Abstract
The conservation of cultivated plants in ex-situ collections is essential for the optimal management and use of their genetic resources. For the olive tree, two world germplasm banks (OWGB) are presently established, in Córdoba (Spain) and Marrakech (Morocco). This latter was recently founded and includes 561 accessions from 14 Mediterranean countries. Using 12 nuclear microsatellites (SSRs) and three chloroplast DNA markers, this collection was characterised to examine the structure of the genetic diversity and propose a set of olive accessions encompassing the whole Mediterranean allelic diversity range. We identified 505 SSR profiles based on a total of 210 alleles. Based on these markers, the genetic diversity was similar to that of cultivars and wild olives which were previously characterised in another study indicating that OWGB Marrakech is representative of Mediterranean olive germplasm. Using a model-based Bayesian clustering method and principal components analysis, this OWGB was structured into three main gene pools corresponding to eastern, central and western parts of the Mediterranean Basin. We proposed 10 cores of 67 accessions capturing all detected alleles and 10 cores of 58 accessions capturing the 186 alleles observed more than once. In each of the 10 cores, a set of 40 accessions was identical, whereas the remaining accessions were different, indicating the need to include complementary criteria such as phenotypic adaptive and agronomic traits. Our study generated a molecular database for the entire OWGB Marrakech that may be used to optimise a strategy for the management of olive genetic resources and their use for subsequent genetic and genomic olive breeding.
Electronic supplementary material
The online version of this article (doi:10.1007/s10709-011-9608-7) contains supplementary material, which is available to authorized users.
Keywords: Olea europaea, SSR characterisation, Chloroplast DNA markers, Model-based Bayesian clustering, Allelic diversity capture
Introduction
The olive tree (Olea europaea L.) is one of the most important fruit crops of the Mediterranean area (Zohary and Spiegel-Roy 1975). Palynological and anthracological (fossil charcoal) studies have shown that wild olive populations were present in eastern and western Mediterranean zones before the Neolithic (Terral et al. 2004; Carrión et al. 2010). Early domesticated forms were probably disseminated during successive human migrations (especially from east to west) throughout the Mediterranean Basin, but olive selection from local western populations has also been revealed by genetic studies (Besnard et al. 2002; Breton et al. 2006; Khadari et al. 2008).
Today, the area devoted to olive growing worldwide is estimated at 8.8 Mha (IOC 2007). It is one of the most economically important trees in Mediterranean areas, largely due to its multiple uses (e.g. oil, canned fruit, wood, ornamental uses, medicinal applications). Over 2,000 cultivars have been described, exhibiting significant levels of variation in oil content, fruit size and adaptation to local environmental conditions (Bartolini et al. 1998), but this number is probably underestimated since there is a lack of information on minor local varieties (Cantini et al. 1999).
Olive growing is undergoing a sharp transition from traditional to modern orchards, with a reduced number of main varieties featuring interesting agronomic traits such as yield, oil quality, and adaptive traits related to biotic and abiotic stresses. For instance, the “Picual” and “Arbequina” varieties have been massively planted over the last two decades in Andalusia and Catalonia, respectively (Belaj et al. 2010). A similar trend was noted in Portugal, with the main cultivar “Galega” grown in about 80% of the olive groves (Gemas et al. 2004), and in Morocco where “Picholine Marocaine” is the dominant cultivar throughout the country (Khadari et al. 2008). Hence, despite the high initial varietal diversity, a recent trend towards establishing modern orchards based on the most productive cultivars leads to high erosion of this germplasm. Several olive germplasm collections have been created at national and regional levels to manage ex-situ olive genetic resources for conservation purposes and eventual use in subsequent breeding programs (Bartolini 2008). The first major attempt to conserve and characterise the most important cultivars from all olive growing countries led to the establishment of the World Olive Germplasm Bank in Córdoba, Spain (OWGB Córdoba). This bank was initiated by FAO-INIA in 1970, with the contribution of the International Olive Oil Council (IOOC; Caballero et al. 2006). It includes Spanish cultivars that were collected by Barranco and Rallo (2000) and varieties originating from other Mediterranean countries. OWGB Córdoba has served for many studies using morphological descriptors (Caballero and del Río 2002) and molecular markers such as random amplified polymorphism DNA (RAPDs) and single sequence repeat (SSRs; Belaj et al. 2003, 2004).
Over the course of the RESGEN project, numerous surveys and studies on conservation and characterisation have been conducted by each of the following 15 participating countries: Algeria, Croatia, Cyprus, Egypt, France, Greece, Israel, Italy, Morocco, Portugal, Serbia-Montenegro, Slovenia, Spain, Syria, Tunisia (Caballero et al. 2006). These partners have completed their sampling by collecting local olive cultivars based on a morphological characterisation. As genetic redundancies, homonymy and synonymy cases are common problems in the management of ex-situ collections (Engels and Visser 2003), morphological description has been complemented by the use of molecular markers. Recently, the development of SSRs in olive (e.g. Sefc et al. 2000; de La Rosa et al. 2002) has significantly enhanced the possibility of individual olive cultivar identification. After testing 37 SSR loci for their reproducibility and discriminating power in four independent laboratories, Baldoni et al. (2009) proposed a consensus list of SSRs for genotyping of cultivated olive.
More recently, a second international germplasm bank was set up at the experimental orchard of Tessaout [National Institute of Agronomic Research, Marrakech, Morocco; (OWGB Marrakech)] in 2003. Compared to OWGB Córdoba, OWGB Marrakech was set up in a different scientific context with more knowledge available about the plant material. To optimise olive germplasm sampling, local genetic resources had been characterised in several Mediterranean countries using standardised morphological descriptors. The bank was established by introducing previously characterised genetic resources from each Mediterranean area. For some partner countries (e.g. Spain; Barranco and Rallo 2000), a set of accessions representative of the local diversity was proposed following morphological or molecular characterisation. In 2010, OWGB Marrakech included 561 accessions originating from 14 Mediterranean countries and further introduction of additional olive germplasm is ongoing.
To optimise the management and use of the large olive ex-situ collections, it is essential to select a sub-sample of accessions, so-called core collections, displaying the overall genetic diversity and phenotypic variability, as first proposed by Frankel and Brown (1984). Several strategies have been proposed to facilitate the construction of core collections, which can be classified into two groups according to the allocation methods. The first one is based on maximising the variability, including the M-Method strategy developed by Schoen and Brown (1993) and implemented in the MSTRAT software package (Gouesnard et al. 2001); and the second group, known as the stratified method, is based on similarity clustering (Escribano et al. 2008). Among numerous potential applications, core collections can be used as a first step in genetic association studies for detecting quantitative trait loci (QTLs) related to agromonic traits (Barnaud et al. 2006; Le Cunff et al. 2008; Aranzana et al. 2010).
