Abstract
In spite of the well-established understanding of the phenotypic lesions occurring in the shift from native epithelia to invasive (adeno) carcinoma, the molecular typing of the precancerous changes in the gastrointestinal tract remains unreliable. In recent years, no biomarkers have aroused as much interest as the miRNAs, a class of non-coding RNA molecules that function as endogenous silencers of numerous target genes. Aberrant miRNA expression is a hallmark of human disease, including cancer. Unlike most mRNAs, miRNAs are both long-living in vivo and very stable in vitro. Such characteristics allow their testing in paraffin-embedded tissue samples, which is essential in the biological profiling of small (phenotypically characterized) preneoplastic lesions of the gastrointestinal tract (as well as in other fields of human pathology). The upcoming challenge lies in the reliable identification of disease-specific targets of dysregulated miRNAs, to enable miRNA testing in the clinical management of the secondary prevention of gastrointestinal cancer.
Keywords: miRNA, Dysplasia, Barrett’s esophagus, Atrophic gastritis, Inflammatory bowel disease
INTRODUCTION
The molecular mechanisms involved in the multistep processes of gastrointestinal (GI) cancers are one of the most intriguing research fields in pathology.
In spite of our well-established understanding of the phenotypic lesions occurring in the shift from native epithelia to invasive (adeno)carcinoma, the molecular typing of the precancerous changes in the GI tract remains unreliable due to: (1) discrepancies in their histological classification; (2) their inherent biological heterogeneity; and (3) their variability on molecular biology testing. As a result, no reliable molecular data are currently available that can be implemented with confidence in GI cancer secondary prevention strategies[1,2].
Molecular profiling, as achievable using well-characterized histology samples, would ideally be done using formalin-fixed, paraffin-embedded (FF-PE) tissue samples. Unfortunately, molecular profiling from FF-PE samples is patchy, which makes it difficult to integrate molecular data (gene expression arrays, among others) with histological information consistently[3-5].
Hence the priority to overcome this methodological impasse. Despite efforts made by the scientific community to identify biomarkers for human cancer, no such biomarkers have aroused as much interest as the miRNAs, a class of endogenous, small, non-coding RNAs that modulate gene expression by causing target mRNA degradation or inhibiting their translation[6-10]. Since their initial discovery in Caenorhabditis elegans (C. elegans) in 1993[11], an enormous amount of research has been published, indicating that the biological function of miRNAs is crucial to most cellular processes. In humans, aberrant miRNA expression is a hallmark of various diseases, including cancer[9,10]. Unlike most mRNAs (due to their molecular structure), miRNAs are long-living in vivo and very stable in vitro[3-5]. These particular features are fundamental to their analysis in FF-PE samples, supporting a potentially central role for miRNAs in the molecular study of preneoplastic GI lesions.
Focusing on the similar, multistep carcinogenic cascade occurring in both esophageal and gastric adenocarcinomas, and on the colorectal carcinogenic processes, this Editorial briefly discusses the role of miRNAs in preneoplastic GI pathology.
miRNAs AS DIAGNOSTIC TOOLS
miRNA expression profiling has the potential for differentiating between normal and pathological lesions, and among preneoplastic lesions, it may be able to classify different subtypes[9,10].
Several reports have already demonstrated the excellent reproducibility and accuracy of miRNA expression profiling in archived FF-PE specimens. In gastroenterology, as in other fields of human pathology, integrating genome-wide profiling with the functional characterization of miRNAs (their overexpression or downregulation) and the identification of miRNA-specific gene targets currently represents the approach most likely to yield advances in the new field of non-coding RNA research[6-8,12].
In FF-PE specimens, miRNA expression could also be visualized at cellular/sub-cellular level (in situ hybridization) and this particular characteristic makes miRNAs potentially suitable for supporting routine diagnostic surgical pathology practice[13,14].
Aberrant miRNA expression signatures have been extensively investigated in preneoplastic GI diseases and several key oncogenic miRNAs have consistently been found dysregulated (Table 1)[6-8,12,15-21]. In some cases, specific miRNA expressions have been linked to cancer-associated pathways, indicating a role for them in GI carcinogenesis. The systematic molecular evaluation of the GI mucosa (always supported by the “advanced” histological and clinical characterization of the specimens) not only provides new basic biological information, but can also pave the way to risk-stratified patient management programs and innovative therapeutic measures (Figure 1).
Table 1.
