon a homonym that was so heavily used with different meanings by large segments of the neuroscience community that selecting it as the standard for one of the concepts could not be expected to reduce confusion in oral and written communication. The term ‘hippocampus’ is a homonym used by various authors to represent either: 1) the combination of the CA1, CA2 and CA3 fields (Carpenter, 1983); 2) the combination of subiculum, CA fields and dentate gyrus (Crosby, 1962), or 3) those structures plus the fasciolar gyrus, the supracallosal gyrus and the paraterminal gyrus (Stephan, 1975; Schiebler et al., 1999). When we first worked on this problem 20 years ago it appeared that Stephan’s use of ‘Hippocampus’, which was based on the comparative anatomy of the region, was not gaining traction. The terminology was in Latin, the supporting text was in German, and thus, inaccessible to most English-reading neuroscientists, and the same set of structures had a well-established anglicized Latin synonym in ‘archicortex’. So we adopted ‘archicortex’ for that concept.
The structure whose sea horse-like appearance was the historical basis for the term ‘hippocampus’ was increasingly referred to as the ‘hippocampal formation’ by authors who reserved ‘hippocampus’ to refer just to the CA1-3 fields located between the dentate gyrus and the subiculum (Carpenter, 1983). Thus, we adopted ‘hippocampal formation’ for the composite structure. None of the options for the intervening CA1-3 portion was perfect. ‘Cornu ammonis’ was Latin and awkward to pronounce in English; ‘Ammon’s horn’ was an eponym and contained an apostrophe, which was an illegal character for the alphanumeric-oriented software of the day; the PubMed use frequency of the synonym ‘hippocampus proper’ was 300 times less than that of ‘hippocampus’. So despite the fact that ‘hippocampus’ was still used as a homonym for the hippocampal formation, we adopted it as the standard name for the CA fields with the expectation that over time it would come to be used most commonly with that meaning.
Our prediction turned out wrong. Germans studying neuroanatomy today learn that ‘hippocampus’ refers to the archicortex (Schiebler et al., 1999). Most English-speaking neuroscientists who work with MRI use ‘hippocampus’ to refer to the hippocampal formation. And most brain atlases now label the CA1-3 portion of the hippocampal formation with some variation on the ‘ammon’ root. In light of this evolution we concluded that the better strategy, for BrainInfo’s purposes, was to avoid assigning the homonym ‘hippocampus’ to any of the contending concepts. It was preferable to create a unique, short, mnemonic term, ‘CA fields’, for the concept and to classify the homonym, ‘hippocampus’, as a synonym of the standard names of all three contending concepts. In principle, we believe that once a standard name has been adopted, it should not be changed. But if, on a time base of decades, it turns out to be incompatible with the active ontology of the domain one must change it. Otherwise the portal using it will generate more confusion than clarity in responding to queries and in defining other terms.
Entity ambiguity was the final challenge. This form of ambiguity occurs when uncertainty exists in the discipline as to whether several concepts defined by different methods and by different investigators, represent a single entity. This is common in a developing research area where scientists have not reached consensus on definitions and names for entities hypothesized to exist. In such a conceptual environment the informaticist’s best strategy is to create standard names for all of the contending concepts. A current example is the definition of cortical areas on the basis of architecture, gene expression and neurochemical characteristics. To resolve this kind of ambiguity we found it useful to create names in a standard format. We encode into the standard name the acronym given by the author, the author’s surname and, if necessary to achieve uniqueness, other distinguishing features of the concept, such as the species in which the area was reported. Examples: ‘area 10 of Walker’, but ‘area 10 of Brodmann (guenon)’ because Brodmann defined area 10s in several species without declaring whether he regarded them as equivalent structures.