Several molecular characterisation studies have focused on olive germplasm at local or national levels (Belaj et al. 2004; Banilas et al. 2003), but also at the scale of the Mediterranean Basin (Besnard et al. 2001a; Sarri et al. 2006). Olive cultivars analysed in these latter studies were considered as representative of local or Mediterranean genetic diversity, but to date no genetic studies of the entire OWGB have been published. Different aspects have been addressed in previous studies and, for instance, an analysis of genetic relationships among Mediterranean olive cultivars revealed a correlation between genetic structure and geographical origin of cultivars, suggesting multilocal olive selection from different genetic pools (Besnard et al. 2001a). More recently, Sarri et al. (2006) conducted a study of genetic relationships based on SSRs among 118 cultivars sampled in several Mediterranean countries and showed that Mediterranean olive germplasm was structured into three main gene pools, corresponding to the western, central and eastern Mediterranean regions. Studies on genetic structure have also been conducted on wild olive trees (or oleasters) or for investigating genetic relationships between wild and cultivated olives (Besnard et al. 2002, 2007; Breton et al. 2006; Belaj et al. 2010).
The purpose of this study was to investigate the genetic structure of the entire OWGB Marrakech, including all 561 accessions, using both nuclear SSRs and chloroplast DNA markers. The genetic structure of Mediterranean olive germplasm was investigated using model-based Bayesian clustering method to assign individuals into defined gene pools according to genetic and geographic criteria. This study also provides critical baseline information for the development of core collections to maximise the representativeness of olive genetic diversity. Our results represent an essential step towards optimised conservation of olive genetic resources and subsequently for genetic association studies to detect QTLs of adaptive and agronomic interest.
Materials and methods
Plant material
A total of 561 accessions maintained in the ex-situ World Olive Germplasm Bank at the experimental orchard of Tessaout (OWGB, INRA Marrakech, Morocco) were analysed. These accessions were derived from 14 olive-growing countries: Algeria (43 accessions), Croatia (16), Cyprus (28), Egypt (19), France (12), Greece (13), Italy (167), Lebanon (16), Morocco (40), Portugal (14), Slovenia (9), Spain (89), Syria (71) and Tunisia (24) (Table 1 and Supplementary file, Table S1). This second OWGB was launched in 2003 under IOOC supervision (see website: http://www.internationaloliveoil.org/), approximately 20 years after the first one was set up at IFAPA Córdoba (OWGB Córdoba, Spain). OWGB Córdoba mainly includes Spanish accessions (271) and 172 accessions from the following countries: Albania (7 accessions), Algeria (2), Argentina-Chile (1), Croatia (5), Egypt (4), France (10), Greece (19), Israel (3), Italy (32), Lebanon (3), Morocco (3), Portugal (12), Slovenia (1), Syria (36) Tunisia (9), Turkey (23), USA (2) (http://www.oleadb.it/olivodb.html; Bartolini et al. 1998). Among the 384 and 491 cultivars identified in OWGB Córdoba and OWGB Marrakech, respectively; only 153 are present in both collections, while most of the accessions are exclusively present in one collection (338 only present in OWGB Marrakech and 231 present in OWGB Córdoba including 157 Spanish cultivars; Supplementary file, Table S1).
Table 1.
Locality, sample size, multilocus SSR profiles and maternal lineages of olive accessions maintained in the OWGB collection Marrakech (INRA, Morocco)
| Geographical origin | Sample size | SSR profile | Maternal lineagea |
|---|---|---|---|
| Western Mediterraneanb | |||
| Morocco | 40 | 31 | 31 E |
| Portugal | 14 | 14 | 14 E1 |
| Spain | 89 | 89 | 80 E1, 6 E2, 3 E3 |
| Central Mediterraneanb | |||
| Algeria | 43 | 40 | 32 E1, 8 E3 |
| Tunisia | 24 | 24 | 24 E1 |
| France | 12 | 12 | 10 E1, 1 E2, 1 E3 |
| Italy | 167 | 165 | 161 E1, 2 E2, 2 E3 |
| Croatia | 16 | 14 | 14 E1 |
| Slovenia | 9 | 9 | 9 E1 |
| Greece | 13 | 13 | 11 E1, 2 E2 |
| Eastern Mediterraneanb | |||
| Egypt | 19 | 19 | 19 E1 |
| Cyprus | 28 | 23 | 23 E1 |
| Lebanon | 16 | 13 | 13 E1 |
| Syria | 71 | 47 | 47 E1 |
| Total | 561 | 513 | 488 E1, 11 E2, 14 E3 |
aNumber of individuals classified into the three maternal lineages as defined by Besnard et al. (2007): E1 from the eastern Mediterranean; E2 and E3 from the western Mediterranean
bOlive accessions were classified into three main gene pools according to model-Bayesian clustering: western, central and eastern Mediterranean gene pools (see Fig. 1)
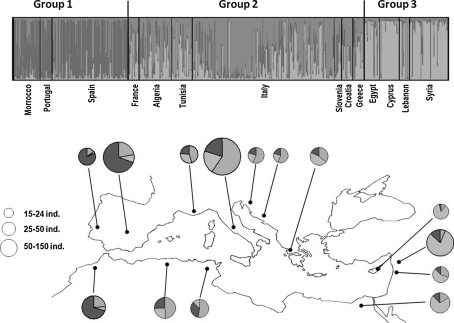
OWGB Marrakech was set up following the ResGen project, which included most Mediterranean olive-growing countries. Olive genetic resources for each partner Mediterranean country have been characterised using standardised morphological descriptors. Hence, OWGB Marrakech included different national olive germplasm previously characterised using morphological descriptors alone, or both morphological descriptors and molecular markers. The composition of national olive genetic resources refers to the number of accessions, the representativeness of ancient and modern olive orchards in different areas and agro-ecosystems, and the ratio between main, minor and local cultivars, as supervised by each national institute in charge of their olive genetic resource management. According to a recent study (Sarri et al. 2006), cultivated olive accessions were clustered into three main gene pools respectively distributed in the eastern, central and western Mediterranean Basin. Based on this study, the accessions analysed here are classified into three distinct regional groups: (1) a western Mediterranean group, including accessions originating from Morocco, Portugal, and Spain; (2) a central Mediterranean group, with accessions from Algeria, Tunisia, France, Italy, Slovenia, Croatia, and Greece; and (3) an eastern Mediterranean group, including accessions from Egypt, Cyprus, Lebanon, and Syria (Table 1).
DNA preparation and genotyping procedure
Genomic DNA was extracted from 100 mg of fresh leaf tissue, as described in Khadari et al. (2008). DNA quality was checked on 2% agarose gel and the DNA concentration was estimated using spectrofluorometry.