miRNAs associated with different histological lesions of the gastrointestinal tract
| Organ | Pathology | Overexpressed miRNAs | Downregulated miRNAs | Ref. |
| Esophagus | Barrett’s mucosa | miR-15b; miR-21; miR-25; miR-143; miR-145; miR-192; miR-194; miR-196a; miR-200c; miR-215 | let-7a; let-7c; miR-125b; miR-203; miR-205; miR-486-5p | [30,31-33,35, 37] |
| LG-IEN | miR-192; miR-196a; miR-215 | let-7c; miR-203; miR-205 | [32,35] | |
| HG-IEN | miR-15b; miR-21; miR-125b; miR-192;miR-196a; miR-200a*; miR-215 | let-7a; let-7c; miR-181b; miR-193b; miR-203; miR-205; miR-486-5p | [30,31-33,35, 36] | |
| Neosquamous epithelium | miR-143 | - | [38] | |
| Stomach | H. pylori-related gastritis | miR-21; miR-146a; miR-155; miR-223 | let-7f; miR-34b; miR-34c; miR-124a-1; miR-124a-2; miR-124a-3; miR-141; miR-200a; miR-203; miR-204; miR-455 | [43,45,47,49, 50,53,56,58] |
| Atrophic gastritis | - | let-7a | [55] | |
| Colon | Traditional adenoma | miR-21; miR-135 | miR-143; miR-145 | [65] |
| Serrated adenoma | miR-21; miR-181b | - | [70,71] | |
| Hyperplastic polyp | miR-181b | - | [71] | |
| Crohn's disease | miR-9; miR-9*; miR-21; miR-22; miR-23b; miR-26a; miR-29b; miR-29c; miR-30a; miR-30b; miR-30c; miR-31; miR-34c-5p; miR-106a; miR-126; miR-126*; miR-127-3p; miR-130a; miR-133b; miR-146a; miR-146b-5p; miR-150; miR-155; miR-181c miR-191; miR-196a; miR-223; miR-324-3p; miR-375 | miR-19b; miR-629 | [75,79] | |
| Ulcerative colitis | let-7f; mIR-7; miR-16; miR-21; miR-23a; miR-24; miR-29a; miR-29b; miR-31; miR-126; miR-126*; miR-127-3p; miR-135b; miR-223; miR-324-3p; miR-195; miR-196 | miR-188-5p; miR-192; miR-215; miR-320a; miR-346; miR-375; miR-422b | [75,79] | |
| IBD dysplasia | miR-31 | - | [80] |
LG-IEN: Low-grade-intraepithelial neoplastic lesions; HG-IEN: High-grade-intraepithelial neoplastic lesions; H. pylori: Helicobacter pylori; IBD: Inflammatory bowel disease.
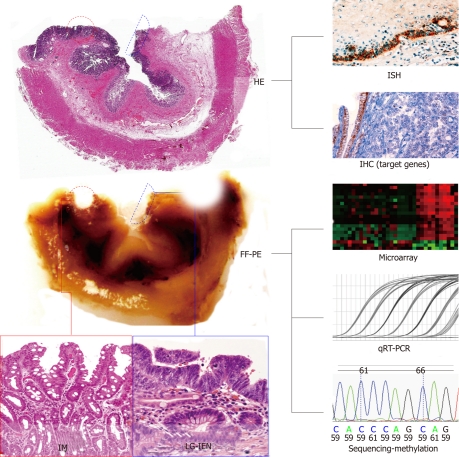
Figure 1.
miRNA analysis in formalin-fixed, paraffin-embedded samples. Routine histological analysis (hematoxylin and eosin) enables adequate selection of preneoplastic lesions (in this case, intestinal metaplasia and low-grade-intraepithelial neoplastic lesions obtained from a resected Barrett’s adenocarcinoma specimen). Either miRNA-specific in situ hybridization or immunohistochemical analysis of the miRNA gene targets could be performed starting from the original paraffin block. To obtain total RNA or DNA, lesions are macro-dissected from the paraffin block (or micro-dissected from unstained formalin-fixed, paraffin-embedded sections). The genome material can be further analyzed using modern genomic techniques. HE: Hematoxylin and eosin; FF-PE: Formalin-fixed, paraffin-embedded; IM: Intestinal metaplasia; LG-IEN: Low-grade-intraepithelial neoplastic lesions; ISH: in situ hybridization; IHC: Immunohistochemistry; qRT-PCR: Quantitative reverse transcription-polymerase chain reaction.
Therefore, miRNAs represent both potential cancer prognostic markers and therapeutic targets for the treatment of human malignancies. Further understanding of the biological roles of miRNAs in cancer is required, particularly the experimental validation of miRNA targets.
ONCOGENIC CASCADES IN GASTROESOPHAGEAL CARCINOMA
In the last 25 years, it has been demonstrated that full-blown gastroesophageal cancers are the final outcome of long-standing biological processes that include a progressive accumulation of genotypic and phenotypic changes triggered by persistent inflammatory conditions, i.e., chronic acid/bile reflux in the esophagus and longstanding gastritis, due primarily to Helicobacter pylori (H. pylori) infection, in the stomach[1,2].
Such natural histories have been described as Barrett’s model of carcinogenesis for esophageal adenocarcinoma and Pelayo Correa’s multistage cascade for gastric adenocarcinoma[1,2].
At first, both conditions lead to the replacement of native by metaplastic epithelia [columnar epithelia in Barrett’s esophagus (BE) and intestinal metaplasia of the native gastric glands][1,2]. The phenotypic shift involved in these processes is due to the combined effect of changes in the expression of genetic factors, epigenetic silencing, transcription factors, signaling pathways and growth factors. None of the biological mechanisms underlying the metaplastic transformation have been clarified in detail as yet, however, and genome-wide microarray expression analysis seems to be the most promising line of investigation.
There are significant differences in the pathogenetic, morphological (histochemical and immunohistochemical) and clinical impact of metaplasia in the two above-mentioned anatomical settings. Intestinal metaplasia (IM), however, is known as the “carcinogenic field” in which both esophageal and gastric intraepithelial neoplastic lesions (IENs), be it low-grade-IENs or high-grade (HG)-IENs and subsequent adenocarcinomas may develop[22-27].
The Barrett’s cascade
BE is characterized by the native stratified squamous epithelium lining the esophagus being replaced by a columnar epithelium with intestinal differentiation [Barrett’s mucosa (BM)][2]. From the clinical point of view, only a histological diagnosis of IEN is currently considered in the definition of high-risk BE populations, confirming the need for prognostically useful, novel molecular biomarkers[28]. In this scenario, several reports have investigated miRNA expression profiling in Barrett’s carcinogenesis[15,29-33].