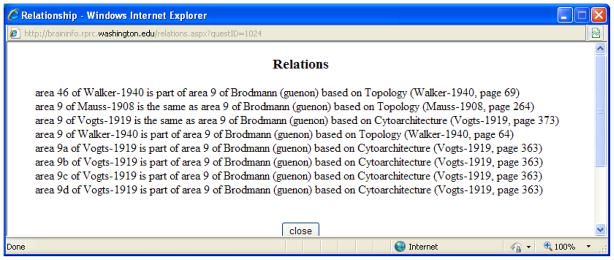
While the neuroanatomical community has yet to reach consensus on the equivalence of the concepts, investigators usually record their judgments as to the relations of their area to areas previously reported by others. The informaticist can save future investigators an immense amount of time in literature review by making that information available to users. In BrainInfo such information is displayed in a Relations table (Fig. 5). The table contains a series of statements in the format: ‘area 8 of Brodmann (guenon) is the same as area 8 of Mauss based on topology (Mauss-1908, page 264)’. Note that the format includes the method, topology, by which the author judged two areas to be the same. If in the future scientists come to consensus that several concepts in fact represent the same entity the definitions of the concepts can be merged into a single definition and their names can be made synonyms of the standard name for that concept in NeuroNames.
Figure 5. Relations of area 9 of Brodmann (guenon).
to eight areas reported by other investigators to be the same as, or to overlap, that area of cortex. Note that the Vogts based their judgment as to equivalence of their structures on cytoarchitecture, i.e., similarity of internal structure. Walker and Mauss judged their structures equivalent on the basis of topology, i.e., similarity of shape and location. The method used to identify a structure is an essential component of its scientific definition.
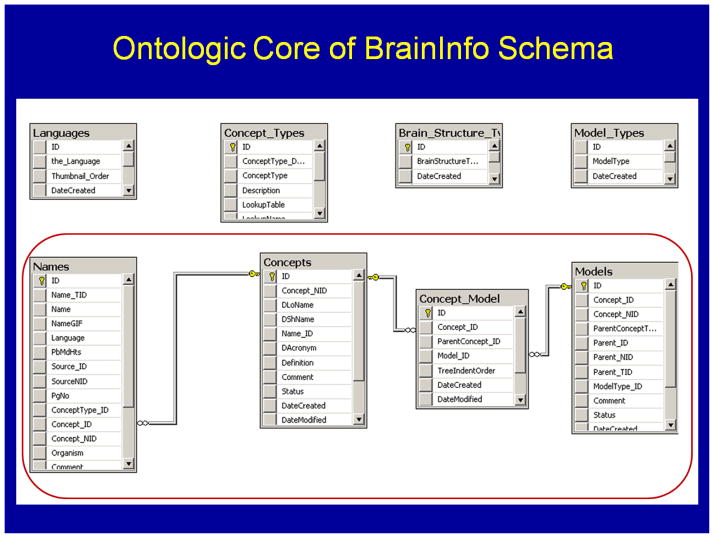
The BrainInfo/NeuroNames database schema
The relations between names, concepts and entities are maintained in BrainInfo’s SQL database. The database schema for the NeuroNames ontology is built on three core tables: Names, Concepts and Models (Fig. 6). The relations coded between columns in these tables enable the Portal to interact with users to interpret synonyms and disambiguate homonyms.
Figure 6. Names, Concepts and Models Tables.
form the ontologic core of some 80 tables in the database schema of BrainInfo.
The Names Table
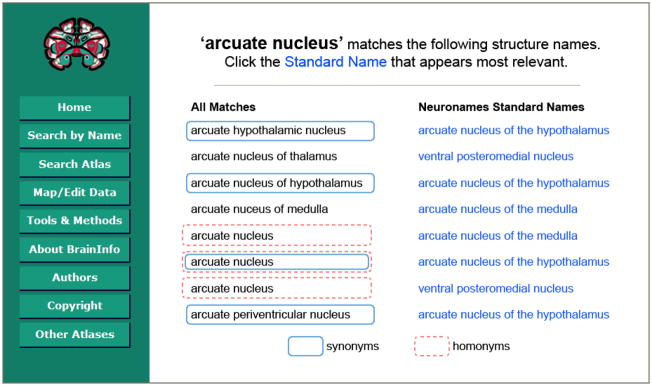
The Names table contains some 16,000 terms in eight languages denoting brain structures found in the four species most studied by neuroscientists. In the database schema (Fig. 6) the homonyms problem is resolved by devoting multiple rows of the Names table to the same character string. For example, the character string ‘arcuate nucleus’ appears three times in the Names column of the Names Table. The three entries have different numeric IDs and link through different entries in the Concept ID column to different concepts, whose standard names are ‘arcuate nucleus of the hypothalamus’, ‘arcuate nucleus of the medulla’ and ‘ventral posteromedial nucleus’ (Fig. 7). When a user submits the query string ‘arcuate nucleus’, BrainInfo displays a list of all Names that contain that string and the standard name of the corresponding structure. This enables the user to disambiguate homonyms by selecting the appropriate concept/entity from the NeuroNames Standard Names column.