Twelve nuclear DNA SSR loci were selected according to their level of polymorphism and reproducibility, as well as by an easy and accurate scoring method (data not shown): DCA03, DCA04, DCA05, DCA08, DCA09, DCA11, DCA15, DCA18 (Sefc et al. 2000); GAPU59, GAPU71B (Carriero et al. 2002); UDO36 (Cipriani et al. 2002) and EMO90 (de La Rosa et al. 2002). In addition, the three maternal lineages (E1, E2, and E3; Besnard et al. 2007) were detected using a combination of three chloroplastic indel markers; psbK-trnS-polyT-A, trnS-G-indel-1, and trnS-G-indel-2 (Besnard et al. 2003). PCR amplification was carried out in a total volume of 20 μl containing 20 or 10 ng of genomic DNA for nuclear SSR or chloroplast indel genotyping, respectively, 1.5 mM MgCl2, 0.2 M of each dNTP, 0.1 units of Taq DNA polymerase, and 2 pmol of forward (fluorescent labelled; Table 2) and reverse primers. Reactions were performed in a thermal cycler (Mastercycler ep gradient S). PCR products were separated using an automatic capillary sequencer (ABI 3130 Genetic Analyzer Applied Biosystems) and chromatograms were then analysed with the GenMapper 3.7 software (Applied Biosystems).
Table 2.
Locus name, fluorescent dye-labelled, size range of amplified fragments (bp), number of alleles per locus (N a), observed heterozygosity (H O), discriminating power (D) and probability of identity (PI)
| Locus name | Fluorescent dye | Allele size range (bp) | Alleles | Profile | D | PI | |||
|---|---|---|---|---|---|---|---|---|---|
| N a | Rare alleles* (%) | Once observed alleles (%) | H O | ||||||
| DCA04a,1 | HEX | 117–193 | 30 | 24 (80) | 2 (6) | 0.62 | 95 | 0.95 | 0.034 |
| DCA09a,1 | NED | 162–219 | 24 | 17 (70) | 2 (8) | 0.93 | 92 | 0.96 | 0.025 |
| DCA15a,1 | FAM | 231–266 | 08 | 4 (50) | 3 (37) | 0.65 | 14 | 0.77 | 0.191 |
| DCA05a,2 | FAM | 192–218 | 14 | 10 (71) | 2 (14) | 0.49 | 31 | 0.71 | 0.273 |
| DCA08a,3 | FAM | 125–168 | 19 | 14 (73) | 4 (21) | 0.90 | 50 | 0.92 | 0.054 |
| DCA03a,2 | NED | 229–265 | 14 | 7 (50) | 1 (7) | 0.89 | 49 | 0.95 | 0.032 |
| DCA18a,3 | NED | 155–207 | 19 | 13 (68) | 1 (5) | 0.83 | 58 | 0.93 | 0.044 |
| DCA11a,4 | HEX | 126–196 | 25 | 20 (80) | 3 (12) | 0.71 | 83 | 0.93 | 0.054 |
| GAPU59b,3 | HEX | 207–241 | 12 | 8 (67) | 2 (16) | 0.68 | 23 | 0.81 | 0.185 |
| GAPU71Bb,4 | FAM | 117–166 | 14 | 9 (64) | 1 (7) | 0.90 | 38 | 0.93 | 0.051 |
| UDO36c,2 | HEX | 134–168 | 16 | 13 (81) | 1 (6) | 0.69 | 41 | 0.89 | 0.110 |
| EMO90d,4 | FAM | 160–210 | 15 | 11 (73) | 2 (15) | 0.79 | 31 | 0.85 | 0.132 |
| Mean | – | – | 17.5 | 12.5 | 2 | 0.76 | 50.41 | 0.88 | 2.55 × 10−14 |
| Total | – | – | 210 | 150 (70) | 24 (11) | – | – | – | – |
Data analysis
Genetic diversity and multivariate analysis
Based on the scored-size SSR allele dataset, we computed the following genetic diversity parameters: for each SSR locus, the number of alleles (N a) and the observed heterozygosity (H O) were calculated using Genetix 4.05 (Belkhir et al. 2004). The probability of identity PI (Paetkau et al. 1995) was computed using IDENTITY 4.0 (Wagner and Sefc 1999). The discriminating power (D) was computed as defined by Tessier et al. (1999): D j = ∑p i [(Np i − 1)/N − 1)], where p i is the frequency of the ith molecular profile revealed at locus j, and N is the number of identified genotypes.
Genetic relationships among the single genetic profiles (i.e. 505 genotypes were distinguished based on SSRs, see below) were studied via principal component analysis (PCA) using the Genalex 6 program (Peakall and Smouse 2005). Genotypes were plotted on the first two principal axes to visualise genetic affinities.
Bayesian model-based clustering approach
To identify the genetic structure in Mediterranean olive germplasm, a model-based analysis was performed using STRUCTURE 2.2.3 (Pritchard et al. 2000) based on SSR data. This program assumes Hardy–Weinberg equilibrium and linkage equilibrium within clusters. The analysis was done without prior information concerning the geographic origin of the accessions. The STRUCTURE algorithm was run using the “admixture model”, assuming a “correlation among allele frequencies”, with 10 independent replicate runs per K value (number of clusters) ranging from 1 to 10. Each run involved a burning period of 100,000 iterations, and a post burning simulation length of 1,000,000.
Validation of the most likely number of clusters K was performed using the statistics proposed by Evanno et al. (2005). Q-matrix values for individual runs for each K were analysed by the CLUMPP 1.1 program (Jakobsson and Rosenberg 2007). Matrixes of individuals are represented as colored histograms of q values were constructed using DISTRUCT 1.1 (Rosenberg 2004).
Core collection sampling
The M-strategy (Maximisation strategy) proposed by Schoen and Brown (1993) and implemented in the MSTRAT software (Gouesnard et al. 2001) was used to generate core olive collections that maximise the number of observed alleles in the data set. The M-strategy consists in searching, among all potential core collections, for the best sample size that can capture all observed alleles with the highest genetic diversity score. After having determined the optimal size of the core subsets, 1,000 core collections were generated independently using the redundancy option with 10 independent runs and 1,000 iterations. Based on the 1,000 core collections obtained, we selected the reference core collection according to its Nei diversity index (Nei 1987) as first criterion, and then on the basis of its composition (representativeness among three gene pools and maternal lineages of accessions). Core subsets were constructed using all the observed alleles and without private alleles (observed once) to limit the impact of the later on genetic structure and on linkage disequilibrium (LD; Barnaud et al. 2006; Aranzana et al. 2010). The distribution of the selected core accessions was plotted on the first two PCA axes.