In their seminal work, Feber et al[29] investigated miRNA expression profiling in a small series of frozen specimens (nine native squamous esophageal mucosa, five BM, one HG-IEN, and 10 Barrett’s adenocarcinoma). Among the miRNA expression signatures observed, miR-203 and miR-205 were downregulated and miR-21, miR-192 and miR-194 were overexpressed in esophageal adenocarcinoma samples[29]. It is important to bear in mind that miR-194 is involved in the lineage commitment decisions of the intestinal epithelium system, with a major role in the maturation of the intestinal epithelium[34]. The authors also considered 10 squamous cell carcinoma specimens in their analysis, showing that specific miRNA expression profiles correspond to specific esophageal tumor histotypes[29].
Two further papers with consistent results focused on the miRNA expression signatures associated with disease progression[30,31]. Our group provided comprehensive information on the miRNA profile coupled with each single step in the natural history of Barrett’s carcinogenesis[32]. To confirm our microarray and quantitative RT-PCR data, we also performed miRNA-specific in situ hybridization analyses on FF-PE tissues and identified HMGA2 (a let-7c mRNA target, which is a small, non-histone chromosomal protein that can modulate transcription by altering chromatin architecture) as a promising biomarker of BM transformation on immunohistochemical analysis. Based on these microarray findings, Bansal et al[33] recently have investigated the prognostic impact of miRNA expression in determining BE progression, demonstrating that miRNAs could pinpoint high-risk BE patients with a reasonable clinical accuracy.
In exploring the molecular mechanisms implicated in BM transformation and progression, Maru et al[35] have particularly focused on the role of miR-196a. As previously demonstrated by the same research group, miR-196a is overexpressed in neoplastic samples, and its expression levels increase proportionally with the dedifferentiation of the IEN[35]. Moreover, they have shown that miR-196a plays a part in the downregulation of the SPRR2C, S100A9 and KRT5 genes, the expression of which is characteristically decreased or lost during the neoplastic transformation of esophageal tissue[35]. These findings strongly support a role for miR-196a as a novel therapeutic target in the treatment of esophageal cancers and as a valuable early marker of cancer-prone BE.
Another important oncomiR is miR-21, which is upregulated in various human tumors, and has been found to be involved in Barrett’s adenocarcinoma[29,31]. Recent studies have shown that miR-21 promotes cell transformation by repressing tumor suppressor genes such as PTEN, PDCD4, RECK and TPM1[3]. Our group has demonstrated significant miR-21 upregulation in samples of HG-IENs and adenocarcinoma, consistent with PDCD4 downregulation[36].
The dysregulation of not just one, but multiple miRNAs could be implicated in cancer progression. miRNAs could be organized as a cluster of genes expressed by a single transcription unit (i.e., polycistron), which goes to show how complex miRNA research may be. The miR-106b-25 polycistron on chromosome 7q22.1 (i.e., miR-25, miR-93 and miR-106b) has been found to be increasingly activated in successive stages of Barrett’s carcinogenesis, with potentially proliferative, antiapoptotic, and cell cycle promoting effects in vitro and tumorigenic effects in vivo by targeting p21 and Bim[37].
From the therapeutic standpoint, argon plasma coagulation (thermal ablation) is one of the options for the surgical treatment of BE patients. BM ablation usually eventually results in the formation of neosquamous epithelium. IM can recur, however (so cancer could subsequently develop too) even after apparently complete BM ablation. Biomarkers might therefore be helpful in clinical decision-making for BE patients, by providing information on the likely clinical behavior of the mucosa after ablative therapy. That is why Dijckmeester et al[38] investigated miR-143 and miR-205 expression in neosquamous epithelium and BM. Only miR-143 expression was significantly higher in neosquamous and native squamous esophageal mucosa than in samples of BM[38]. It is worth adding that miR-143 is highly expressed in colonic tissues and has a significant role in suppressing colorectal cancer cell growth by inhibiting KRAS translation[39].
Different genetic polymorphisms could be implicated in miRNA-related carcinogenesis. The biogenesis of miRNAs is complex and involves multiple proteins and RNAs. Although miRNA expression profiles have frequently been reported to correlate with the etiology, classification, progression and prognosis of numerous human cancers, the effect of common genetic variants [i.e., single nucleotide polymorphisms (SNP)] of miRNA-related genes on susceptibility to cancer remains unclear[9,10]. Ye et al[40] have demonstrated that seven SNPs were significantly associated with the risk of esophageal adenocarcinoma, pointing to intriguing new fields of translational research.
Correa’s cascade
Despite a steady decline in the related mortality in the past few decades, gastric cancer is still the second cause of death due to cancer worldwide[41]. H. pylori-associated gastritis is the most common gastric adenocarcinoma precursor (leading to intestinal-type, or the so-called “epidemic”gastric cancer)[42].
Matsushima et al[43] have investigated the miRNA expression profiling of H. pylori-positive mucosa samples, finding 31 significantly dysregulated miRNAs. The severity of both active and chronic inflammatory infiltration significantly correlated with the expression of several miRNAs, supporting a biological role for miRNAs in host immune response to H. pylori infection[44]. Among others, miR-155 has been suggested as a central effector of H. pylori-induced immune response: H. pylori infection raises miR-155 expression levels in gastric epithelial cell lines and gastric mucosal tissues by activating nuclear factor-κB and the activator protein-1 pathway, and via the Foxp3 transcription factor in T cells[45,46]. The overexpressed miR-155 could also negatively modulate the release of the proinflammatory cytokines interleukin-8 and GRO-a, leading to chronic H. pylori-related infection[47].
Another important H. pylori-induced miRNA is miR-146a, which seems to have a role in a negative feedback loop that modulates the inflammatory damage by targeting IRAK1 and TRAF6[47]. Two common SNPs in its pre-miRNA sequence (rs2910164 and rs11614913) have also been associated with a greater susceptibility to gastric cancer in the Chinese and Japanese populations[48].