Figure 7. BrainInfo Response to Query ‘arcuate nucleus’.
Six names for three concepts. Red (dashed) ovals: ‘arcuate nucleus’ as homonym for three different concepts. Blue (solid) ovals: Five synonyms for the same concept ‘arcuate nucleus of the hypothalamus’. [Page modified from BrainInfo (2010d).]
In neuroanatomy there are on average six different English, Latin and anglicized Latin synonyms for the same brain structure. Since different Names can refer to the same concept (blue solid ovals in Fig. 7), the relation of the Concept ID column in the Names Table to the Concepts Table is many to one (Fig. 6). This allows users to interpret synonyms. Note that the one-to-many relation of names to concepts addressed by multiple rows for the same name combined with the many-to-one relation of names to concepts gives the many-to-many relation of names to concepts illustrated in Fig. 3. By interacting with users to disambiguate homonyms and interpret synonyms before initiating retrieval, BrainInfo is able to narrow the search and greatly reduce false positive and false negative items responses to users’ queries.
From an applied ontology point of view, the most important attributes of names in the Names table are: 1) the concept to which the name applies, which is used to resolve ambiguities, and 2) the name’s use-frequency. The most important attributes recorded for each concept include the NeuroNames standard name and acronym (labeled ‘Default Name’ and ‘Default Acronym’ in the Concepts Table), which are selected from the set of names for the concept in the Names Table. These are used in composing text for the BrainInfo website, including the definition of the concept and the source of the definition.
The Concepts and Models Tables
The Concepts Table (Fig. 6), which, through the Concept_Model junction table, stands in many-to-many relation to the Models Table (Fig. 3) contains the names and definitions of both concepts and models. Rows in the Models Table identify a model, a structural concept and, if the model is hierarchical, the parent of the concept. The most important attribute of a concept in a hierarchical model is its parent, which BrainInfo uses to assemble hierarchical displays of the model.
Use cases: disambiguating homonyms and interpreting synonyms
The following use cases illustrate how the ontologic principles built into BrainInfo support precise retrieval of information when non-standard terminologies are involved. The first illustrates BrainInfo’s interpretation of a query that a user poses in non-standard terminology. The second illustrates how BrainInfo assists a user’s interpretation of information at a website that uses a different terminology.
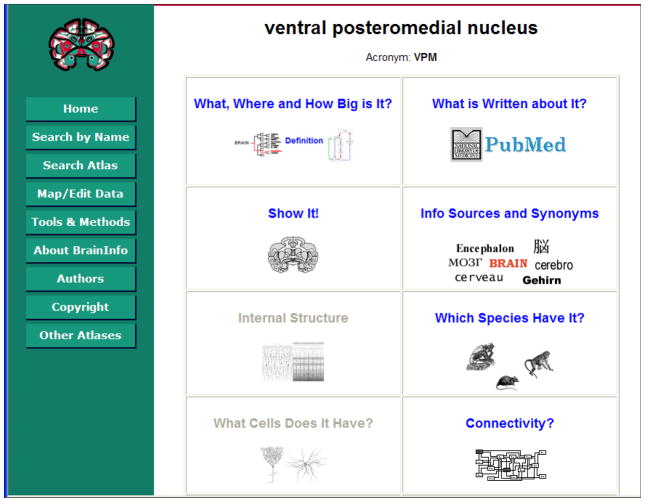
A user submits a query to BrainInfo by keying a character string, ‘arcuate nucleus’, into a search box. BrainInfo compares the input character string to names in the Names table (Fig. 6) and presents a list of matches with the standard name(s) of the concept(s) to which each name corresponds (Fig. 7). The user clicks the standard name that corresponds to the concept he has in mind, ‘ventral posteromedial nucleus’, and BrainInfo displays the Central Directory for that concept (Fig. 8). The Central Directory has iconic buttons for most kinds of information about brain structures that are likely to interest a user. Each button hyperlinks to a selection of pages in websites containing pertinent information.