Results
SSR polymorphism
Based on SSR data (i.e. 210 alleles for the 12 loci; Table 2), the analysis of 561 accessions revealed 505 genotypes. The number of alleles detected per locus ranged from 8 at the DCA15 locus to 30 at the DCA04 locus. The number of single genotypes identified per locus ranged from 14 to 95, with an average of 50.4 genotypes. Frequencies observed for the 210 alleles ranged from 0.001 to 0.71, with an average of 0.05. There were 151 alleles (71.9%) with frequencies equal to or less than 5%, which were considered as rare alleles (Supplementary file, Table S2). The reliability of the 24 alleles observed once was also checked by examining their occurrence in Mediterranean oleaster populations (unpublished data). The observed heterozygosity (H O) ranged from 0.49 at the DCA05 locus to 0.93 at the DCA09 locus, with a mean of 0.76. The highest probability identity (PI) value (0.273) was at the DCA05 locus and the lowest (0.025) was at the DCA03 locus (Table 2). Based on the 12 SSR loci, the cumulative probability identity was 2.55 × 10−14, indicating that the probability of two randomly sampled olive trees having the same genotype was extremely low.
Characterisation of olive accessions
Among the identified 505 multilocus SSR profiles, 26 synonymous cases involving 82 accessions were detected (Supplementary file, Table S3). For instance, some accessions shared the same genotype: accessions “PM4 5116”, “PM3 5112”, “ZDH1”, “ZDH3”, “ZDH7”, “Zsb10” and “ZZ1” in Morocco shared SSR profile no. 002, and “Humaissi”, “Adgam” and “Souri” in Syria shared SSR profile no. 464. Otherwise, some accessions from different countries shared the same SSR profile, such as the “Zmj1” accession from Morocco and the “Zabarka” accession from Croatia (see Supplementary file, Table S3).
In the 127,260 pairwise comparisons among the 505 identified genotypes, only 366 comparisons (0.28%) represented closely related genotypes that differed by one to seven dissimilar alleles, whereas the remaining pairwise genotypes were distinguished by 8–44 dissimilar alleles (Supplementary file, Fig. S1). The highest SSR dissimilarity (44 dissimilar SSR alleles) was noted in only one genotype pair, i.e. “Souidi” from Algeria and “Baladi” from Lebanon.
Genetic structure of Mediterranean olive tree accessions
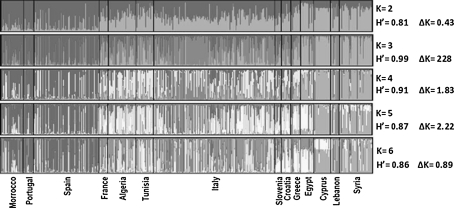
Using model-based Bayesian clustering, the genetic structure of Mediterranean olive genotypes was examined under the models with K = 2–6 clusters (Fig. 1). According to the K = 2 model, most olive accessions from Morocco, Portugal and Spain were distinguished from other Mediterranean olive trees. At K = 3, cultivars from France, Algeria, Tunisia, Italy, Slovenia, Croatia and Greece (central Mediterranean group) were mostly assigned to cluster 3 and distinguished from cluster 2, which mostly included accessions from the eastern Mediterranean region. Actually, cluster 3 also included several accessions with mixed inferred ancestry from the western and eastern Mediterranean gene pools (Table 3, Fig. 1, and Supplementary file Table S4). At K = 4–6, the Mediterranean olive germplasm, structured into three gene pools, i.e. eastern, western and central Mediterranean groups, was not modified since the fourth, fifth and sixth inferred ancestry gene pools were detected mainly in accessions of the central Mediterranean group (Fig. 1).
Fig. 1.
Inferred population structure for K = 2 to K = 6 as the presumed number of subpopulations within the Mediterranean ex situ collection, including 561 accessions classified into 505 multilocus SSR profiles. CLUMPP H’ (Jakobsson and Rosenberg 2007) represents the similarity coefficient between runs for each K, and ∆K represents the ad-hoc measure of (Evanno et al. 2005)
Table 3.
Number and proportion of genotypes from different countries assigned to the three clusters identified by structure
| Country | Genotypes | Assigned genotypesa | Cluster 1 | Cluster 2 | Cluster 3 | |||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |||
| Morocco | 31 | 16 | 15 | 94 | 1 | 6 | -b | 0 |
| Portugal | 14 | 12 | 11 | 92 | 1 | 8 | -b | 0 |
| Spain | 89 | 69 | 59 | 85 | 9 | 13 | 1 | 2 |
| Total for the group 1 | 134 | 97 | 85 | 88 | 11 | 11 | 1 | 1 |
| Algeria | 40 | 15 | 4 | 27 | 11 | 73 | -b | 0 |
| Tunisia | 24 | 10 | -b | 0 | 8 | 80 | 2 | 20 |
| France | 12 | 2 | -b | 0 | 2 | 100 | -b | 0 |
| Italy | 165 | 89 | 6 | 7 | 75 | 84 | 8 | 9 |
| Slovenia | 9 | 2 | -b | 0 | 2 | 100 | -b | 0 |
| Croatia | 14 | 5 | -b | 0 | 5 | 100 | -b | 0 |
| Greece | 13 | 2 | 1 | 50 | -b | 0 | 1 | 50 |
| Total for the group 2 | 277 | 125 | 11 | 9 | 103 | 82 | 11 | 9 |
| Egypt | 19 | 11 | 1 | 9 | -b | 0 | 10 | 91 |
| Cyprus | 23 | 22 | -b | 0 | 2 | 9 | 20 | 91 |
| Lebanon | 13 | 12 | 1 | 8 | 3 | 25 | 8 | 67 |
| Syria | 47 | 42 | 3 | 7 | -b | 0 | 39 | 93 |
| Total for the group 3 | 102 | 87 | 5 | 6 | 5 | 6 | 77 | 88 |
aUnder the assignation probability P (qI) ≥ 80%
bNo genotype assigned
Based on the highest ∆K and H′ values, K = 3 appeared to be the best model for olive genetic structure (Fig. 1; and see Supplementary file, Fig. S2), supporting the existence of the three gene pools described above. For groups 1 and 3, most genotypes were classified into one cluster based on the shared ancestry values, which were higher than 0.80; while for group 2, most genotypes were admixed (Fig. 2, Table 3, and Supplementary file Table S4).
Fig. 2.
Distribution of different gene pools from different countries in the Mediterranean Basin at K = 3. The three main groups were determined according to model-Bayesian clustering (see Fig. 1); Group 1 western Mediterranean, Group 2 central Mediterranean, and Group 3 eastern Mediterranean
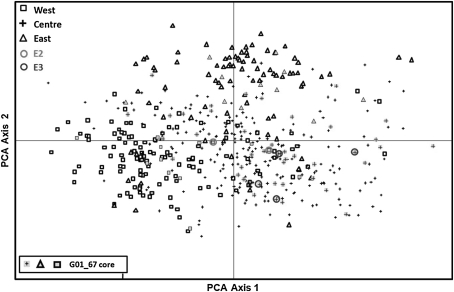
The three groups defined by model-based Bayesian clustering were plotted on the two first PCA axes (Fig. 3). The western and eastern groups were distinguished by both of the axes, while the central Mediterranean group was in an intermediate position, as shown by the admixed inferred ancestry origins of their genotypes (Fig. 2).