Among healthy volunteers, individuals with H. pylori infection show higher methylation levels of miR-34b, miR-34c, miR-124a-1, miR-124a-2 and miR-124a-3[49,50]. These data strongly suggest that H. pylori infection (in addition to protein-coding genes) induces DNA methylation of miRNA genes, also indicating that miRNA silencing is an early event in the oncogenic process and the result of a global epigenetic field defect.
Another important miRNA that is dysregulated in chronic gastritis is miR-27a. Arisawa et al[51] have shown a close correlation between miR-27a genome polymorphism and the onset of advanced gastric mucosa atrophy in Japanese men, suggesting a definitive role for miR-27a in the development of the “cancerization field. These findings are further supported by the fact that miR-27a has been found upregulated in gastric cancer samples; it correlates with gastric cancer lymph node metastasis; and functions as an oncogene by targeting the tumor suppressor gene prohibitin[52].
miR-21 seems to play a fundamental part in gastric carcinogenesis, being constantly upregulated in gastric adenocarcinoma (as already demonstrated in others miscellaneous solid tumors, as well as Barrett’s adenocarcinoma)[53]. In vitro, miR-21 upregulation has been shown to enhance significantly migration and capacity for invasion of gastric cancer cell lines[53]. On the other hand, miR-21 knockdown causes a significant reduction in cell proliferation and a significant increase in apoptosis[53]. miR-21 is also significantly overexpressed in H. pylori-infected gastric mucosa, suggesting an early, important involvement of miR-21 in the development of gastric cancer[53].
The tumor suppressor let-7a is one of the founder members of the miRNA family, which was first identified in C. elegans. In gastric cancer, let-7a is downregulated and negatively regulates HMGA2 expression (like let-7c)[54,55]. let-7a downregulation has already been identified in chronic atrophic gastritis samples[56].
The oncogenic miR-106a, a member of the miR-106a-92 cluster, has been found upregulated in gastric cancer tissues and it has been significantly associated with negative clinicopathological parameters[56,57]. The miR-106a-92 cluster has a marked gene structure homology with other miRNA clusters recognized as being oncogenic, i.e., miR-17-92 and miR-106b-25. On this point, Petrocca et al[58] have observed dysregulation of the miR-106b-25 cluster (i.e., miR-106b, miR-92 and miR-25) determined by transcription factor E2F1 expression in H. pylori-related gastric carcinogenesis and gastric cancer cell lines. miR-206b and miR-93 directly regulate E2F1 expression, establishing an miRNA-related negative feedback loop[58]. The upregulation of these miRNAs interferes with the transforming growth factor (TGF)-β tumor suppressor pathway, hindering the expression of CDKN1A and BCL2L11[58]. TGF-β controls the turnover of intestinal, as well as gastric, cells leading them to cell cycle arrest followed by apoptosis. By downregulating two of the most important downstream effectors in the TGF-β pathway, the miR-106b-25 cluster ensures the development of a resistance to TGF-β-mediated cell cycle arrest and apoptosis, and may represent an interesting novel therapeutic target for the treatment of gastric cancer. In the same study, the authors identified a seven-miRNA signature associated with chronic H. pylori-related gastritis, which included the above-mentioned miR-155[58].
ONCOGENIC CASCADES IN COLORECTAL ADENOCARCINOMA
The historically well-established pathway of sporadic colorectal cancer is the so-called adenoma-carcinoma sequence, formally described by Morson in 1962[59]. Approximately 60% of sporadic colorectal cancers (CRCs) are consistent with this phenotypic sequence. The lesions included in this “cascade” are tubular, tubulovillous and villous adenomas (with different IEN grades)[60].
More recently, however, molecular genetic findings have highlighted two further pathways: (1) serrated carcinogenesis, in which the sessile serrated adenoma is considered the precursor lesion; and (2) the mixed-type sequence, combining the molecular features of both the traditional and the serrated pathways[60,61].
A fourth route is the one resulting from longstanding/relapsing inflammation associated with inflammatory bowel disease (IBD), Crohn’s disease (CD) and ulcerative colitis (UC)[62,63]. The first report of CRC in IBD came from Crohn and Rosenberg in 1925[64]. From a molecular point of view, the IBD-related carcinogenic process seems to follow a temporal sequence of genetic alterations different from the situation seen in sporadic cancers. The IBD-related CRC risk increases in long-standing colitis and parallels the anatomical extent and severity of the related inflammation. From the pathological standpoint, IBD-associated CRC can arise from: (1) raised dysplastic lesions or dysplasia-associated lesions or masses; and (2) endoscopically flat dysplastic lesions. In both cases, the lesions can blend easily with the gross inflammatory abnormalities commonly encountered in IBDs, making their endoscopic detection difficult even for the experienced endoscopist, resulting in the need for innovative dysplasia-specific biomarkers[61].
Sporadic colorectal cancer
A whole constellation of experimental studies on CRC has provided insight into the miRNA-mediated, regulatory links to well-known oncogenic and tumor suppressor signaling pathways[20,65-67].
Global miRNA downregulation is a common feature of colorectal carcinogenesis. As described previously for gastroesophageal diseases, CpG island hypermethylation has been described as a mechanism for miRNA silencing in CRC samples too. Balaguer et al[68] also have demonstrated high rates of miR-137 CpG island methylation in colorectal adenomas, suggesting that the epigenetic silencing of specific tumor-suppressor miRNAs is an early event in the multistep colorectal carcinoma cascade.