Figure 8. BrainInfo’s Central Directory for ventral posteromedial nucleus.
(standard name for arcuate nucleus of the thalamus). Central Directories for some 2500 structures provide hyperlinks to most kinds of information that a user may seek.
If BrainInfo does not have links to information in a given category, the icon is grayed to save the user’s navigating to a dead end. BrainInfo can then help the user search PubMed for information on the topic. When the user clicks the button labeled ‘What is Written about It?’ BrainInfo uses the ontology to compose a query that includes the standard name of the structure with its synonyms and sends the sometimes lengthy query string to PubMed. For example, the query submitted by BrainInfo for the ventral posteromedial nucleus reads: “arcuate nucleus-3” OR “Nucleus ventralis posteromedialis” OR “semilunar nucleus” OR “thalamic gustatory nucleus” OR “ventral posterior medial nucleus” OR “ventral posteromedial nucleus” OR “ventral posteromedial thalamic nucleus”. The value added by this application of the ontology is to eliminate false negative omission of citations resulting from authors’ use of terms other than ‘ventral posteromedial nucleus’ in referring to the structure.
If BrainInfo retrieves a page of information from a website that uses different terminology from the NeuroNames standard, BrainInfo uses the ontology to provide clarification in the format “Look for [terms used by the website authors]” (Fig. 9). By disambiguating homonyms and interpreting synonyms BrainInfo achieves one of its major purposes, viz., to eliminate terminology as an obstacle to effective communication.
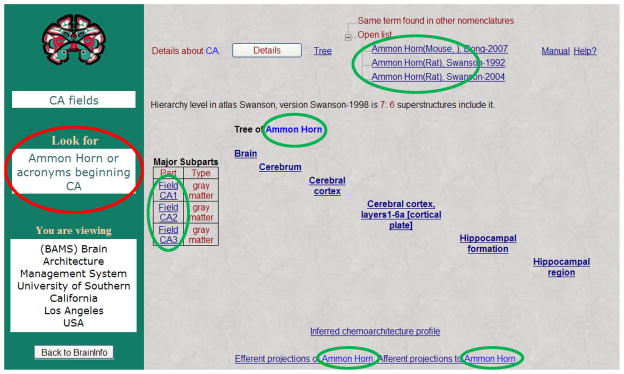
Figure 9. The NeuroNames ontology aids interpretation of other websites.
The user sought information about the hippocampus (as a synonym of ‘CA fields’). BrainInfo has retrieved the page at USC’s Brain Information Management System (BAMS, 2010), which provides information about the structure using the terminology ‘Ammon Horn’, ‘CA1’, ‘CA2’ and ‘CA3’ (green dark ovals). BrainInfo indicates terms for which the user should look (red light oval).
The BrainInfo Portal was established in 2001. It now links to several thousand pages in more than 50 of the most informative neuroscience sites on the Web. Several observations suggest that BrainInfo is providing useful service to the neuroscience community. In recent years an average of 400 unique visitors have viewed an average of 2000 pages per day. The top 20 institutional affiliations of identifiable users include universities and governmental agencies with large concentrations of neuroscientists, such as Harvard University, the National Institutes of Health (US), Oxford University, Washington University St. Louis, the National Health Service (UK), McGill University and the University of California at Los Angeles (UCLA). At least 20% of users return to the site one or more times during a given month, and the total number of unique visitors per year exceeds 100,000.
Translation of NeuroNames into OWL
A large portion of the NeuroNames brain hierarchy has been translated into OWL for the Neuroscience Information Framework (Bug et al., 2008). The NeuroNames hierarchy provides the core anatomical ontology for the Neuroscience Information Framework Standard (NIFSTD) gross anatomy module. All ontology modules in the NIFSTD are normalized to the same upper ontology, the Basic Formal Ontology (BFO). The initial volumetric partonomy of NeuroNames was refactored to an “is-a” hierarchy through the creation of categories such as “Predominantly gray part of hypothalamus” and listing the NeuroNames parts underneath. As the reasoning of OWL over partonomies became more powerful, these somewhat contrived and artificial categories were replaced through the assignment of the “part of” relationship from the OBO relations ontology. As part of the NIFSTD infrastructure, each term within the NIFSTD is presented as its own page on the NeuroLex Wiki (NeuroLex, 2011). The NIFSTD has progressively added more bridging relationships among modules, e.g., defining cell types according to the brain region in which the cell soma lies, through the definition of bridge files.