Fig. 3.
PCA plot of 505 olive genotypes based on the first two principal axes accounting for 46.7% of the total genetic variation (first axis = 25.67% and the second = 21.11% of the total genetic variation). Open square western Mediterranean accessions, plus central Mediterranean accessions, and open triangle eastern Mediterranean accessions. Yellow color indicates G01_67 core accessions within the overall distribution. Groups were defined on the basis of Bayesian model clustering using Structure software (Pritchard et al. 2000; see Fig. 1). E2 and E3 are the western Mediterranean maternal lineages, while E1 is the Eastern lineage as defined by Besnard et al. (2007). (Color figure online)
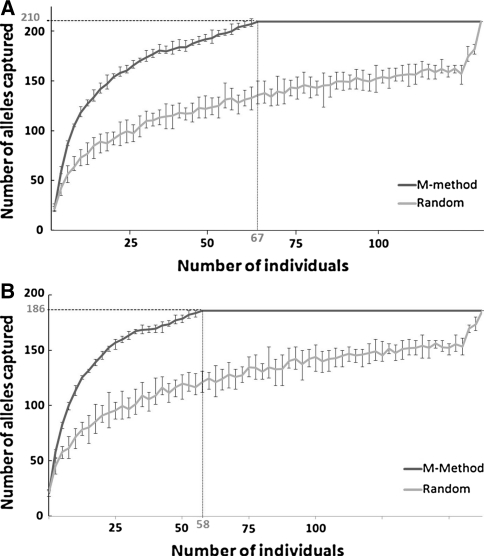
Construction of nested core collections maximising diversity
When comparing the two strategies to capture the maximum genetic diversity, the sampling efficiency of the M-strategy was always superior to a random strategy and the relative efficiency was highest for small-size samples (Fig. 4). Based on the M-strategy, the total allelic diversity could be captured with 67 genotypes (Fig. 4A). After exclusion of alleles observed only once, a minimum of 58 genotypes was necessary to capture the allelic diversity (based on 186 alleles; Fig. 4B).
Fig. 4.
Sampling efficiency based on the ability to capture the genetic diversity via the M-strategy (M-method) and a random strategy: A based on the total number of alleles (210), and B after exclusion of observed alleles once. In each case, the size of the proposed core collection was determined
Under the optimal size of a core collection (67 accessions) capturing all 210 alleles, 10 cores (G01 to G10) obtained by the M-strategy were classified according to their observed heterozygosity and Nei’s index diversity. The genetic diversity of the best 10 cores ranged from 0.77 to 0.78 for the observed heterozygosity and from 9.55 to 9.73 for the Nei’s index diversity (Table 4). Each of the 10 cores (G01 to G10; Table 4) consisted of accessions from the western Mediterranean (group 1;14.40 ± 2.17), central Mediterranean (group 2; 38.10 ± 2.68) and eastern Mediterranean gene pools (group 3; 14.5 ± 1.90; Table 4). Within each of the 10 cores, 60–63 accessions displayed cpDNA lineage E1, while western cpDNA lineages E2 and E3 were both represented by two to four accessions (Table 4). To illustrate the representativeness of the reference core collection, the position of the G01_67 core accessions is presented on the first two axes of the PCA (Fig. 3).
Table 4.
SSR diversity within samples of the G-core classified according to their observed heterozygosity and their Nei’s index diversity
| Sample name | Observed heterozygosity | Nei’s index | Group 1a (N = 134) | Group 2a (N = 277) | Group 3a (N = 102) | Maternal lineage |
|---|---|---|---|---|---|---|
| G01_67 core | 0.78 | 9.73 | 13 | 43 | 11 | 61E1, 2E2, 4E3 |
| G02_67 core | 0.77 | 9.72 | 13 | 40 | 14 | 61E1, 4E2, 2E3 |
| G03_67 core | 0.77 | 9.70 | 13 | 39 | 15 | 62E1, 2E2, 3E3 |
| G04_67 core | 0.77 | 9.68 | 12 | 38 | 17 | 62E1, 2E2, 3E3 |
| G05_67 core | 0.78 | 9.66 | 17 | 33 | 17 | 62E1, 2E2, 3E3 |
| G06_67 core | 0.76 | 9.65 | 17 | 36 | 14 | 62E1, 3E2, 2E3 |
| G07_67 core | 0.76 | 9.63 | 13 | 39 | 15 | 62E1, 2E2, 3E3 |
| G08_67 core | 0.76 | 9.62 | 13 | 38 | 16 | 60E1, 3E2, 4E3 |
| G09_67 core | 0.77 | 9.58 | 18 | 36 | 13 | 63E1, 2E2, 2E3 |
| G10_67 core | 0.77 | 9.55 | 15 | 39 | 13 | 63E1, 2E2, 2E3 |
| Mean | 0.77 | 9.65 | 14.40 | 38.10 | 14.5 | – |
| SD | – | – | 2.17 | 2.68 | 1.90 | – |
aGroups defined on the basis of Bayesian model clustering using Structure software (see Fig. 2)
Under the optimal size of a core collection (58 accessions) capturing alleles scored at least twice (186 alleles), the genetic diversity of the best 10 cores ranged from 0.75 to 0.77 for the observed heterozygosity and from 9.55 to 9.73 for the Nei’s index diversity (Supplementary file; Table S5).
Discussion
Is the ex-situ OWGB Marrakech representative of the Mediterranean olive germplasm?
All olive accessions analysed in this study are maintained in an ex-situ collection considered as being representative of Mediterranean olive germplasm since it was set up with genetic resources from 14 Mediterranean countries. Each national institute in charge of olive genetic resource management has defined the composition of their national germplasm. However, sampling criteria such as the number of accessions, the representativeness of ancient and modern olive orchards in different areas and agro-ecosystems, and the ratio between main, minor and local cultivars, have not been uniformly used by the different Mediterranean partner countries (Caballero et al. 2006). Because of this strategy for setting up the studied ex-situ collection, we noted discrepancies in the composition of different national olive genetic resource sets, e.g. Spanish and Italian germplasm included 89 and 167 accessions, respectively, while Moroccan germplasm included only 40 accessions. Despite such discrepancies, is the ex-situ OWGB Marrakech representative of Mediterranean olive germplasm? We observed no significant independence between the number of alleles detected in 118 Mediterranean cultivars (Sarri et al. 2006) and the allelic richness in OWGB Marrakech, which was computed for 118 individuals using the standardised G value (see Supplementary file; Table S6). Further, we noted 24 alleles (11.4%) detected once and 151 alleles (71.9%) were considered as rare. Compared to wild olives sampled around the Mediterranean Basin and genotyped by Breton et al. (2006), the number of alleles at eight SSR loci (that were shared with the present study) was not significantly different from the allelic richness observed in the OWGB Marrakech computed for 166 individuals using the standardised G value (see Supplementary file, Table S6). Otherwise, although not statistically tested, we noted a relatively lower observed heterozygosity in Mediterranean wild olives (H O = 0.67) than in OWGB Marrakech. Hence, the genetic diversity observed in OWGB Marrakech may be similar to the existing Mediterranean wild and domesticated olive diversity.