Several investigations on paired samples of colorectal neoplasia and normal mucosa have demonstrated reduced levels of miR-143 and miR-145 in both colonic adenoma and carcinoma[39]. These two miRNAs reveal a tumor suppressor-like activity in vitro by targeting KRAS (miR-143) and the insulin receptor substrate 1 (IRS-1; miR-145), among others[39].
Among the overexpressed miRNAs, miR-21 (an important regulator of PDCD4 expression) has frequently been found dysregulated in CRC samples[3,69]. Consistent with these data, we found that miR-21 was significantly upregulated in IEN and CRC biopsy samples, suggestive of a diagnostic role for this miRNA in discriminating between neoplastic and non-neoplastic lesions[70]. We observed a similar PDCD4 dysregulation in serrated adenomas too, suggesting a major oncogenic function for miR-21 that is acquired early in different pathways of colorectal carcinogenesis. Similar results were have been reported by Schmitz et al[71], who have found significant miR-21 overexpression in sessile serrated adenomas by comparison with both normal mucosa and hyperplastic polyps (though miR-21 expression alone failed to distinguish between the histological lesions considered).
Sequence variations in the miRNA-binding sites of CD86 and INSR have been associated with a higher risk of CRC (with ORs of 2.74 and 1.94, respectively)[72,73]. These data, integrated with genome-wide association studies supported by high-resolution SNP arrays and next-generation sequencing technology, could help clinicians to assess CRC genetic susceptibility more accurately, with the prospect of stratifying patients according to their “molecular” cancer risk.
Inflammatory bowel diseases
IBDs result from an abnormal immune response to environmental factors in genetically susceptible hosts. miRNAs have been increasingly recognized as important in the development of both the innate and the adaptive immune system, and dysregulated miRNA expression has already been described in several immune-related diseases. miRNA expression profiling in active CD and UC has been the object of several studies, which have consistently shown unique disease-specific miRNA signatures[74-77]. Supporting the role of miRNAs in immune system dysregulation, Wu et al[78] have demonstrated that miR-192 (which is significantly upregulated in UC) blocks tumor necrosis factor α-induced stimulation by targeting the chemotactic cytokine MIP-2α. Fasseu et al[79] have studied both active and inactive IBD samples, and have demonstrated that miRNAs are crucial players in the onset/relapse of active inflammation. In particular, they have found two subsets of 14 (UC) and 23 (CD) miRNAs with a significant dysregulation in comparison to healthy controls (Table 1), and among these, eight that were commonly dysregulated in non-inflammed UC and CD (miR-26a, miR-29a, miR-29b, miR-30c, miR-126*, miR-127-3p, miR-196a, and miR-324-3p)[79]. What is more, underlying the importance of miRNA dysregulation in IBD onset, several miRNA genes with altered expression co-localized with acknowledged IBD-susceptibility loci[79].
miR-31a has been indicated as a specific marker of IBD-related dysplasia[80]. It is overexpressed in both IBD-related dysplasia and IBD-associated CRC (and also significantly increased in sporadic CRC, although to a lesser degree than in IBD-related neoplasia). Preliminary studies have demonstrated the angiogenic potential of miR-31 via the targeting of FIH-1 in CRC-derived cell lines[80].
Beyond their histopathological applications, miRNAs offer a number of practical advantages due to their relatively high stability in vivo, and in the circulation in particular. In addition, they do not require the use of specific antibody-linked detection reagents of protein biomarkers, and they offer the specificity of nucleic acid detection methods such as RT-PCR. In IBD patients, differences in circulating immune cells in CD and UC are reflected by altered miRNA expression, supporting the usefulness of peripheral blood miRNA analysis as an innovative non-invasive class of biomarkers[81].
CONCLUSION
Preneoplastic lesions within the GI tract include a broad spectrum of phenotypic alterations, which may be difficult to assess only on the basis of their phenotypical features and are hard to stratify in different prognostic classes.
As this review shows, miRNAs and miRNA-related gene expression and polymorphism have a central role in assessing the individual (patient-specific) cancer susceptibility and cancer progression. The upcoming challenge lies in the reliable identification of disease-specific targets of dysregulated miRNAs, to enable miRNA testing in clinical practice. The miRNA revolution is only just beginning!