Limitations of the BrainInfo portal as a web textbook
A serious challenge to development of BrainInfo as a web-resource compared to conventional publications is the copyright constraint. While images of most brain structures appear on the Web in one form or another, the original photomicrographs illustrating the definitions of the several hundred primary structures of the brain reside in copyrighted publications. We have been able to scan and display the original images of cortical areas from publications that are out of copyright, from Brodmann (1909) up to sources from the early 1960s. We are generally unable to display original images from later publications, because publishers who readily grant permission to republish images in a conventional textbook are hesitant to grant permission to publish them on the Web.
Another challenge arises from the great variability in care for accuracy exercised by the authors of neuroanatomy websites. Some of the best images for illustrating some structures show erroneous labels for others. We address this issue by linking to such images only for the structures that are correctly labeled and for which no other image is available. If later we find an equivalent image that is more accurately labeled, we eliminate the link to the first and link to the new image.
A third challenge, which fortunately occurs infrequently, is the disappearance or reorganization of a website that results in the loss of access to informative pages. In six years we have lost or discontinued contact with three websites on those bases.
Perhaps the most serious limitation of NeuroNames and BrainInfo in the eyes of its users is the failure to achieve comprehensive coverage of the neuroanatomical domain. While the NeuroNames vocabulary is estimated to contain 90% of English and Latin names of neuroanatomical structures a person encounters in the neuroscientific literature, the NeuroNames ontology only includes the text definitions of about 70% of the structures. And it provides even less complete information about the connectivity, cells, genes expressed, models, function and other features of specific structures. The main reason for less than comprehensive coverage is that in the beginning we populated the knowledge base with a focus on information not readily available in standard English-language textbooks. It is apparent, however, that the Web is becoming the first line of inquiry, even for basic information about the classical structures. As a result we are currently incorporating text definitions of all concepts in the NeuroNames ontology.
A further challenge to NeuroNames and BrainInfo is common to all web-based resources. That is the challenge of gaining access to constructive peer review. The NeuroNames brain hierarchy was subjected to, and improved by, peer review when it was first published (Bowden and Martin, 1995). In the subsequent 15 years the number of concepts in the ontology has more than tripled without review. For several years we sought critique through a ‘Feedback’ button on the home page and a ‘Comments’ button on every informational page. Both attracted little other than e-graffiti.
In the long run the greatest limiting factor to NeuroNames development may prove to be its dependence on the continuous effort of a single individual. At this time it is unclear whether the web portal and textbook format of BrainInfo can merge into an institutional framework in a way that maintains its growth and development. Continuous development requires long-term expenditure of scholarly time and effort. It requires at least one individual who, like a textbook author, uses semiautomated informatics tools to modify and extend the ontology as the domain evolves. In the world of conventional publication, when Author A of a successful textbook XYZ moves on, the publisher recruits a new Author B and the series continues as A’s textbook of xyz by B. In the world of the noncommercial internet, when an author moves on the resource risks deterioration as untended links break and lack of updating leads to obsolescence of its content. In 2009 the International Neuroinformatics Coordinating Facility (INCF, 2011 and the Center for Research in Biological Systems (CREBS, 2010) assumed sponsorship of BrainInfo to maintain current functions. We are pursuing several potential mechanisms to assure that the system will continue to grow in usefulness to the neuroscience community.
Information Sharing
The NeuroNames nomenclature and links to the Central Directories of neuroanatomical concepts in BrainInfo are available for download as an Excel workbook, NeuroNames Ontology of Mammalian Neuroanatomy: NN2010, at http://braininfo.rprc.washington.edu/Nnont.aspx. The workbook contains the standard English names for 2350 structures of the human, macaque, rat and mouse brains, as well as almost 10,000 English and Latin names for them. The tables provide web addresses to pages in BrainInfo for structures that can be used to make neuroanatomic web sites interoperable with each other and with BrainInfo. A second Excel workbook contains the nomenclatures used in the rat and mouse atlases of Paxinos and Franklin (2001), Hof et al. (2000), Swanson (2004), and Dong (2004). Its tables provide web addresses to pages in BrainInfo that correspond to the authors’ terms. The text definitions of most structures can be obtained separately by contacting: dmbowden@uw.edu. Access by the public to these resources is free of charge or registration. The OWL translation of the NeuroNames Brain Hierarchy is made available as a component of NIFSTD at the NeuroLex website (NeuroLex, 2011).