SSR markers as Mediterranean olive characterisation tools
Among the 12 SSR loci used in this study, the three loci (DCA09, DCA04, and DCA03) with the highest discriminating power were able to distinguish about 80% of the 505 defined genotypes (see Supplementary file, Table S7). This level of discrimination could be even higher without the closely related genotypes, which likely correspond to mutants of clones, as previously shown (Khadari et al. 2008; Baali-Cherif and Besnard 2005). Olive discrimination based only on these three most discriminating loci was validated by the low probability of classifying two random accessions under the identical SSR profile (PI = 2.72 × 10−5). This probability decreased substantially when all 12 SSR loci used were taken into account (PI = 2.55 × 10−14). Six of these loci were included in the best consensus set of SSR markers (Baldoni et al. 2009) that has already been used for genetic structure studies (Sarri et al. 2006), and our study again confirmed that these markers are reliable tools for olive characterisation.
When considering accessions from the different Mediterranean countries, the olive cultivars were not distinguished with the same efficiency. All Spanish accessions were characterised as distinct genotypes (89 accessions corresponding to 89 distinct SSR profiles). Similar results were noted for Italian olive germplasm since 167 accessions were classified into 165 genotypes. These results may be attributed to previous characterisation, based on morphological descriptors (Barranco et al. 2005), and molecular markers such as RAPDs (Belaj et al. 2003) and SSRs (Belaj et al. 2003, 2004), before selecting accessions for introduction into OWGB Marrakech. In contrast, Syrian germplasm displayed 14 of the 26 cases of synonymy detected in OWGB Marrakech and a relatively higher number of Syrian accessions pairs displayed closely related genotypes (Supplementary file, Table S3). Beyond the fact that these related genotypes may be derived from local selection with a narrow genetic base, these observations could be explained by the scattered and partial olive characterisation (Baldoni et al. 2009). However, it is also very likely that mutations on SSR alleles have led to the distinction of very similar profiles for accessions belonging to the same original genotype, particularly when a clone is vegetatively multiplicated for a very long time (Khadari et al. 2008; Lopes et al. 2004). Even ramets of the same individual have been distinguished in relict Laperrine’s olive populations, demonstrating that SSR mutations can be relatively frequent under some environmental conditions (Baali-Cherif and Besnard 2005). One can also expect that the oldest cultivars have accumulated more mutations than more recently selected cultivars, and the occurrence of ancient clones in the Near East (e.g. Zohary and Spiegel-Roy 1975) could thus explain our observations. Furthermore, some accessions from different countries were classified under the same multilocus SSR profile, e.g. the “Zmj1” accession from Morocco and the “Zabarka” accession from Croatia. Such cases of cultivar identity from different origins might be related to their dissemination to different cropping areas where growers might have given them different local names (Trujillo et al. 1995; Besnard et al. 2001b; Sarri et al. 2006). In olive germplasm collections, over the last two decades, substantial efforts have been focused on identifying these redundant accessions using morphological and molecular data (Belaj et al. 2004; Barranco et al. 2005).
Mediterranean olive germplasm is structured into three main gene pools
Our study showed that Mediterranean olive germplasm was structured into three main gene pools, which strongly matched three distinct geographic areas, i.e. western, central and eastern Mediterranean regions. First, we showed that K = 3 was the best genetic structure model, as also reported by Baldoni et al. (2009) and Sarri et al. (2006). Second, on the basis of this genetic structure, most accessions clearly clustered according to their geographic origin. Numerous genetic studies have reported genetic differentiation between western and eastern Mediterranean areas (Besnard et al. 2002, 2007; Breton et al. 2006; Sarri et al. 2006). Furthermore, Mediterranean cultivars analysed by RAPD markers showed relative differentiation among Spanish and Italian varieties (Besnard et al. 2001a), and a clear distinction between Spanish varieties and those from Greece and Turkey (Owen et al. 2005). Such a genetic structure indicates a correlation between the genetic differentiation of olive trees and their geographic distribution. Despite this genetic structure, we observed dominance of the eastern maternal lineage (95% of E1 vs. only 5% of western maternal lineages E2 and E3). As previously shown in Mediterranean wild and domesticated olives (Besnard et al. 2002, 2007; Breton et al. 2006), all eastern Mediterranean accessions carried E1, whereas lineages E2 and E3 were only observed in the western Mediterranean Basin, but with a relatively low frequency in cultivars (16%; Besnard et al. 2001a). These cpDNA lineages again confirm that cultivated olive has been selected from different gene pools from both eastern and western regions of the Mediterranean Basin (Besnard et al. 2001c).
A set of 67 accessions was sufficient to capture the allelic diversity
The purpose of core collections is to facilitate the use of germplasm by providing a set of accessions displaying the genetic diversity available in the larger collection (Brown 1989). Our approach was based exclusively on genetic criteria, by capturing most of the diversity in a small set of accessions (58) after the exclusion of alleles observed only once. This sampling could be extended to a larger set of 67 accessions to capture the total allelic diversity (210 alleles). When examining the cultivar composition in each of the 10 best core accessions, we noted that about 40 cultivars were in common, whereas the remaining were different in each core. These investigations indicated the need to include complementary criteria such as phenotypic, agronomic and adaptive traits, but also the cultivar value and their importance at historical, economic and sociocultural levels to ensure optimal management. Beyond this variability, it is also essential to assess genetic structures and pedigree relationships within the core collection used for genetic association studies and for identifying QTLs related to phenotypic traits (Barnaud et al. 2006; Aranzana et al. 2010).