Footnotes
Supported by The AIRC grant Veneto Region 2009; the “G. Berlucchi” and “G.B. Morgagni” Foundations
Peer reviewer: Wendy Wilhelmina Johanna de Leng, PhD, Department of Pathology, Internal address H04.312, Heidelberglaan 100, Postbox 85500, Utrecht, 3508 GA, The Netherlands
S- Editor Tian L L- Editor Kerr C E- Editor Xiong L
References
- 1.Rugge M, Pennelli G, Pilozzi E, Fassan M, Ingravallo G, Russo VM, Di Mario F. Gastritis: the histology report. Dig Liver Dis. 2011;43 Suppl 4:S373–S384. doi: 10.1016/S1590-8658(11)60593-8. [DOI] [PubMed] [Google Scholar]
- 2.Fiocca R, Mastracci L, Milione M, Parente P, Savarino V. Microscopic esophagitis and Barrett’s esophagus: the histology report. Dig Liver Dis. 2011;43 Suppl 4:S319–S330. doi: 10.1016/S1590-8658(11)60588-4. [DOI] [PubMed] [Google Scholar]
- 3.Baffa R, Fassan M, Volinia S, O’Hara B, Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 4.Fassan M, Baffa R, Palazzo JP, Lloyd J, Crosariol M, Liu CG, Volinia S, Alder H, Rugge M, Croce CM, et al. MicroRNA expression profiling of male breast cancer. Breast Cancer Res. 2009;11:R58. doi: 10.1186/bcr2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehmann U. MicroRNA-profiling in formalin-fixed paraffin-embedded specimens. Methods Mol Biol. 2010;667:113–125. doi: 10.1007/978-1-60761-811-9_8. [DOI] [PubMed] [Google Scholar]
- 6.Visone R, Petrocca F, Croce CM. Micro-RNAs in gastrointestinal and liver disease. Gastroenterology. 2008;135:1866–1869. doi: 10.1053/j.gastro.2008.10.074. [DOI] [PubMed] [Google Scholar]
- 7.O’Hara SP, Mott JL, Splinter PL, Gores GJ, LaRusso NF. MicroRNAs: key modulators of posttranscriptional gene expression. Gastroenterology. 2009;136:17–25. doi: 10.1053/j.gastro.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albulescu R, Neagu M, Albulescu L, Tanase C. Tissular and soluble miRNAs for diagnostic and therapy improvement in digestive tract cancers. Expert Rev Mol Diagn. 2011;11:101–120. doi: 10.1586/erm.10.106. [DOI] [PubMed] [Google Scholar]
- 9.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 12.Liu W, Mao SY, Zhu WY. Impact of tiny miRNAs on cancers. World J Gastroenterol. 2007;13:497–502. doi: 10.3748/wjg.v13.i4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nuovo GJ. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods. 2008;44:39–46. doi: 10.1016/j.ymeth.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Nuovo GJ, Elton TS, Nana-Sinkam P, Volinia S, Croce CM, Schmittgen TD. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc. 2009;4:107–115. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kan T, Meltzer SJ. MicroRNAs in Barrett’s esophagus and esophageal adenocarcinoma. Curr Opin Pharmacol. 2009;9:727–732. doi: 10.1016/j.coph.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belair C, Darfeuille F, Staedel C. Helicobacter pylori and gastric cancer: possible role of microRNAs in this intimate relationship. Clin Microbiol Infect. 2009;15:806–812. doi: 10.1111/j.1469-0691.2009.02960.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Wang Q, Liu H, Hu B, Zhou W, Cheng Y. MicroRNA expression and its implication for the diagnosis and therapeutic strategies of gastric cancer. Cancer Lett. 2010;297:137–143. doi: 10.1016/j.canlet.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Smith CM, Watson DI, Michael MZ, Hussey DJ. MicroRNAs, development of Barrett’s esophagus, and progression to esophageal adenocarcinoma. World J Gastroenterol. 2010;16:531–537. doi: 10.3748/wjg.v16.i5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J, Sung JJ. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29:5761–5771. doi: 10.1038/onc.2010.352. [DOI] [PubMed] [Google Scholar]
- 20.Wu WK, Law PT, Lee CW, Cho CH, Fan D, Wu K, Yu J, Sung JJ. MicroRNA in colorectal cancer: from benchtop to bedside. Carcinogenesis. 2011;32:247–253. doi: 10.1093/carcin/bgq243. [DOI] [PubMed] [Google Scholar]
- 21.Dong Y, Wu WK, Wu CW, Sung JJ, Yu J, Ng SS. MicroRNA dysregulation in colorectal cancer: a clinical perspective. Br J Cancer. 2011;104:893–898. doi: 10.1038/bjc.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cassaro M, Rugge M, Tieppo C, Giacomelli L, Velo D, Nitti D, Farinati F. Indefinite for non-invasive neoplasia lesions in gastric intestinal metaplasia: the immunophenotype. J Clin Pathol. 2007;60:615–621. doi: 10.1136/jcp.2006.040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaninotto G, Minnei F, Guirroli E, Ceolin M, Battaglia G, Bellumat A, Betetto G, Bozzola L, Cassaro M, Cataudella G, et al. The Veneto Region’s Barrett’s Oesophagus Registry: aims, methods, preliminary results. Dig Liver Dis. 2007;39:18–25. doi: 10.1016/j.dld.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 24.Rugge M, Meggio A, Cataudella G, Parente P, Salvagnini G, de Pretis G, Zaninotto G. Barrett’s esophagus: still much to learn, but “Yes, we can!”. Am J Gastroenterol. 2008;103:2944–2946; author reply 2944-2946. doi: 10.1111/j.1572-0241.2008.02094_9.x. [DOI] [PubMed] [Google Scholar]
- 25.Rugge M, Fassan M, Battaglia G, Parente P, Zaninotto G, Ancona E. Intestinal or gastric? The unsolved dilemma of Barrett’s metaplasia. Hum Pathol. 2009;40:1206–1207; author reply 1206-1207. doi: 10.1016/j.humpath.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 26.Correa P, Piazuelo MB, Wilson KT. Pathology of gastric intestinal metaplasia: clinical implications. Am J Gastroenterol. 2010;105:493–498. doi: 10.1038/ajg.2009.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rugge M, de Boni M, Pennelli G, de Bona M, Giacomelli L, Fassan M, Basso D, Plebani M, Graham DY. Gastritis OLGA-staging and gastric cancer risk: a twelve-year clinico-pathological follow-up study. Aliment Pharmacol Ther. 2010;31:1104–1111. doi: 10.1111/j.1365-2036.2010.04277.x. [DOI] [PubMed] [Google Scholar]