Acknowledgments
The authors wish to thank Maryann Martone for current information on the translation of NeuroNames into OWL and Erik McArthur for assistance in the production of figures for this report. The work was supported by the International Neuroinformatics Coordinating Facility (INCF) and by grants LM-008247, MH-069259 and RR-000166 from the U.S. National Institutes of Health, to the University of Washington.
References
- Anthoney TR. Similar Sounding Non-synonyms and Terms of Variable Meaning. CRC; Boca Raton: 1994. Neuroanatomy and the Neurologic Exam: a Thesaurus of Synonyms. [Google Scholar]
- Arbib MA, Amari S-I, editors. Research Notes in Neural Computing. Vol. 1. Springer; New York: 1988. Dynamic Interactions in Neural Networks: Models and Data. [Google Scholar]
- Berners-Lee T, Hendler J, Lassila O. The semantic web. Scientific American. 2001;284(5):34–44. [Google Scholar]
- Bowden DM, Martin RF, Bowden DM. Primate Brain Maps. Elsevier Science; Amsterdam: 2000. Atlases for informatics and quantitative neuroanatomy. [Google Scholar]
- Bowden DM, Dubach MF. Neuroanatomical nomenclature and ontology. In: Koslow S, Subramaniam S, editors. Databasing the Brain: Data to Knowledge. Chapter 2. Wiley; New York: 2004. [Google Scholar]
- Bowden DM, Martin RF. NeuroNames Brain Hierarchy. Neuroimage. 1995;2:63–83. doi: 10.1006/nimg.1995.1009. [DOI] [PubMed] [Google Scholar]
- BrainInfo. NeuroMaps Mapper: Macaque. University of Washington; Seattle, USA: 2010a. [accessed 19 October 2010]. http://neuromaps.braininfo.org/cgi-bin/NeuroMaps.py?species=Macaque. [Google Scholar]
- BrainInfo. Location in English Hierarchy of Brain Structures. University of Washington; Seattle, USA: 2010b. [accessed 30 September, 2010]. http://braininfo.rprc.washington.edu/whereisit.aspx?language=0&requestID=ID21&parentID=ID3111&originalID=ID21. [Google Scholar]
- BrainInfo. NeuroNames Ontology of Mammalian Neuroanatomy: NN2010. University of Washington; Seattle, USA: 2010c. [accessed 5 September, 2010]. http://braininfo.rprc.washington.edu/Nnont.aspx. [Google Scholar]
- BrainInfo. Arcuate nucleus of the hypothalamus. University of Washington; Seattle, USA: 2010d. [accessed 31 March, 2011]. http://braininfo.rprc.washington.edu/Search.aspx?searchstring=arcuate%20nucleus&fromwhere=main. [Google Scholar]
- Bug WJ, Ascoli GA, Grethe JS, Gupta A, Fennema-Notestine C, Laird AR, Larson SD, Rubin D, Shepherd GM, Turner JA, Martone ME. The NIFSTD and BIRNLex vocabularies: Building comprehensive ontologies for neuroscience. Neuroinform. 2008;6:175–194. doi: 10.1007/s12021-008-9032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MB, Sutin J. Human Neuroanatomy. Williams & Wilkins; Baltimore: 1983. [Google Scholar]
- CRBS. Center for Research in Biological Systems. University of California; San Diego: 2010. [accessed 8 September, 2010]. http://crbs.ucsd.edu/ [Google Scholar]
- Crosby EC, Humphrey T, Lauer EW. Correlative Anatomy of the Nervous System. Macmillan; New York: 1962. [Google Scholar]
- Dong HW. Allen Reference Atlas. Allen Institute for Brain Science; Seattle: 2004. [accessed 31 March, 2011]. http://mouse.brain-map.org/atlas/index.html. [Google Scholar]
- Google. Google; Mountainview, CA: 2011. [accessed 20 April, 2011]. http://google.com. [Google Scholar]