Conclusion
The present study demonstrated that the OWGB Marrakech collection gives an accurate picture of Mediterranean olive germplasm diversity. We provided a molecular database that should facilitate management of this germplasm. In addition, core collections will also be very useful for developing new breeding strategies for adaptive and agronomic traits through genome-wide association studies (Myles et al. 2009).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Frequency distribution of genetic dissimilarity for all pairwise comparisons between 505 olive genotypes. (JPEG 137 kb)
Mean posterior probabilities, averaged across 10 runs, (a) LK, (b) LnK’ (c) LnK”, (d) ∆K. (JPEG 22 kb)
Acknowledgments
The authors would like to thank Dr. Ph. Chatelet for his kind remarks on the early version of the manuscript, and M. Latreille for her help in the genotyping. They also acknowledge the International Olive Oil Council and INRA Morocco for their contribution to founding and management of OWGB Marrakech. This study was supported by PRAD 08-01, Volubilis Ma-08-197, Agropolis Foundation FruitMed no. 0901-007. The PhD student, H. Haouane, was supported by fellowships from Erasmus-Averroes and PRAD 08-01.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Aranzana MJ, Abassi EK, Howad W, Arús P. Genetic variation, population structure and linkage disequilibrium in peach commercial varieties. BMC Genet. 2010;11:69. doi: 10.1186/1471-2156-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baali-Cherif D, Besnard G. High genetic diversity and clonal growth in relict populations of Olea europaea subsp. laperrinei (Oleaceae) from Hoggar, Algeria. Ann Bot. 2005;96:823–830. doi: 10.1093/aob/mci232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldoni L, Cultrera NG, Mariotti R, Ricciolini C, Arcioni S, Vendramin GG, Buonamici A, Porceddu A, Sarri V, Ojeda MA, Trujillo I, Rallo L, Belaj A, Perri E, Salimonti A, Muzzalupo I, Casagrande A, Lain O, Messina R, Testolin R. A consensus list of microsatellite markers for olive genotyping. Mol Breed. 2009;24:213–231. doi: 10.1007/s11032-009-9285-8. [DOI] [Google Scholar]
- Banilas G, Minas J, Gregoriu C, Demoliou C, Kourti A, Hatzopoulos P. Genetic diversity among accessions of an ancient olive variety of Cyprus. Genome. 2003;46:370–376. doi: 10.1139/g03-011. [DOI] [PubMed] [Google Scholar]
- Barnaud A, Lacombe T, Doligez A. Linkage disequilibrium in cultivated grapevine, Vitis vinifera L. Theor Appl Genet. 2006;112:708–716. doi: 10.1007/s00122-005-0174-1. [DOI] [PubMed] [Google Scholar]
- Barranco D, Rallo L. Olive cultivars in Spain. HortTechnology. 2000;10:107–110. [Google Scholar]
- Barranco D, Trujillo I, Rallo L (2005) Libro I. Elaiografia hispanica. In: Rallo L, Barranco D, Caballero JM, Del Río C, Martín A, Tous J, Trujillo I (eds) Variedades del olivo en España. Junta de Andalucia, MAPA y Ediciones Mundi-Prensa, Madrid, pp 45–229
- Bartolini G (2008) Olive germplasm (Olea europaea L.). Available at: http://www.oleadb.it/olivodb.html
- Bartolini G, Prevost G, Messeri C, Carignani C, Menini UG. Olive germplasm: cultivars and world-wide collections. Rome, Italy: FAO; 1998. [Google Scholar]
- Belaj A, Satovic Z, Cipriani G, Baldoni L, Testolin R, Rallo L, Trujillo I. Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theor Appl Genet. 2003;107:736–744. doi: 10.1007/s00122-003-1301-5. [DOI] [PubMed] [Google Scholar]
- Belaj A, Cipriani G, Testolin R, Rallo L, Trujillo I. Characterization and identification of the main Spanish and Italian olive cultivars by simple-sequence-repeat markers. HortScience. 2004;39:1557–1561. [Google Scholar]
- Belaj A, Muñoz-Diez C, Baldoni L, Satovic Z, Barranco D. Genetic diversity and relationships of wild and cultivated olives at regional level in Spain. Sci Hortic. 2010;124:323–330. doi: 10.1016/j.scienta.2010.01.010. [DOI] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows™ pour la génétique des populations. Laboratoire Génome, Populations, Interactions, Centre National de la Recherche Scientifique, Unité Mixte de Recherche 5000. Montpellier, France: Université de Montpellier II; 2004. [Google Scholar]
- Besnard G, Baradat P, Bervillé A. Genetic relationships in the olive (Olea europaea L.) reflect multilocal selection of cultivars. Theor Appl Genet. 2001;102:251–258. doi: 10.1007/s001220051642. [DOI] [Google Scholar]
- Besnard G, Breton C, Baradat P, Khadari B, Bervillé A. Cultivar identification in the olive (Olea europaea L.) based on RAPDS. J Am Soc Hortic Sci. 2001;126:668–675. [Google Scholar]
- Besnard G, Baradat P, Breton C, Khadari B, Bervillé A. Olive domestication from structure of oleasters and cultivars using nuclear RAPDs and mitochondrial RFLPs. Genet Sel Evol. 2001;33:S251–S268. [Google Scholar]
- Besnard G, Khadari B, Baradat P, Bervillé A. Olea europaea (Oleaceae) phylogeography based on chloroplast DNA polymorphism. Theor Appl Genet. 2002;104:1353–1361. doi: 10.1007/s00122-001-0832-x. [DOI] [PubMed] [Google Scholar]
- Besnard G, Rubio de Casas R, Vargas P. A set of primers for length and nucleotide-substitution polymorphism in chloroplastic DNA of Olea europaea L. (Oleaceae) Mol Ecol Notes. 2003;3:651–653. doi: 10.1046/j.1471-8286.2003.00547.x. [DOI] [Google Scholar]
- Besnard G, Rubio de Casas R, Vargas P. Plastid and nuclear DNA polymorphism reveals historical processes of isolation and reticulation in the olive tree complex (Olea europaea L.) J Biogeogr. 2007;34:736–752. doi: 10.1111/j.1365-2699.2006.01653.x. [DOI] [Google Scholar]
- Breton C, Tersac M, Bervillé A. SSR genetic diversity in wild olive (oleaster, Olea europaea L.) suggests several Plio-Pleistocene refuge zones in the Mediterranean basin and gene flow with olive. J Biogeogr. 2006;33:1916–1928. doi: 10.1111/j.1365-2699.2006.01544.x. [DOI] [Google Scholar]
- Brown AHD. The case for core collection. Genome. 1989;31:818–824. doi: 10.1139/g89-144. [DOI] [Google Scholar]
- Caballero JM, del Río C. Caracterización del banco de germoplasma de olivo por caracteres agronomicos. Procedimientos de Jornadas de Investigación y Transferencía de Technología al Sector Oleicola, Programa de Mejora de la Calidad de la Producción de Aceite de Oliva. España: Córdoba; 2002. pp. 13–15. [Google Scholar]
- Caballero JM, Del Río C, Barranco D, Trujillo I. The olive world germplasm of Córdoba, Spain. Olea. 2006;25:14–19. [Google Scholar]
- Cantini C, Cimato A, Sani G. Morphological evaluation of olive germplasm present in Tuscany region. Euphytica. 1999;109:173–181. doi: 10.1023/A:1003728800464. [DOI] [Google Scholar]