- 28.Rugge M, Fassan M, Zaninotto G, Pizzi M, Giacomelli L, Battaglia G, Rizzetto C, Parente P, Ancona E. Aurora kinase A in Barrett’s carcinogenesis. Hum Pathol. 2010;41:1380–1386. doi: 10.1016/j.humpath.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–260; discussion 260. doi: 10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Gu J, Wang KK, Zhang W, Xing J, Chen Z, Ajani JA, Wu X. MicroRNA expression signatures in Barrett’s esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:5744–5752. doi: 10.1158/1078-0432.CCR-09-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ. MicroRNA profiling of Barrett’s oesophagus and oesophageal adenocarcinoma. Br J Surg. 2010;97:853–861. doi: 10.1002/bjs.7000. [DOI] [PubMed] [Google Scholar]
- 32.Fassan M, Volinia S, Palatini J, Pizzi M, Baffa R, De Bernard M, Battaglia G, Parente P, Croce CM, Zaninotto G, et al. MicroRNA expression profiling in human Barrett’s carcinogenesis. Int J Cancer. 2011;129:1661–1670. doi: 10.1002/ijc.25823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bansal A, Lee IH, Hong X, Anand V, Mathur SC, Gaddam S, Rastogi A, Wani SB, Gupta N, Visvanathan M, et al. Feasibility of mcroRNAs as biomarkers for Barrett’s Esophagus progression: a pilot cross-sectional, phase 2 biomarker study. Am J Gastroenterol. 2011;106:1055–1063. doi: 10.1038/ajg.2011.37. [DOI] [PubMed] [Google Scholar]
- 34.Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T, Watanabe M. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA. 2008;14:1433–1442. doi: 10.1261/rna.810208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maru DM, Singh RR, Hannah C, Albarracin CT, Li YX, Abraham R, Romans AM, Yao H, Luthra MG, Anandasabapathy S, et al. MicroRNA-196a is a potential marker of progression during Barrett’s metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–1948. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fassan M, Pizzi M, Battaglia G, Giacomelli L, Parente P, Bocus P, Ancona E, Rugge M. Programmed cell death 4 (PDCD4) expression during multistep Barrett’s carcinogenesis. J Clin Pathol. 2010;63:692–696. doi: 10.1136/jcp.2010.078253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dijckmeester WA, Wijnhoven BP, Watson DI, Leong MP, Michael MZ, Mayne GC, Bright T, Astill D, Hussey DJ. MicroRNA-143 and -205 expression in neosquamous esophageal epithelium following Argon plasma ablation of Barrett’s esophagus. J Gastrointest Surg. 2009;13:846–853. doi: 10.1007/s11605-009-0799-5. [DOI] [PubMed] [Google Scholar]
- 39.Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 40.Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, Wu X. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1:460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 42.Correa P, Houghton J. Carcinogenesis of Helicobacter pylori. Gastroenterology. 2007;133:659–672. doi: 10.1053/j.gastro.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 43.Matsushima K, Isomoto H, Inoue N, Nakayama T, Hayashi T, Nakayama M, Nakao K, Hirayama T, Kohno S. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer. 2011;128:361–370. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- 44.Tang B, Xiao B, Liu Z, Li N, Zhu ED, Li BS, Xie QH, Zhuang Y, Zou QM, Mao XH. Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation. FEBS Lett. 2010;584:1481–1486. doi: 10.1016/j.febslet.2010.02.063. [DOI] [PubMed] [Google Scholar]
- 45.Xiao B, Liu Z, Li BS, Tang B, Li W, Guo G, Shi Y, Wang F, Wu Y, Tong WD, et al. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J Infect Dis. 2009;200:916–925. doi: 10.1086/605443. [DOI] [PubMed] [Google Scholar]
- 46.Fassi Fehri L, Koch M, Belogolova E, Khalil H, Bolz C, Kalali B, Mollenkopf HJ, Beigier-Bompadre M, Karlas A, Schneider T, et al. Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PLoS One. 2010;5:e9500. doi: 10.1371/journal.pone.0009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z, Xiao B, Tang B, Li B, Li N, Zhu E, Guo G, Gu J, Zhuang Y, Liu X, et al. Up-regulated microRNA-146a negatively modulate Helicobacter pylori-induced inflammatory response in human gastric epithelial cells. Microbes Infect. 2010;12:854–863. doi: 10.1016/j.micinf.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Okubo M, Tahara T, Shibata T, Yamashita H, Nakamura M, Yoshioka D, Yonemura J, Ishizuka T, Arisawa T, Hirata I. Association between common genetic variants in pre-microRNAs and gastric cancer risk in Japanese population. Helicobacter. 2010;15:524–531. doi: 10.1111/j.1523-5378.2010.00806.x. [DOI] [PubMed] [Google Scholar]
- 49.Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, Sugiyama T, Ushijima T. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124:2367–2374. doi: 10.1002/ijc.24219. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki H, Yamamoto E, Nojima M, Kai M, Yamano HO, Yoshikawa K, Kimura T, Kudo T, Harada E, Sugai T, et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis. 2010;31:2066–2073. doi: 10.1093/carcin/bgq203. [DOI] [PubMed] [Google Scholar]
- 51.Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Hasegawa S, Takagi T, Wang FY, et al. A polymorphism of microRNA 27a genome region is associated with the development of gastric mucosal atrophy in Japanese male subjects. Dig Dis Sci. 2007;52:1691–1697. doi: 10.1007/s10620-006-9648-5. [DOI] [PubMed] [Google Scholar]