- Gray H. Anatomy of the Human Body. Bartleby.com; 1918. [accessed 20 October 2010]. http://www.bartleby.com/107/183.html. [Google Scholar]
- Grillner S, Graybiel AM, editors. Microcircuits: The Interface between Neurons and Global Brain Function. MIT; Cambridge: 2006. [Google Scholar]
- Hof PR, Young WG, Bloom FE, Belichenko PV, Celio MR. Comparative Cytoarchitectonic Atlas of the C57BL/6 and 129/Sv Mouse Brains. Elsevier; Amsterdam: 2000. [Google Scholar]
- IANC. Nomina Anatomica, International Anatomical Nomenclature Committee. Williams & Wilkins; Baltimore: 1983. [Google Scholar]
- INCF. International Neuroinformatics Coordinating Facility. Karolinska Institute and Royal Institute of Technology; Stockholm, Sweden: 2011. [accessed 11 June 2011]. http://www.incf.org. [Google Scholar]
- Kahle W, Frotscher M. Nervensystem und Sinnesorgane. Vol. 3. Georg Thieme Verlag; Stuttgart: 2001. Taschenatlas der Anatomie. [Google Scholar]
- NeuroLex. NeuroLex: The Neuroscience Lexicon. University of California; San Diego, CA: 2011. [accessed 9 June, 2011]. http://neurolex.org/wiki/Main_Page. [Google Scholar]
- NLM. Medical Subject Headings. National Library of Medicine; 2010. [accessed 6 October 2010]. http://www.nlm.nih.gov/mesh/meshhome.html. [Google Scholar]
- NIF. Neuroscience Information Framework, version 3.5. University of California; San Diego: 2011. [accessed 30 May 2011]. http://www.neuinfo.org/about/index.shtm. [Google Scholar]
- Ono M, Kubik S, Abernathey CD. Atlas of the Cerebral Sulci. Thieme; Stuttgart: 1990. [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. 2. Academic; San Diego: 2001. [Google Scholar]
- Paxinos G, editor. The Human Nervous System. Academic Press; San Diego: 1990. [Google Scholar]
- PubMed. PubMed. National Library of Medicine; Bethesda, MD: 2009. [accessed 20 April, 2011]. http://pubmed.gov. [Google Scholar]
- Puelles L, Martinez-de-la-Torre M, Paxinos G, Watson C, Martinez S. An atlas featuring neuromeres and mammalian homologies. San Diego: Elsevier Academic Press; 2007. The Chick Brain in Stereotaxic Coordinates. [Google Scholar]
- Riley HA. Brain Stem and Spinal Cord. Baltimore: Williams & Wilkins; 1943. An Atlas of the Basal Ganglia. [Google Scholar]
- Schiebler TH, Schmidt W, Zilles K. Auflage. Vol. 8. Springer; Berlin: 1999. Anatomie. [Google Scholar]
- Stephan H. Allocortex. In: Bargmann W, editor. Handbuch der mikroskopischen Anatomie des Menschen. Part 9. Vol. 4. Berlin: Springer-Verlag; 1975. Nervensystem. [Google Scholar]
- Swanson LW. What is the brain? Trends in Neuroscience. 2000;23:519–527. doi: 10.1016/s0166-2236(00)01639-8. [DOI] [PubMed] [Google Scholar]
- Swanson LW. Brain Maps: Structure of the Rat Brain. 3. Elsevier Academic; Oxford: 2004. [Google Scholar]
- UMLS. Unified Medical Language System. National Library of Medicine; Bethesda, Maryland: 2011. [accessed 31 March, 2011]. http://www.nlm.nih.gov/research/umls/new_users/index.html. [Google Scholar]
- Wikipedia. [accessed 31 March, 2011];Ontology (information science) 2010a http://en.wikipedia.org/wiki/Ontology_(information_science)
- Wikipedia. [accessed 6 October, 2010];List of regions in the human brain. 2010b http://en.wikipedia.org/wiki/Talk:List_of_regions_in_the_human_brain.
- Wikipedia. Wikipedia. Wikipedia.com; San Francisco, CA: 2011. [accessed 20 April, 2011]. http://wikipedia.org. [Google Scholar]