- Carriero F, Fontanazza G, Cellini F, Giorio G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.) Theor Appl Genet. 2002;104:301–307. doi: 10.1007/s001220100691. [DOI] [PubMed] [Google Scholar]
- Carrión Y, Ntinou M, Badal E. Olea europaea L. in the North Mediterranean Basin during the Pleniglacial and the early-middle holocene. Quat Sci Rev. 2010;29:952–968. doi: 10.1016/j.quascirev.2009.12.015. [DOI] [Google Scholar]
- Cipriani G, Marrazzo MT, Marconi R, Cimato A, Testolin R. Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theor Appl Genet. 2002;104:223–228. doi: 10.1007/s001220100685. [DOI] [PubMed] [Google Scholar]
- de La Rosa R, James CM, Tobutt KR. Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Mol Ecol Notes. 2002;2:265–267. doi: 10.1046/j.1471-8286.2002.00217.x. [DOI] [Google Scholar]
- Escribano P, Viruel MA, Hormaza JI. Comparison of different methods to construct a core germplasm collection in woody perennial species with simple sequence repeat markers. A case study in cherimoya (Annona cherimola, Annonaceae), an underutilised subtropical fruit tree species. Ann Appl Biol. 2008;153:25–32. doi: 10.1111/j.1744-7348.2008.00232.x. [DOI] [Google Scholar]
- Engels JMM, Visser L (eds) (2003) A guide to effective management of germplasm collections. IPGRI (ITA), Rome, pp 157–161
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Frankel OH, Brown AHD (1984) Plant genetic resources today: a critical appraisal. Crop genetic resources. In: Holden JHW, Williams JT (eds) Conservation and evaluation. Georges Allen & Unwin Ltd, London, pp 249–257
- Gemas VJV, Almadanim MC, Tenreiro R, Martins A, Fevereiro P. Genetic diversity in the Olive tree (Olea europaea L. subsp. europaea) cultivated in Portugal revealed by RAPD and ISSR markers. Genet Resourc Crop Evol. 2004;51:501–511. doi: 10.1023/B:GRES.0000024152.16021.40. [DOI] [Google Scholar]
- Gouesnard B, Bataillon TM, Decoux G, Rozale C, Schoen DJ, David JL. MSTRAT: an algorithm for building germplasm core collections by maximizing allelic or phenotypic richness. J Hered. 2001;92:93–94. doi: 10.1093/jhered/92.1.93. [DOI] [PubMed] [Google Scholar]
- International Olive Council (IOC) (2007) Olive oil production and consumption in the season 2006/07. Available at: www.internationaloliveoil.org/downloads/
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Khadari B, Charafi J, Moukhli A, Ater M. Substantial genetic diversity in cultivated Moroccan olive despite a single major cultivar: a paradoxical situation evidenced by the use of SSR loci. Tree Genet Genomes. 2008;4:213–221. doi: 10.1007/s11295-007-0102-4. [DOI] [Google Scholar]
- Le Cunff L, Fournier-Level A, Laucou V, Vezzulli S, Lacombe T, Adam-Blondon AF, Boursiquot JM, This P (2008) Construction of nested genetic core collections to optimize the exploitation of natural diversity in Vitis vinifera L. subsp. sativa. BMC Plant Biol 8:31 [DOI] [PMC free article] [PubMed]
- Lopes MS, Mendonça D, Sefc KM, Sabino GF, da Câmara Machado A. Genetic evidence of intra-cultivar variability within Iberian olive cultivars. Hort Science. 2004;39:1562–1565. [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang ZW, Cotich DE, Buckler ES. Association mapping: critical considerations shift from genotyping to expiremental design. Plant Cell. 2009;21:2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M (1987) Molecular evolutionary genetics. Columbia University Press, Columbia
- Owen CA, Bita EC, Banilas G, Hajjar SE, Sellianakis V, Aksoy U, Hepaksoy S, Chamoun R, Talhook S, Metzidakis I, Hatzopoulos P, Kalaitzis P. AFLP reveals structural details of genetic diversity within cultivated olive germplasm from the Eastern Mediterranean. Theor Appl Genet. 2005;110:1169–1176. doi: 10.1007/s00122-004-1861-z. [DOI] [PubMed] [Google Scholar]
- Paetkau D, Calvert W, Stirling I, Strobeck C. Microsatellites analysis of population structure in Canadian polar bears. Mol Ecol. 1995;4:347–354. doi: 10.1111/j.1365-294X.1995.tb00227.x. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6: genetic analysis in Excel. population genetic software for teaching and research. Mol Ecol Notes. 2005;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population. Mol Ecol Notes. 2004;4:137–138. doi: 10.1046/j.1471-8286.2003.00566.x. [DOI] [Google Scholar]
- Sarri V, Baldoni L, Porceddu A, Cultrera NGM, Contento A, Frediani M, Belaj A, Trujillo I, Cionini PG. Microsatellite markers are powerful tools for discriminating among olive cultivars and assigning them to geographically defined populations. Genome. 2006;49:1606–1615. doi: 10.1139/g06-126. [DOI] [PubMed] [Google Scholar]
- Schoen DJ, Brown AHD. Conservation of allelic richness in wild crop relatives is aided by assessment of genetic markers. Proc Natl Acad Sci USA. 1993;90:10623–10627. doi: 10.1073/pnas.90.22.10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefc KM, Lopes MS, Mendonça D, Rodrigues Dos Santos M, Da Cámara Machado L. Identification of microsatellites loci in Olive (Olea europaea L.) and their characterization in Italian and Iberian trees. Mol Ecol. 2000;9:171–1193. doi: 10.1046/j.1365-294x.2000.00954.x. [DOI] [PubMed] [Google Scholar]
- Szpiech ZA, Jakobsson M, Rosenberg NA. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics. 2008;24:2498–2504. doi: 10.1093/bioinformatics/btn478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terral JF, Alonso N, Capdevila RBI, Chatti N, Fabre L, Fiorentino G, Marinval P, Jordá GP, Pradat B, Rovira N, Alibert P. Historical biogeography of olive domestication (Olea europaea L.) as revealed by geometrical morphometry applied to biological and archaeological material. J Biogeogr. 2004;31:63–77. doi: 10.1046/j.0305-0270.2003.01019.x. [DOI] [Google Scholar]
- Tessier C, David J, This P, Boursiquot JM, Charrier A. Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theor Appl Genet. 1999;98:171–177. doi: 10.1007/s001220051054. [DOI] [Google Scholar]
- Trujillo I, Rallo L, Arús P. Identifying olive cultivars by isozyme analysis. J Am Soc Hortic Sci. 1995;120:318–324. [Google Scholar]
- Wagner HW, Sefc KM (1999) Identity 1.0. Centre for applied genetics. University of Agricultural Sciences, Vienna
- Zohary D, Spiegel-Roy P. Beginnings of fruit growing in the Old World. Science. 1975;187:319–327. doi: 10.1126/science.187.4174.319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frequency distribution of genetic dissimilarity for all pairwise comparisons between 505 olive genotypes. (JPEG 137 kb)
Mean posterior probabilities, averaged across 10 runs, (a) LK, (b) LnK’ (c) LnK”, (d) ∆K. (JPEG 22 kb)