- 52.Liu T, Tang H, Lang Y, Liu M, Li X. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett. 2009;273:233–242. doi: 10.1016/j.canlet.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 54.Zhu YM, Zhong ZX, Liu ZM. Relationship between let-7a and gastric mucosa cancerization and its significance. World J Gastroenterol. 2010;16:3325–3329. doi: 10.3748/wjg.v16.i26.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang HH, Wang XJ, Li GX, Yang E, Yang NM. Detection of let-7a microRNA by real-time PCR in gastric carcinoma. World J Gastroenterol. 2007;13:2883–2888. doi: 10.3748/wjg.v13.i20.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L, Zhang WG, Nan KJ, Song TS, Huang C. MicroRNA profiling of human gastric cancer. Mol Med Report. 2009;2:963–970. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- 57.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Morson BC. Precancerous lesions of the colon and rectum. Classification and controversial issues. JAMA. 1962;179:316–321. doi: 10.1001/jama.1962.03050050006002. [DOI] [PubMed] [Google Scholar]
- 60.East JE, Saunders BP, Jass JR. Sporadic and syndromic hyperplastic polyps and serrated adenomas of the colon: classification, molecular genetics, natural history, and clinical management. Gastroenterol Clin North Am. 2008;37:25–46, v. doi: 10.1016/j.gtc.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 61.Lanza G, Messerini L, Gafà R, Risio M. Colorectal tumors: the histology report. Dig Liver Dis. 2011;43 Suppl 4:S344–S355. doi: 10.1016/S1590-8658(11)60590-2. [DOI] [PubMed] [Google Scholar]
- 62.Xie J, Itzkowitz SH. Cancer in inflammatory bowel disease. World J Gastroenterol. 2008;14:378–389. doi: 10.3748/wjg.14.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahmadi A, Polyak S, Draganov PV. Colorectal cancer surveillance in inflammatory bowel disease: the search continues. World J Gastroenterol. 2009;15:61–66. doi: 10.3748/wjg.15.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crohn BB, Rosenberg H. The sigmoidoscopic picture of chronic ulcerative colitis. Am J Med Sci. 1925;170:220–228. [Google Scholar]
- 65.Faber C, Kirchner T, Hlubek F. The impact of microRNAs on colorectal cancer. Virchows Arch. 2009;454:359–367. doi: 10.1007/s00428-009-0751-9. [DOI] [PubMed] [Google Scholar]
- 66.Agostini M, Pucciarelli S, Calore F, Bedin C, Enzo M, Nitti D. miRNAs in colon and rectal cancer: A consensus for their true clinical value. Clin Chim Acta. 2010;411:1181–1186. doi: 10.1016/j.cca.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 67.Shah MS, Schwartz SL, Zhao C, Davidson LA, Zhou B, Lupton JR, Ivanov I, Chapkin RS. Integrated microRNA and mRNA expression profiling in a rat colon carcinogenesis model: effect of a chemo-protective diet. Physiol Genomics. 2011;43:640–654. doi: 10.1152/physiolgenomics.00213.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balaguer F, Link A, Lozano JJ, Cuatrecasas M, Nagasaka T, Boland CR, Goel A. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010;70:6609–6618. doi: 10.1158/0008-5472.CAN-10-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allgayer H. Pdcd4, a colon cancer prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol. 2010;73:185–191. doi: 10.1016/j.critrevonc.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 70.Fassan M, Pizzi M, Giacomelli L, Mescoli C, Ludwig K, Pucciarelli S, Rugge M. PDCD4 nuclear loss inversely correlates with miR-21 levels in colon carcinogenesis. Virchows Arch. 2011;458:413–419. doi: 10.1007/s00428-011-1046-5. [DOI] [PubMed] [Google Scholar]
- 71.Schmitz KJ, Hey S, Schinwald A, Wohlschlaeger J, Baba HA, Worm K, Schmid KW. Differential expression of microRNA 181b and microRNA 21 in hyperplastic polyps and sessile serrated adenomas of the colon. Virchows Arch. 2009;455:49–54. doi: 10.1007/s00428-009-0804-0. [DOI] [PubMed] [Google Scholar]
- 72.Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, Vodickova L, Novotny J, Försti A, Hemminki K, Canzian F, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–584. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 73.Landia D, Morenob V, Guinob E, Vodickac P, Pardinic B, Naccaratic A, Canziand F, Baralea R, Gemignania F, Landia S. Polymorphisms affecting micro-RNA regulation and associated with the risk of dietary-related cancers: A review from the literature and new evidence for a functional role of rs17281995 (CD86) and rs1051690 (INSR), previously associated with colorectal cancer. Mutat Res. 2010;717:109–115. doi: 10.1016/j.mrfmmm.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 74.Dalal SR, Kwon JH. The Role of MicroRNA in Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y) 2010;6:714–722. [PMC free article] [PubMed] [Google Scholar]
- 75.Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, Brant SR, Kwon JH. Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm Bowel Dis. 2010;16:1729–1738. doi: 10.1002/ibd.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okubo M, Tahara T, Shibata T, Yamashita H, Nakamura M, Yoshioka D, Yonemura J, Kamiya Y, Ishizuka T, Nakagawa Y, et al. Association study of common genetic variants in pre-microRNAs in patients with ulcerative colitis. J Clin Immunol. 2011;31:69–73. doi: 10.1007/s10875-010-9461-y. [DOI] [PubMed] [Google Scholar]
- 77.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hébuterne X, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 78.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635.e24. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 79.Fasseu M, Tréton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, Aparicio T, Daniel F, Soulé JC, Moreau R, et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5:1–12. doi: 10.1371/journal.pone.0013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olaru AV, Selaru FM, Mori Y, Vazquez C, David S, Paun B, Cheng Y, Jin Z, Yang J, Agarwal R, et al. Dynamic changes in the expression of MicroRNA-31 during inflammatory bowel disease-associated neoplastic transformation. Inflamm Bowel Dis. 2011;17:221–231. doi: 10.1002/ibd.21359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu F, Guo NJ, Tian H, Marohn M, Gearhart S, Bayless TM, Brant SR, Kwon JH. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2011;17:241–250. doi: 10.1002/ibd.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]